Abstract
The Enterococcus faecalis virulence plasmid pAD1 encodes a mating response induced by exposure to an octapeptide sex pheromone, cAD1, secreted by plasmid-free enterococci. The determinant for the pheromone in E. faecalis FA2-2, designated cad, was found to encode a 309-amino-acid lipoprotein precursor with the last 8 residues of its 22-amino acid signal sequence representing the cAD1 moiety. The lipoprotein moiety contained two 77-amino-acid repeats (70% identity) separated by 45 residues. The nonisogenic E. faecalis strain V583 determinant encodes a homologous precursor protein, but it differs at two amino acid positions, both of which are located within the pheromone peptide moiety (positions 2 and 8). Construction of a variant of strain FA2-2 containing the differences present in V583 resulted in cells that did not produce detectable cAD1. The mutant appeared normal under laboratory growth conditions, and while significantly reduced in recipient potential, when carrying pAD1 it exhibited a normal mating response. A mutant of FA2-2 with a truncated lipoprotein moiety appeared normal with respect to recipient potential and, when carrying plasmid DNA, donor potential. A gene encoding a protein designated Eep, believed to be a zinc metalloprotease, had been previously identified as required for pheromone biosynthesis and was believed to be involved in the processing of a pheromone precursor. Our new observation that the pAD1-encoded inhibitor peptide, iAD1, whose precursor is itself a signal sequence, is also dependent on Eep is consistent with the likelihood that such processing occurs at the amino terminus of the cAD1 moiety.
Certain conjugative plasmids in Enterococcus faecalis encode a mating response to sex pheromones secreted by plasmid-free enterococci (19, 20). The response is characterized by the synthesis of a surface protein aggregation substance which is able to bind to enterococcal binding substance on the surfaces of both recipient and donor cells. The response of plasmid-containing donors to nearby recipient (plasmid-free) cells results in the initiation of mating aggregate formation. However, if donors are exposed to a culture supernatant of recipients, a self-aggregation (clumping) is observed, a phenomenon that serves as the basis for quantitative assay for pheromone activity. For recent reviews of the enterococcal pheromone systems, see references 12 and 15.
pAD1 (11, 17, 50) is a highly conjugative pheromone-responding plasmid that has been studied extensively; its nucleotide sequence has recently been completed (24). pAD1 is a member of a widely disseminated family of mobile enterococcal elements that encode a hemolysin/bacteriocin (cytolysin) and resistance to UV light (29, 32, 35, 44); its hemolysin and aggregation substance have been shown to contribute to virulence (10, 30, 34, 36, 37, 47). The cognate sex pheromone cAD1 is an octapeptide with the sequence LFSLVLAG (42).
When a plasmid-free, pheromone-producing bacterium acquires a plasmid by conjugation, pheromone activity in culture supernatants of the transconjugant can no longer be detected. This is because of plasmid-encoded functions that involve “masking” and, in some cases, a “shutdown” of endogenous pheromone. Masking relates to the secretion of specific octa- or heptapeptides that desensitize the cells to exogenous pheromone (16, 33, 43). They act as competitive inhibitors of the pheromones and serve to prevent self-induction of conjugation functions in the absence of recipient cells. While a given plasmid-bearing cell does not emit the cognate pheromone, it continues to produce unrelated pheromones specific for other families of plasmids. In the case of pAD1, the inhibitor iAD1 has the structure LFVVTLVG, which is 50% identical to cAD1 (41).
Exogenous cAD1 is believed to bind to a plasmid-encoded surface lipoprotein, TraC (48), that enhances donor sensitivity and participates in uptake of the peptide via a host-encoded ABC peptide transport system (38). There is evidence that once inside the cell the peptide binds directly to a DNA-binding, negative regulator protein, TraA, which in turn releases its binding to DNA, allowing induction of the mating response (26). The inhibitor iAD1 probably competes with cAD1 for binding to TraC; there is currently no evidence that secreted inhibitor reenters the cell.
The known enterococcal sex pheromones (cAD1, cPD1, cCF10, cAM373, and cOB1) (15) and related inhibitors are all relatively hydrophobic, linear octa- or heptapeptides that are active at nanomolar concentrations. Interestingly, some of them have relatively strong neutrophil chemotaxis activity (22, 46). With the recent availability of enterococcal genome sequence data, it was noted that they correspond to part of the signal sequences of precursors of certain lipoproteins (13). In most cases, the signal sequences correspond to 21- or 22-amino-acid segments, with the last 7 or 8 residues representing the specific pheromone. Typical lipoprotein signal peptidase target sites are appropriately located such that cleavage results in separation of the signal sequence, which in turn needs only to be processed at a second location seven or eight residues from the other processing site to generate a mature pheromone peptide. It is not known if there is a functional relationship between the activity of the putative lipoproteins and the pheromone component of their precursor structures or whether the lipoprotein connection is simply fortuitous.
Interestingly, the plasmid-encoded inhibitors are synthesized as 20- to 23-amino-acid precursors which resemble a signal sequence. Such precursors must be processed to generate the mature inhibitor peptide, perhaps by a mechanism resembling the processing system utilized by pheromone precursors. We have recently characterized a gene on the E. faecalis chromosome that is necessary for the production of cAD1 as well as certain other sex pheromones (3). This gene (eep) encodes a 46.5-kDa membrane protein initially identified based on its ability to enhance expression of cAD1 (eightfold) when present on a plasmid vector (pAM401) in E. faecalis. In addition, enterococcal eep mutants did not produce detectable pheromone. It was therefore suggested that it might be involved in processing of pheromone precursors.
In this communication we present data relating to the gene for the cAD1 precursor (cad) in E. faecalis FA2-2 and other strains of E. faecalis, as well as a strain which differs significantly only in the pheromone component of the precursor. The effects of mutations within cad are also presented, as well as data relating to the likely involvement of Eep in processing.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The bacterial strains and plasmids used in this study are listed in Table 1. pAM211 and pAM714 are derivatives of pAD1 containing insertions of Tn916 (28) and Tn917 (2, 31), respectively. The transposon insertions are in locations that are not involved in transfer and are believed to exhibit wild-type conjugation. pAM4110 is a derivative of pAM373 with a Tn917-lac insertion (18); it is readily transferable but produces an elevated level of the inhibitor iAM373.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. faecalis | ||
| FA2-2 | rif fus | 17 |
| OG1X | str gel | 33 |
| OG1SS | str spc | 25 |
| JH2SS | str spc | 49 |
| V583 | Clinical isolate (resistant to vancomycin, gentamicin, erythromycin, and tetracycline) | 45 |
| FA373 | FA2-2 carrying pAM373; Tn 918 (tet) on chromosome | 14 |
| FA3328 | Eep-negative mutant of OG1X | 3 |
| FA3333 | FA2-2 defective in cAD1 | This study |
| FA3335 | FA2-2 with pAM3334 integrated in chromosome | This study |
| DS16 | Clinical isolate carrying pAD1 and pAD2 | 17 |
| 39-5 | Clinical isolate carrying pPD1 | 53 |
| E. coli DH5α | F−φ80 lacZΔM15 endA1 recA1 hsdR17 (rK− mk+) supE44 thi-1 ΔgyrA96 (ΔlacZYA-argF) U169 | Promega Research Laboratories |
| Plasmids | ||
| pAM211 | pAD1::Tn 916 (tet) | 27 |
| pAM714 | pAD1::Tn 917 | 31 |
| pAM4110 | pAM373::Tn917-lac | 18 |
| pCF10 | Carries Tn 925 (tet); responds to cCF10 | 21 |
| pAM88 | pSU18 with additional (gram-positive) cat determinant | Victoria Francia |
| pAM401 | E. coli-E. faecalis shuttle vector; cat tet | 51 |
| pVA891 | E. coli plasmid vector; erm cat | 39 |
| pAM3332 | PCR fragment with part of cad (V583) in pVA891 | This study |
| pAM3334 | PCR fragment of cad (internal) in pAM88 | This study |
Media, reagents, and assays.
The media used were Todd-Hewitt broth (THB) (Difco Laboratories, Detroit, Mich.) and N2GT (nutrient broth no. 2 [Oxoid Ltd., London, United Kingdom] supplemented with 0.2% glucose and 0.1 M Tris-HCl [pH 7.5]) for E. faecalis and Luria-Bertani broth (40) for Escherichia coli. Cell density was determined using a Spectronic colorimeter at a wavelength of 600 nm. The solid medium used was Todd-Hewitt broth with 1.5% agar. General reagents were essentially as previously described (6, 7). Matings were performed as detailed elsewhere (14). Antibiotics were used at the following concentrations: chloramphenicol, 20 μg/ml; ampicillin, 100 μg/ml; erythromycin, 20 μg/ml for E. faecalis and 200 μg/ml for E. coli; streptomycin, 1 mg/ml; rifampin, 25 μg/ml; fusidic acid, 25 μg/ml; and spectinomycin, 500 μg/ml. Synthetic cAD1 peptide was prepared at the University of Michigan peptide synthesis core facility. The synthetic vAD1, peptides used for the cAD1 alanine substitution experiments, and cAD1 derivatives containing additional residues at the N terminus were purchased from PeptidoGenic Research & Co. (Livermore, Calif.). Pheromone response (clumping) assays, including preparation of culture fluids, were performed as previously described (20). Restriction enzymes were purchased from Invitrogen Life Technologies (Grand Island, N.Y.), and reactions were carried out under the conditions recommended by the manufacturers.
DNA and RNA methodology.
Cloning, electrophoretic analyses, plasmid isolation, and electroporation techniques were as previously described (1, 3, 4). PCR was performed with a Perkin-Elmer Cetus apparatus under conditions recommended by the manufacturer. Specific primers, in some cases containing specifically added restriction sites, were synthesized at the Biomedical Research DNA Core Facility of the University of Michigan or Invitrogen Life Technologies. PCR products were handled as previously described (3). The primers used for the generation of the reverse PCR product were 5′ GCCACAGCCTGCTAATACTAAACTAAATAA (rev-pcr-1) and 5′ TGGTAACACAGACACAATCGAAATCGACAAT (rev-pcr-2). The primers used to generate the mutated (V583) sequence were 5′ ggaattccGTCATTGACAATCCGCTCCTTAA (891-top; contains an incorporated EcoRI site [lowercase]) and 5′ ggaattccTTCTTTTGGTCCAACACCAGA (891-btm; contains an incorporated EcoRI site [lowercase]). The primers used to generate the segment representing an internal portion of cad were 5′ acatgcatgcatgtGGTTACCGTGCAGTCTTTGAA (su-top; contains an incorporated SphI site [lowercase]) and 5′ gctctagagcTTCAACACCTGCAGGTGAGCC) (su-btm; contains an incorporated XbaI site [lowercase]).
DNA sequencing made use of the Biomedical Research DNA Core Facility of the University of Michigan, and sequences were analyzed using a MacVector software package from Eastman Kodak. Primer extension was performed as described elsewhere (7) and made use of the primer rev-pcr-1 described above.
Identification of cad.
Information that played a role in the identification of cad came initially from a partial enterococcal sequence database from AstraZeneca (13). However, sequence information utilized in the current study made use of that available for E. faecalis strain V583 located on the public domain website of The Institute for Genomic Research (TIGR; http://www.tigr.org/tdb/mdb/mdbcomplete.html). The latter differed from the AstraZeneca data at two of the eight amino acid residues of cAD1, but downstream sequences were essentially the same. Oligonucleotides based on the TIGR (V583) sequence were designed to amplify (via PCR) various segments using template DNA from E. faecalis FA2-2. The sequences upstream and downstream of cad were determined using inverse PCR with outwardly reading primers, rev-pcr-1 and rev-pcr-2 (Fig. 1); chromosomal DNA that had been cleaved with HindIII and self-ligated was used as a template. (Note that the inverse PCR made use of a different E. faecalis strain [OG1X], but sequences adjacent to cad ultimately proved to be the same for FA2-2 and OG1X)
FIG. 1.
Key regions of the nucleotide sequence of cad along with related amino acids in E. faecalis FA2-2. The region representing vAD1 of E. faecalis V583 is also marked (box). The arrows approximate the locations representing specific primers used for PCR and for the primer extension experiment shown in Fig. 3. The locations of the −10 and −35 hexamers of the promoter, as well as the transcription start site (bent arrow) and the ribosome binding site (S. D.), are noted.
Generation of cad mutants.
Using chromosomal DNA from E. faecalis V583 as a template, a 655-bp PCR product was generated with primers that flanked the cAD1 component of cad by more than 300 nucleotides on each side. (The approximate locations of the PCR primers, 891-top and 891-btm, are shown in Fig. 1.) The segment corresponding to cAD1 of strain V583 is designated vAD1 because it differs by two amino acids from cAD1. EcoRI sites introduced by the primers were used to clone the PCR fragment into the E. coli vector pVA891. A resulting chimera in E. coli strain DH5α, designated pAM3332, was introduced into E. faecalis strain FA2-2 by electroporation and selection on plates containing erythromycin. Because the plasmid cannot replicate in E. faecalis, transformation resulted in integration of the element via homologous recombination. Several transformants were subcultured in the absence of erythromycin for five or more passages in broth at 42°C, resulting in several derivatives that had become sensitive to erythromycin. Of eight strains tested, four no longer produced detectable cAD1 while four produced a normal level of cAD1. DNAs from two of each type were sequenced and confirmed to be the desired mutant or the wild type. One of the cAD1-negative strains was designated FA3333.
The generation of a mutation affecting the mature portion of cad involved the PCR amplification of a product representing a 588-bp internal segment of the gene, which was then cloned into the E. coli vector pAM88. (pAM88 was kindly provided by V. Francia and is a derivative of pSU18 [5] with a cat determinant that expresses in gram-positive bacteria [M. V. Francia and D. B. Clewell, unpublished data].) The primers used, su-top and su-btm, are indicated in Fig. 1 and were designed to add SphI and XbaI restriction sites, respectively, for cloning. A plasmid chimera, pAM3334, was generated in DH5α and used to transform E. faecalis FA2-2 by electroporation and selection with chloramphenicol. A strain confirmed to have the plasmid inserted into cad via PCR was designated FA3335.
Nucleotide sequence accession number.
The GenBank accession number for the sequence shown in Fig. 1 is AF421355.
RESULTS AND DISCUSSION
Nucleotide sequence of cad in E. faecalis FA2-2.
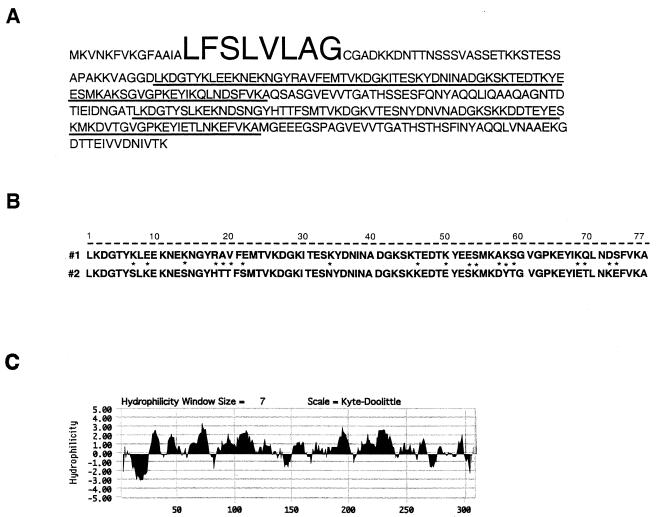
The cad coding sequence in the chromosome of E. faecalis FA2-2 was identified as described in Materials and Methods; key regions are shown in Fig. 1 and 2. It encodes a 309-amino-acid protein representing an apparent lipoprotein precursor (Cad) with a typical“lipobox” associated with a cysteine residue (52) at the signal sequence junction. (This polypeptide is larger than that originally found in the preliminary sequence data provided earlier by AstraZeneca [13].) The only homologue noted in the database is a lipoprotein of Treponema pallidum, and the resemblance is not particularly strong. The signal sequence corresponds to 22 amino acids, the last 8 of which constitute cAD1. The resemblance of the region to the iAD1 precursor sequence (16) is striking in that the latter appeared to represent a “lone” signal sequence, with the last 8 residues corresponding to the mature peptide. Although the regions upstream of the cAD1 and iAD1 moieties differ from each other, both can be represented as amphipathic helices (not shown). The hydrophilicity plot (Fig. 2) shows an absence of transmembrane regions in the presumed mature or processed lipoprotein, suggesting that the latter is an extracellular or surface molecule. It has a net negative charge (pI, 4.9); the precursor has a pI of 5.1. A 77-amino-acid segment is repeated within the sequence with a 45-amino-acid separation; the repeated regions exhibit 70% amino acid identity (Fig. 2).
FIG. 2.
Amino acid sequence of Cad and hydrophilicity plot. (A and B) Sequences and comparison, respectively, of the 77-residue repeats. (C) Hydrophilicity plot. In panel A, large letters represent cAD1 and repeated sequences are underlined. In panel B, asterisks indicate where the amino acids differ.
The nonisogenic E. faecalis strain V583 contains an almost identical lipoprotein (TIGR database) precursor differing in only three amino acids, two of which are located within the pheromone-related segment of the signal sequence (shown in bold type) (LSSLVLAA [designated vAD1 here] rather than LFSLVLAG [cAD1]). The related codons in the pheromone segment involve a change in 2 nucleotides in each case (Fig. 1), and we were not able to detect a cAD1 activity in supernatants of the V583 strain. The third amino acid difference is at position 37, where V583 has an alanine but FA2-2 has a valine. A synthetic peptide representing vAD1 of V583 (i.e., LSSLVLAA) had no cAD1 activity (using E. faecalis DS16 as a responder) at concentrations as high as 10 μg/ml (more than 10,000 times the concentration at which cAD1 activity is generally detectable), nor was it able to induce a clumping response by V583. Furthermore, synthetic cAD1 was not able to induce clumping of V583 at concentrations as high as 100 μg/ml. Interestingly, an iAD1 activity with a titer of 16 was detectable in V583 culture supernatants. This is believed to relate to a pAD1-like element in V583.
Sequence analysis of DNA from the nonisogenic strain OG1X revealed a Cad precursor structure identical to that of strain FA2-2 except that it had an alanine rather than a valine at position 37. Another nonisogenic strain, DS16, also has an alanine at this position but in addition has an asparagine rather than a glycine at position 101.
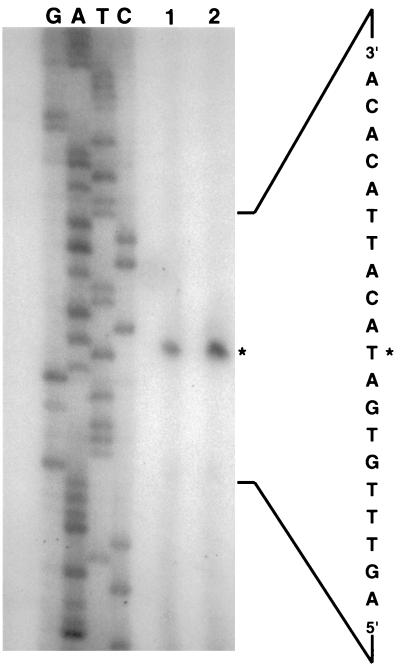
A reasonable ribosome binding site is appropriately located upstream of the translational start site, and a putative promoter sequence consisting of consensus −10 (TATAAT) and −35 (TTGACA) sequences is present upstream. Results of primer extension experiments (Fig. 3) with extracted RNA showed that this site was the cad promoter. The initial nucleotide is an A, which is located 10 residues downstream of the −10 hexamer. An intrinsic termination site is not evident within 90 bp downstream of cad, but a 23-nucleotide sequence that is 91.3% AT spans the translational stop site.
FIG. 3.
Primer extension analysis of RNA representing cad. The primer used was rev-per-1. The asterisk marks the thymine corresponding to the 3′ end of the extended fragment; its complementary adenosine represents the 5′ transcriptional start site that is noted in Fig. 1.
Located 66 nucleotides upstream of the cad translational start site is an oppositely oriented 33-amino-acid reading frame, which is probably nonfunctional, as it does not have a good ribosome binding site. (The open reading frame does not resemble anything in the database.) Similarly, the 90-nucleotide region immediately downstream of cad contains only a similarly oriented 27-amino-acid frame (no homologues) without a significant ribosome binding site.
Generation of mutants.
The differences between the cAD1 and vAD1 Cad sequences prompted a test of the effect substitution of vAD1 for cAD1 in the FA2-2 host would have with regard to not only pheromone production but also the ability to act as a donor when harboring pAD1. This was done by generating a PCR product containing the signal sequence of the V583 homologue (Fig. 1), cloning it onto a plasmid vector (pVA891) in E. coli, and then introducing it into FA2-2 by allelic exchange (see Materials and Methods). FA3333 was one of two candidates that no longer produced cAD1 and was confirmed by sequencing to have the intended modification (the entire allele was sequenced). The sequences upstream of the cad reading frame were identical for more than 200 nucleotides in both FA2-2 and V583, and this region was present intact in the variant. While the variant included the alanine at position 37 in Cad (i.e., from V583), this is not likely to be significant, since the pheromone-encoding strains OG1X and DS16 both have an alanine at this location. Culture filtrates were devoid of cAD1 activity, but normal amounts of cCF10, cPD1, and cAM373 were produced (Table 2). FA3333 had a normal growth rate and appearance of colonies on THB plates; however, they were poor recipients (>2 orders of magnitude fewer transconjugants in 90-min broth matings than FA2-2), as expected (data not shown). When a pAD1::Tn917 derivative, pAM714 (with wild-type conjugation properties), was introduced into the mutant (filter mating), the transconjugants exhibited a typical clumping response to exogenous cAD1. The sensitivity of these transconjugants to the pheromone was essentially the same as that of the wild-type host when carrying pAM714; when donors were compared in short (10-min) matings after a 45-min exposure to synthetic pheromone, the transfer frequencies were essentially the same for both.
TABLE 2.
Pheromone activity of culture filtrates and synthetic peptides
| Strain (filtrate) | Synthetic peptidea | Activity (titer)
|
|||
|---|---|---|---|---|---|
| cAD1 | cPD1 | cCF10 | cAM373 | ||
| FA2-2 | 32 | 64 | 16 | 16 | |
| FA3333 | <2 | 64 | 16 | 16 | |
| FA3335 | 32 | 64 | 16 | 16 | |
| V583 | <2 | <2 | <2 | 8 | |
| cAD1 | >512 | 2 | <2 | <2 | |
| vAD1 | <2 | <2 | <2 | <2 | |
| A-cAD1 | 32 | ||||
| IA-cAD1 | 32 | ||||
| AIA-cAD1 | 32 | ||||
One microgram per milliliter in each case; A, IA, and AIA residues added to amino terminus of cAD1.
In an effort to mutate the lipoprotein component of Cad, we generated a PCR product corresponding to an internal part of cad (see Materials and Methods) (Fig. 1) and introduced it into pAM88 in E. coli DH5α. The chimeric DNA (pAM3334) was then introduced into the plasmid-free FA2-2 with selection (chloramphenicol resistance) for integration. The mutant, FA3335, was examined by PCR (data not shown) to confirm that the integration was correctly positioned (see Materials and Methods). The derivative exhibited a normal growth rate and colony morphology and gave rise to an amount of cAD1 in culture supernatants that was essentially identical to that of wild-type cells (Table 2). The mutant would be expected to have a truncated Cad missing 39 of its own amino acid residues from its carboxyl terminus (about 14% of the mature lipoprotein) but containing some fused missense residues. Because the normal cAD1 component is still present, it was not surprising to observe a normal amount of pheromone. The data also imply that if Cad has a self-regulatory function, the absence of the carboxyl-terminal region does not affect it. Introduction of pAM714 into strain FA3335 resulted in cells that responded normally to exogenous pheromone with respect to both clumping and mating.
Activities of modified synthetic forms of cAD1.
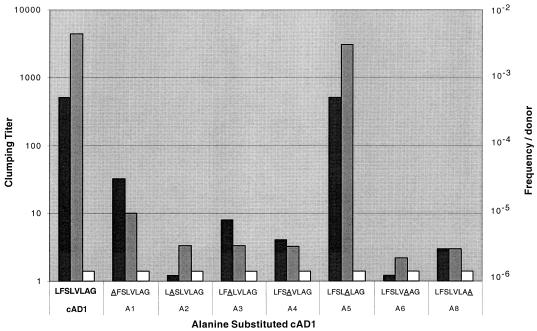
The difference between the lipoprotein precursors of strains FA2-2 and V583 with respect to the pheromone moiety raised the question of the contribution of each amino acid in the mature peptide to pheromone-specific activity. To address this point, we generated a series of synthetic peptides with an alanine substituted at different positions and determined their abilities to induce the clumping and mating responses at concentrations of 1 μg/ml and 50 ng/ml, respectively. The results, shown in Fig. 4, indicated that all but one of the substitutions caused a significant decrease in activity. (Position 7 was not tested, since it is already an alanine). The A1 substitution (alanine substituted at the no. 1 position) had an order of magnitude less activity with respect to induction of clumping and close to 2 orders of magnitude less regarding transfer of plasmid DNA. The others, with the exception of A5, had activities more than 3 orders of magnitude lower than that of cAD1. A5 had an activity that was indistinguishable from that of cAD1, a result consistent with the use of a common “face” of the peptide for induction of both clumping and conjugative transfer. Because peptides A2 and A8 correspond to the two positions in V583 which differ from cAD1, it was not surprising that the synthetic peptides differing at both positions in V583 (i.e., LSSLVLAA [differences in boldface]) had undetectable cAD1 activity (see above). Overall, the induction of clumping and induction of transfer potential were closely parallel for all of the substitutions.
FIG. 4.
Effects of the substitution of alanine in various positions within cAD1 on the clumping and mating response. E. faecalis strain DS16 was used as the responder for the clumping assays. The concentration of synthetic peptide used in each case was 1μg/ml. For the mating induction assays, the peptide concentration used was 50 ng/ml. The donor strain was FA2-2/pAM714, and the recipient was JH2SS. In each case, donors were exposed to the peptide for 45 min and then mixed with recipients for 10 min before being plated on selective medium. Note that a substitution at position 7 was not done because an alanine is already present at that position in cAD1. Solid bars, clumping titer; shaded bars, frequency per donor with induction; open bars, frequency per donor without induction.
We also determined the activities of synthetic forms of cAD1 that contained one, two, or three amino acid residues corresponding to those in the precursor added onto the amino terminus of the pheromone. Peptides ALFSLVLAG, IALFSLVLAG, and AIALFSVLAG (added residues in boldface) were all reduced about 16-fold in ability to induce clumping (Table 2) compared to cAD1. Thus, while the first residue (A) adjacent to the amino terminus affected activity, an extension of two additional residues (AIA) had no further effect. The data are consistent with the importance of processing occurring precisely at the amino terminus of the cAD1 moiety during biosynthesis.
Requirement for Eep in iAD1 in addition to cAD1 production.
The resemblance of the signal sequence of the cAD1 precursor to that of the iAD1 precursor suggests how processing occurs. Since cleavage of Cad at the carboxyl terminus of the cAD1 moiety most likely involves lipoprotein signal peptidase (SPase II), processing at the amino terminus of the segment involves a different proteolytic activity. We have speculated that the product of the recently identified eep determinant may play such a role (3, 13). Since the amino acid residues are similar around this region for both Cad and the iAD1 precursor (i.e., IA-LF and IT-LF, respectively), we determined whether detection of the inhibitor peptide (iAD1) was affected in supernatants of an eep mutant host, FA3328, carrying pAM211 (pAD1::Tn916). Table 3 shows that iAD1 activity was not detectable. This is consistent with both precursors being processed at the same site, although it does not prove unambiguously that Eep acts specifically at this site. Eep is a member of a recently recognized family of zinc metalloproteases that are able to process within membranes of both bacteria and higher systems (9).
TABLE 3.
Inhibitor activity secreted by Eep-negative plasmid-carrying hostsa
| Strain | Eep | Inhibitor | Titer |
|---|---|---|---|
| O1X/pAM211 | + | iAD1 | 8 |
| FA3328/pAM211 | − | iAD1 | <2 |
| OG1X/pAM4110 | + | iAM373 | 16 |
| FA3328/pAM4110 | − | iAM373 | 8 |
| OG1X/pAM351 | + | iPD1 | 4 |
| FA3328/pAM351 | − | iPD1 | <2 |
| OG1X/pCF10 | + | iCF10 | 2 |
| FA3328/pCF10 | − | iCF10 | <2 |
FA3328 is Eep negative. +, present; −, absent.
Eep has been noted previously not to play a role in the production of a different sex pheromone, cAM373 (3). We therefore examined whether an effect could be detected on the corresponding inhibitor, iAM373, encoded by the pAM373 derivative pAM4110. Table 3 shows that there was only a twofold difference (one well in the microtiter dilution assay), which we do not consider significant. This is not surprising, considering that the related processing sites for the cAM373 and iAM373 precursors would be LG-AI and GL-SI, respectively, both of which differ significantly from the respective regions of the cAD1 and iAD1 precursors. Efforts to examine the production of inhibitors encoded by pPD1 and pCF10 were not conclusive, since the normal level of detectable inhibitor in both cases was too low to be able to resolve significant differences using the Eep-negative host.
Concluding remarks.
It is now clear that the determinant of cAD1 corresponds to part of the signal sequence of a lipoprotein precursor. Alterations designed to eliminate expression of cAD1 activity, but presumed to allow continued synthesis of the lipoprotein, did not affect cell growth or colony morphology under the laboratory conditions we normally use for bacterial cultivation. Also, the ability to establish a pAD1 derivative in such a host, although with reduced efficiency, implies that cAD1 is not required for maintenance of the plasmid. Furthermore, the presence of such a mutation in cells carrying the plasmid did not affect the ability to respond and transfer to a recipient strain with normal efficiency. Although deletion of the carboxyl-terminal 14% of the lipoprotein also did not result in a distinguishable phenotype, we are attempting to determine whether a more drastic modification of the lipoprotein might do so.
It is intriguing that E. faecalis V583 has a cad determinant differing from those of FA2-2, OG1X, and DS16 with respect to the two amino acid residues that eliminate cAD1 activity. This would seem to be more than a chance variation, considering that the two corresponding codons each have 2-nucleotide differences. V583 is a clinical isolate resistant to multiple antibiotics, including vancomycin and gentamicin. Among the other three strains, only DS16 carries antibiotic resistance, although this does not include resistance to vancomycin or gentamicin. Whether a connection exists between the peptide difference and the ability to acquire or express certain resistance traits, or other characteristics, remains an open question.
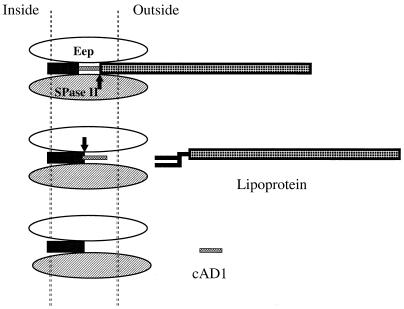
The determinant traH carried on the Staphylococcus aureus plasmid pSK41 has been reported by Firth et al. (23) to encode a lipoprotein precursor with a signal sequence containing a cAD1-like component. It differed in having a threonine rather than a serine at the number 3 position of the peptide, and a low cAD1 activity could be detected in supernatants of a pSK41-bearing strain of S. aureus. A plasmid clone containing the traH determinant was found to express cAD1 activity from E. coli; however, activity was not detected when the construct was present in a lipoprotein signal peptidase (SPase II)-negative host (8). Our efforts to clone a PCR product containing cad in E. coli and to observe expression of cAD1 have not been successful, suggesting that expression may be detrimental to the host. It is noteworthy, however, that we have been successful with regard to a different pheromone determinant, cam373 (camE) (23a). Production of the inhibitor peptide iAD1 has been readily detected in E. coli clones (16), and the fact that appropriate processing occurs suggests that an Eep-like activity, or something able to substitute for such an activity, exists in E. coli. In the E. faecalis host, one can envision an export system that involves a close association of SPase II (a homologue is present in the V583 database) and Eep, with the two membrane proteins working together in the processing of Cad (Fig. 5).
FIG. 5.
View of how Eep and SPase II may act within the cytoplasmic membrane in the processing of the precursor (Cad) protein to generate cAD1 and the related lipoprotein. Note that the processing of the 21-residue inhibitor precursor pAD1-containing cells resembles that of Cad after removal of the lipoprotein moiety from the latter. Arrows point to cleavage sites.
Acknowledgments
This work was supported by National Institutes of Health grant GM33956. The TIGR E. faecalis genome project was supported by NIH grant AI40963.
We thank V. Francia and S. Flannagan for helpful discussions and technical support. We thank Peter Barth, Brian Dougherty, Karen Ketchum, and Linda Banerjei for their help in finding or obtaining information in the databases.
REFERENCES
- 1.An, F. Y., and D. B. Clewell. 1994. Characterization of the determinant (traB) encoding sex pheromone shutdown by the hemolysin/bacteriocin plasmid pAD1 in Enterococcus faecalis. Plasmid 31:215-221. [DOI] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 1997. The origin of transfer (oriT) of the Enterococcal, pheromone-responding, cytolysin plasmid pAD1 is located within the repA determinant. Plasmid 37:87-94. [DOI] [PubMed] [Google Scholar]
- 3.An, F. Y., M. C. Sulavik, and D. B. Clewell. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moor, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 6.Bastos, M. C. F., K. Tanimoto, and D. B. Clewell. 1997. Regulation of transfer of the Enterococcus faecalis pheromone-responding plasmid pAD1: temperature-sensitive transfer mutants and identification of a new regulatory determinant,traD. J. Bacteriol. 179:3250-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastos, M. C. F., H. Tomita, K. Tanimoto, and D. B. Clewell. 1998. Regulation of the Enterococcus faecalis pAD1-related sex pheromone response: analyses of traD expression and its role in controlling conjugation functions. Mol. Microbiol. 30:381-392. [DOI] [PubMed] [Google Scholar]
- 8.Berg, T., N. Firth, and R. A. Skurray. 1997. Enterococcal pheromone-like activity derived from a lipoprotein signal peptide encoded by a Staphylococcus aureus plasmid, p. 1041-1044. In T. Horaud, M. Sicard, A. Bouve, and H. de Montelos (ed.), Streptococci and the host. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 9.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis; a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 10.Chow, J. W., L. A. Thal, M. B. Perri, J. A. Vazquez, S. M. Donabedian, D. B. Clewell, and M. J. Zervos. 1993. Plasmid-associated hemolysin and aggregation substance production contributes to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 37:2474-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clewell, D. B. 1993. Bacterial sex pheromone-induced plasmid transfer. Cell 73:9-12. [DOI] [PubMed] [Google Scholar]
- 12.Clewell, D. B. 1999. Sex pheromone systems in enterococci, p. 47-65 In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 13.Clewell, D. B., F. Y. An, S. F. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35:246-247. [DOI] [PubMed] [Google Scholar]
- 14.Clewell, D. B., F. Y. An, B. A. White, and C. Gawron-Burke. 1985. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J. Bacteriol. 162:1212-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clewell, D. B., and G. M. Dunny. Conjugation and genetic exchange in enterococci. In M. S. Gilmore, P. Courvalin, D. B. Clewell, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci: pathogenesis, molecular biology and antibiotic resistance, in press. ASM Press, Washington, D.C.
- 16.Clewell, D. B., L. T. Pontius, F. Y. An, Y. Ike, A. Suzuki, and J. Nakayama. 1990. Nucleotide sequence of the sex pheromone inhibitor (iAD1) determinant of Enterococcus faecalis conjugative plasmid pAD1. Plasmid 24:156-161. [DOI] [PubMed] [Google Scholar]
- 17.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Boever, E. H., D. B. Clewell, and C. M. Fraser. 2000. Enterococcus faecalis conjugative plasmid pAM373: complete nucleotide sequence and genetic analyses of sex pheromone response. Mol. Microbiol. 37:1327-1341. [DOI] [PubMed] [Google Scholar]
- 19.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunny, G. M., R. A. Craig, R. L. Carron, and D. B. Clewell. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2:454-465. [DOI] [PubMed] [Google Scholar]
- 21.Dunny, G. M., C. Funk, and J. Adsit. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6:270-278. [DOI] [PubMed] [Google Scholar]
- 22.Ember, J. A., and T. E. Hugli. 1989. Characterization of the human neutrophil response to sex pheromones from Streptococcus faecalis. Am. J. Pathol. 134:797-805. [PMC free article] [PubMed] [Google Scholar]
- 23.Firth, N., P. D. Fink, L. Johnson, and R. A. Skurray. 1994. A lipoprotein signal peptide encoded by the staphylococcal conjugative plasmid pSK41 exhibits an activity resembling that of Enterococcus faecalis pheromone cAD1. J. Bacteriol. 176:5871-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Flannagan, S. E., and D. B. Clewell. Identification of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol., in press. [DOI] [PubMed]
- 24.Francia, M. V., W. Haas, R. Wirth, E. Samberger, A. Muscholl-Silberhorn, M. S. Gilmore, Y. Ike, K. E. Weaver, F. Y. An, and D. B. Clewell. 2001. Completion of the nucleotide sequence of the Enterococcus faecalis conjugative virulence plasmid pAD1 and identification of a second transfer origin. Plasmid 46:117-127. [DOI] [PubMed] [Google Scholar]
- 25.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto, S., and D. B. Clewell. 1998. Regulation of the pAD1 sex pheromone response of Enterococcus faecalis by direct interaction between the cAD1 peptide mating signal and the negatively regulating, DNA-binding TraA protein. Proc. Natl. Acad. Sci. USA 95:6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gawron-Burke, M. C., and D. B. Clewell. 1982. A transposon in Streptococcus faecalis with fertility properties. Nature 300:281-284. [DOI] [PubMed] [Google Scholar]
- 28.Gawron-Burke, M. C., and D. B. Clewell. 1984. Regeneration of insertionally inactivated streptococcal DNA fragments following excision of Tn916 in Escherichia coli. A strategy for targeting and cloning genes from gram-positive bacteria. J. Bacteriol. 159:214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilmore, M. S., R. A. Segarra, M. C. Booth, C. P. Bogie, L. R. Hall, and D. B. Clewell. 1994. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 176:7335-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guzman, C. A., C. Pruzzo, M. Plate, M. C. Guardati, and L. Calegari. 1991. Serum dependent expression of Enterococcus faecalis adhesins involved in the colonization of heart cells. Microb. Pathog. 11:399-409. [DOI] [PubMed] [Google Scholar]
- 31.Ike, Y., and D. B. Clewell. 1984. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J. Bacteriol. 158:777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ike, Y., and D. B. Clewell. 1992. Evidence that the hemolysin/bacteriocin phenotype of Enterococcus faecalis subsp. zymogenes can be determined by plasmids in different incompatibility groups as well as by the chromosome. J. Bacteriol. 174:8172-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ike, Y., R. C. Craig, B. A. White, Y. Yagi, and D. B. Clewell. 1983. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. USA 80:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ike, Y., H. Hashimoto, and D. B. Clewell. 1984. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect. Immun. 45:528-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ike, Y., H. Hashimoto, and D. B. Clewell. 1987. High incidence of hemolysin production by Enterococcus (Streptococcus) faecalis strains associated with human parenteral infections. J. Clin. Microbiol. 25:1524-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jett, B. D., H. G. Jensen, R. E. Nordquist, and M. S. Gilmore. 1992. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect. Immun. 60:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreft, B., R. Marre, U. Schramm, and R. Wirth. 1992. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect. Immun. 60:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leonard, B. A. B., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macrina, F. L., R. P. Evans, J. A. Tobian, D. L. Hartley, D. B. Clewell, and K. R. Jones. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145-150. [DOI] [PubMed] [Google Scholar]
- 40.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Mori, M., A. Isogai, Y. Sakagami, M. Fujino, C. Kitada, D. B. Clewell, and A. Suzuki. 1986. Isolation and structure of Streptococcus faecalis sex pheromone inhibitor, iAD 1, that is excreted by donor strains harboring plasmid pAD1. Agric. Biol. Chem. 50:539-541.
- 42.Mori, M., Y. Sakagami, M. Narita, A. Isogai, M. Fujino, C. Kitada, R. Craig, D. Clewell, and A. Suzuki. 1984. Isolation and structure of the bacterial sex pheromone, cAD1, that induces plasmid transfer in Streptococcus faecalis. FEBS Lett. 178:97-100. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama, J., G. M. Dunny, D. B. Clewell, and A. Suzuki. 1995. Quantitative analysis for pheromone inhibitor and pheromone shutdown in Enterococcus faecalis. Dev. Biol. Stand. 85:35-38. [PubMed] [Google Scholar]
- 44.Ozawa, Y., K. Tanimoto, S. Fujimoto, H. Tomita, and Y. Ike. 1997. Cloning and genetic analysis of the UV resistance determinant (uvr) encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pAD1. J. Bacteriol. 23:7468-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahm, D. G., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sannomiya, P., R. A. Craig, D. B. Clewell, A. Suzuki, M. Fujino, G. O. Till, and W. A. Marasco. 1990. Characterization of a class of nonformylated Enterococcus faecalis-derived neutrophil chemotactic peptides: the sex pheromones. Proc. Natl. Acad. Sci. USA 87:66-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sussmuth, S. D., A. Muscholl-Silberhorn, R. Wirth, M. Susa, R. Marre, and E. Rozdzinski. 2000. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect. Immun. 68:4900-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanimoto, K., F. Y. An, and D. B. Clewell. 1993. Characterization of the traC determinant of the Enterococcus faecalis hemolysin-bacteriocin plasmid pAD1: binding of sex pheromone. J. Bacteriol. 175:5260-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomich, P. K., F. Y. An, and D. B. Clewell. 1980. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 141:1366-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomich, P. K., F. Y. An, S. P. Damle, and D. B. Clewell. 1979. Plasmid related transmissibility and multiple drug resistance in Streptococcus faecalis subspecies zymogenes strain DS16. Antimicrob. Agents Chemother. 15:828-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and an Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, H. C. 1996. Biosynthesis of lipoproteins, p. 1005-1014. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella cellular and molecular biology. 2nd ed. ASM Press, Washington, D.C.
- 53.Yagi, Y., R. Kessler, J. Shaw, D. Lopatin, F. An, and D. Clewell. 1983. Plasmid content of Streptococcus faecalis strain 39-5 and identification of a pheromone (cPD1)-induced surface antigen. J. Gen. Microbiol. 129:1207-1215. [DOI] [PubMed] [Google Scholar]