Abstract
A light-inducible promoter (PB) drives the carB operon (carotenoid genes) of the bacterium Myxococcus xanthus. A gene encoding a regulator of carotenoid biosynthesis was identified by studying mutant strains carrying a transcriptional fusion to PB and deletions in three candidate genes. Our results prove that the identified gene, named carA, codes for a repressor of the PB promoter in the dark. They also show that the carA gene product does not participate in the light activation of two other promoters connected with carotenoid synthesis or its regulation in M. xanthus. CarA is a novel protein consisting of a DNA-binding domain of the family of MerR helix-turn-helix transcriptional regulators, directly joined to a cobalamin-binding domain. In support of this, we report here that the presence of vitamin B12 or some other cobalamin derivatives is absolutely required for activation of the PB promoter by light.
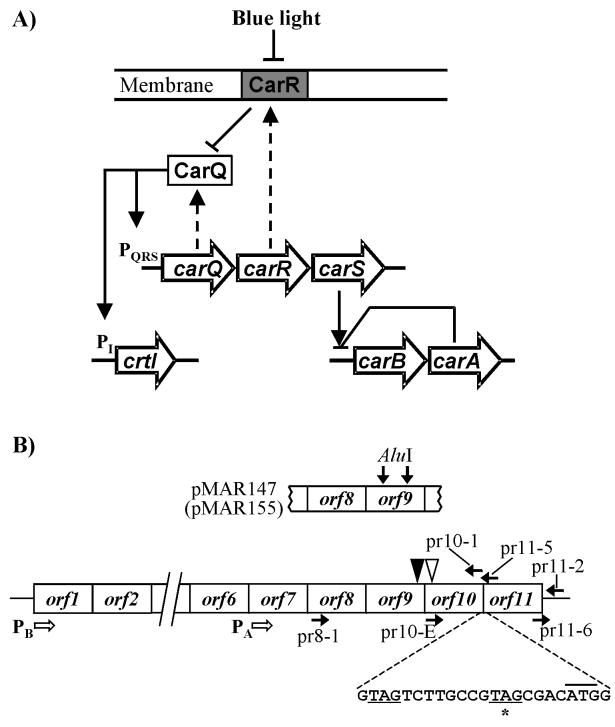
Cells of the gram-negative bacterium Myxococcus xanthus respond to blue light by accumulating significant amounts of carotenoids. This response relies on the transcriptional activation of the structural genes for carotenoid synthesis (Fig. 1A) (see reference 17 for a review). All but one of the carotenoid biosynthesis genes are clustered together at the carB operon. The exception is gene crtI, unlinked to carB. Regulatory genes participating in the activation of the crtI promoter (PI) by blue light have been identified. Two of them, carQ and carR, form part of the carQRS operon, the promoter of which (PQRS) is also regulated by blue light. Protein CarQ is a member of the extracytoplasmic function (ECF) subfamily of σ factors. CarR, a membrane-spanning protein, acts as an anti-σ factor, sequestering protein CarQ to the membrane in the dark. Illumination of the cells somehow results in the loss of CarR, leaving CarQ free to activate the PQRS and PI promoters. These two promoters share two DNA segments, centered at the −31 and −10 positions, which correspond to the binding sites of CarQ. Another protein, CarD, which contains a DNA-binding domain similar to that of eukaryotic high-mobility group A proteins, is required independently for light activation of the PI and PQRS promoters. The latter also requires the normal action of integration host factor (10, 12, 23, 27).
FIG. 1.
(A) Light induction of gene expression in M. xanthus. Three unlinked loci are represented (carQRS, crtI, and carB-carA). Genes are indicated by big arrows, which also indicate the direction of transcription. Discontinuous arrows connect some genes with their gene products. Continuous arrows, positive regulation; blunt-ended lines, negative regulation (see the introduction). The promoters of the carQRS operon (PQRS) and gene crtI (PI) are indicated. The carB and carA gene clusters (represented by single arrows) are expanded in panel B. (B) carB, carA gene clusters. Genes are represented by boxes. The carB operon contains six open reading frames. The steps for carotenoid synthesis controlled by four of them, named crtE (orf1), crtB (orf3), crtD (orf4), and crtC (orf5), are known, but not those controlled by orf2 and orf6 (6). The carB operon is driven by the light-inducible promoter PB. The carA operon contains five genes (orf7 to orf11) and is driven by the light-independent promoter PA, located within orf6. Genes orf9 and orf10 are translationally coupled. The positions of the two nucleotide changes found in the carA1 mutant, a G deletion (solid triangle) and an A→T transversion (open triangle), are shown. Below, the stop codon of orf10 (underlined) and the start codon of orf11 (upper line) are shown. Also shown is the −1-shifted stop codon (asterisk) mentioned in Materials and Methods. On top, the 1.77-kb fragment cloned in plasmid pMAR147 is shown, as well as the two AluI sites that were used to generate the orf9 in-frame deletion (plasmid pMAR155). Horizontal solid arrows indicate the approximate positions of primers used to generate (or check) the orf10 and orf11 deletions (Materials and Methods).
Less is known on the regulation of the carB promoter (PB), but previous lines of evidence indicated that the mechanism of the action of light is different from the one involved in the activation of PQRS and PI. On the one hand, the PB promoter lacks the binding sites for the ECF-σ factor CarQ, showing instead a −35 sequence that perfectly matches the −35 consensus for the major bacterial σ factor (6). On the other hand, the PB promoter, but not PQRS nor PI, is affected by mutations at two unlinked genes. One is carS, the third gene of the carQRS operon mentioned above. The first known carS mutation (carS1) was identified because it caused constitutive expression from the normally light-inducible PB promoter (4). Mutation carS1 is, however, a gain-of-function mutation, as lack of expression of carS blocks the activation of PB by light, whereas expressing carS from a heterologous, light-independent promoter produces high expression of PB in the dark (26). Thus, the carS gene product is a positive regulator of PB.
Mutations affecting the expression of the carQRS operon in different ways, such as those in carQ, carR, or carD, affect the activity of PB in the same way, but that can be explained by the effect of those mutations on the expression of protein CarS. This has been directly proved in the case of carD mutations (28). The second locus specifically involved in the regulation of PB is named carA, and it is located directly downstream of the carB operon. The carA locus was defined by a mutation carA1, which results in high, light-independent expression from the PB promoter (5). The effect of the carA1 mutation is brought about even when the entire carQRS operon is deleted. This confirms that PB is not driven by the σ factor CarQ and leads to a model in which CarA normally acts as a negative regulator of PB in the dark, whose action is somehow counteracted by protein CarS in the light (25, 26).
The carA1 mutation has been sequenced and shown to be a double mutation, which may affect three contiguous genes at the carA locus (6) (see Results). Here, we report the generation of M. xanthus strains carrying nonpolar deletions within each one of these three genes. This allowed us to identify which of the candidate genes encodes the negative regulator CarA. At the time of publication, no similarity was detected between CarA and other known proteins apart from the presence of a DNA-binding domain at the N terminus of CarA (6). We now report that the amino acid sequence and the predicted structural conformation of the carboxyl half of CarA and of another gene product from the carA locus, named Orf11, are strikingly similar to those of the methylcobalamin-binding domain of methionine synthase (9). In accordance with this, we also report here that cyanocobalamin (vitamin B12), or some other cobalamin derivative, is absolutely required for the normal activation of the carB promoter by blue light. These results lead to a model in which cobalamin acts as a CarA prosthetic group that, in conjunction with protein CarS, mediates the inactivation by light of CarA.
MATERIALS AND METHODS
Bacterial strains, transducing phages, and growth conditions.
The M. xanthus strains used in this study are listed in Table 1, together with their phenotype, genotype, and origin. The standard strain DK1050 shows normal light-induced synthesis of carotenoids (Car+ phenotype). Car− stands for lack of carotenoid synthesis, and Carc stands for light-independent production of carotenoids. Some strains carried in vitro-constructed lacZ fusions that were integrated into the M. xanthus chromosome by homologous recombination (see below). The reporter gene retained the normal translation start signal preceded by stop codons in all three reading frames; therefore, it produced transcriptional but not translational fusions. For cloning purposes, Escherichia coli strains DH5α (13) and MC1061 (8) were used.
TABLE 1.
M. xanthus strains used in this study
| Strain | Phenotypea | Genotypeb | Reference or sourcec |
|---|---|---|---|
| DK1050 | Car+ | 31 | |
| MR397 | Car+ LacZi Kmr | carQ::lacZ (pDAH217) | 28 |
| MR401 | Car− LacZi Kmr | carB::Tn5-lac | 4 |
| MR418 | Car− LacZi Tcr | carB::Tn5-132lac | 24 |
| MR553 | Car+ LacZi Kmr | crtI::lacZ (pMAR206) | 10 |
| MR819 | Carc Tcr | crtI::Tn5-132 | M. Fontes |
| MR844 | Carc | Δorf10 | This study |
| MR845 | Carc LacZi Kmr | Δorf10 crtI::lacZ | pMAR206 × MR844 |
| MR846 | Carc LacZc Kmr | Δorf10 carB::Tn5-lac | MR401 × MR844 |
| MR847 | Carc LacZi Kmr | Δorf10 carQ::lacZ | pDAH217 × MR844 |
| MR848 | Car+ | Δorf11 | This study |
| MR856 | Car− LacZi Kmr Tcr | Δorf10 carQ::lacZ crtI::Tn5-132 | MR819 × MR847 |
LacZi, light-inducible synthesis of β-galactosidase; LacZc, high level of β-galactosidase in the dark and in the light.
The integrated plasmid that carries the lacZ transcriptional fusion is indicated in parentheses.
Strains MR845 and MR847 were generated by P1-mediated transduction into MR844 of the integrative plasmids pMAR206 (Kmr crtI::lacZ) and pDAH217 (Kmr crtQ::lacZ), respectively. Strain MR846 was constructed by generalized transduction into MR844 of the Tn5-lac (Kmr) insertion present in strain MR401. Strain MR856 was constructed by generalized transduction into MR847 of the Tn5-132 (Tcr) insertion present in strain MR819.
The rich medium CTT (7) and the exact culture conditions for growth of M. xanthus in the dark and in the light have been described previously (10). Coliphage P1clr100Cm (hereafter called P1) was used to transfer plasmids from E. coli strain MC1061 to M. xanthus (11). For generalized transduction between M. xanthus strains, phage Mx4-LA27 was used. The phages and the conditions used for the transduction of resistance to kanamycin (Kmr) or tetracycline (Tcr) have been described previously (3). Electroporation was also carried out as described earlier (21).
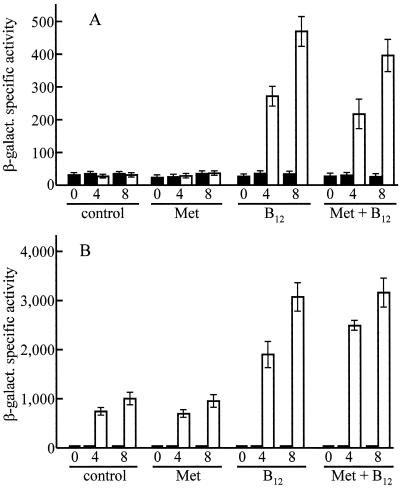
For the vitamin assays, we used CAA medium, containing 5 mg of Vitamin Assay Casamino Acids (Difco) per ml in 10 mM Tris-1 mM KH2PO4-K2HPO4-8 mM MgSO4 buffer (pH 7.6). After autoclaving and cooling to 50°C, filter-sterilized solutions were used to add l-asparagine (0.1 mg/ml), sodium pyruvate (1 mg/ml), and sodium citrate (2 mg/ml). The CAA cultures were inoculated by diluting (10,000-fold) a suspension of cells growing exponentially in CTT (2 × 108 to 5 × 108 cells/ml) that had been washed and resuspended in an equal volume of CAA. The cultures were incubated at 33°C in an orbital shaker (300 rpm) until they reached approximately the same cell density as the original inoculum. This point (time zero in Fig. 5) was attained after a time period that varied between 2.5 and 3.5 days. The same variation was observed for all strains tested.
FIG. 5.
Effect of B12 on activation of PB and PQRS promoters by light. Strains MR401 (PB::lacZ, panel A) and MR397 (PQRS::lacZ, panel B) were grown separately in the dark on vitamin-free CAA medium (control) or CAA supplemented with 100 μg of methionine (Met) or 0.5 μg of cyanocobalamin (B12) or both (Met + B12) per ml. At time zero (exponential phase), each culture was divided in two, one for the dark and the other for the light. Samples for β-galactosidase assays were taken at time zero and after 4 and 8 h of additional growth in the dark (solid bars) or the light (open bars). Enzyme specific activities are given in nanomoles of o-nitrophenol produced per minute per milligram of protein. Bars indicate standard deviations.
Plasmid and DNA manipulation.
Cloning vector pDAH160 (16) carries a Kmr gene and the incompatibility region of P1 for transfer of the plasmid from E. coli to M. xanthus by P1-specialized transduction. Plasmid pDAH217 contains a lacZ transcriptional probe fused to the light-inducible promoter of the carQRS operon. Like all other plasmids used here, pDAH217 cannot replicate in M. xanthus cells but can integrate into the M. xanthus chromosome by homologous recombination. Integration of pDAH217 produces a tandem duplication of the cloned DNA; therefore, a normal copy of the carQRS operon is generated (16). Similarly, plasmid pMAR206 contains a lacZ transcriptional fusion to the crtI promoter (10). Plasmid pMAR976 is a pBJ113 derivative (20) lacking the EcoRI restriction site. It contains a positive-negative cassette, with a Kmr gene for positive screening and a galactose sensitivity (Gals) gene for negative screening (36). For standard cloning procedures, plasmids pUC19 (29) and pUC9 (37) were used. Nucleic acid and enzymatic manipulations were done according to standard procedures (33).
Strain construction.
Plasmids carrying a long deletion within one of three different genes from the carA locus (orf9, orf10, and orf11; see Fig. 1B) were generated in the following way.
(i) orf9 deletion.
Plasmid pMAR147 is a pUC19 derivative carrying a 1.77-kb fragment from the carA locus that extends from a site within orf7 to a site within orf10 (Fig. 1B). The only two AluI sites present in that fragment are located within the coding region of orf9, being separated by 243 bp. The 1.77-kb fragment was isolated from pMAR147 after digestion with BamHI and HindIII (pUC19 polylinker sites) and treated with AluI. The BamHI-AluI and HindIII-AluI subfragments were isolated, ligated by their AluI ends, and recloned into BamHI- and HindIII-digested plasmid pDAH160 to generate plasmid pMAR155. The presence in pMAR155 of the expected deletion was first checked by Southern analysis. Sequencing of the appropriate DNA stretch of pMAR155 confirmed that the correct deletion within orf9 had been generated. The deletion covers the expected 81-codon-long stretch, leaving 59 amino acids from the N terminus and 83 amino acids from the C terminus of the normal orf9 gene product.
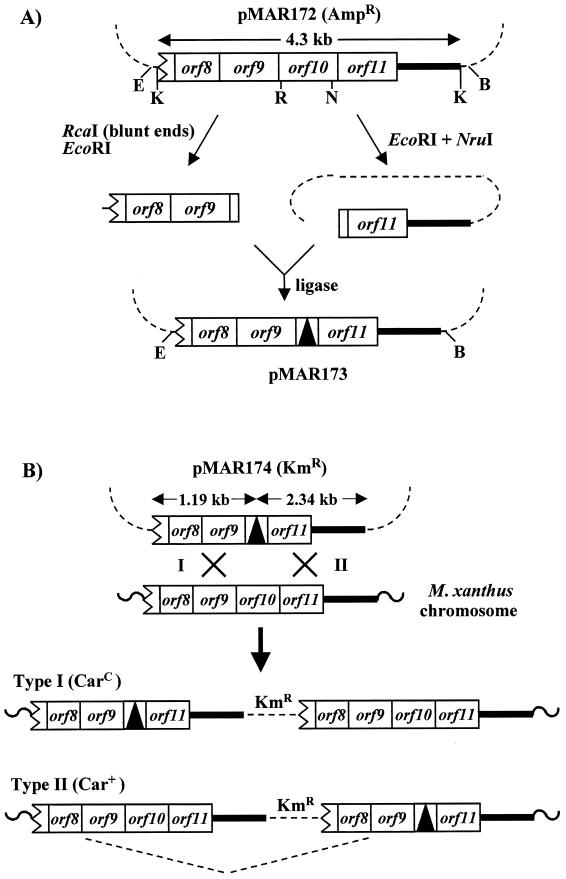
(ii) orf10 deletion.
Plasmid pMAR172 is a pUC19 derivative which carries a KpnI restriction fragment (4.3 kb) from the carA locus. This fragment extends from a site within orf7 to a site located about 1.4 kb downstream of orf11 (see Fig. 2). A two-step procedure was used in this case to remove an RcaI-NruI restriction fragment from pMAR172 to generate the orf10-deleted plasmid pMAR173. For clarity, the procedure is depicted in Fig. 2A. The two steps were required because the NruI site was unique, but several RcaI restriction sites were present in pMAR172. The deletion started right after the initiation codon of orf10 and covered 758 of its 864 bp. So, translation of the mutated gene should initiate normally, but will then immediately suffer a −1 nucleotide shift. This would not, however, compromise the normal translation of the downstream gene orf11. The shifted translation of the deleted orf10 will stop at a new (−1 shifted) UAG triplet located between the normal stop codon of orf10 and the orf11 initiation codon (see Fig. 1B). The M. xanthus DNA present in pMAR173 was recloned into the plasmid vector pDAH160 (EcoRI and BamHI double digestion of both plasmids) to generate plasmid pMAR174. The presence in pMAR174 of the expected deletion was checked by sequencing.
FIG. 2.
Deleting the orf10 gene of M. xanthus. (A) Plasmid pMAR172 is a pUC19 derivative which carries the indicated KpnI fragment from the carA locus. Only part of the vector DNA is shown. Boxes represent genes from the carA operon (see Fig. 1B). A thick line represents DNA downstream of the carA operon. The indicated two-step procedure was used to obtain plasmid pMAR173, which carries the Δorf10 deletion (see the text). B, BamHI; E, EcoRI; K, KpnI; N, NruI; R, RcaI. (B) The EcoRI-BamHI fragment of pMAR173 was transferred to the plasmid vector pDAH160 (kanamycin resistance) to obtain plasmid pMAR174. The figure shows the two types of merodiploids expected from the integration of pMAR174 into the M. xanthus chromosome by a single homologous recombination event to the left (type I) or to the right (type II) of the Δorf10 deletion. Indicated in parentheses are the expected phenotypes (CarC or Car+) of the merodiploids, assuming that the truncated copy of the carA operon lacks a promoter (see Fig. 1B). The discontinuous line shown below represents an intramolecular recombination event that will generate a plasmid-free strain carrying the Δorf10 deletion.
(iii) orf11 deletion.
The same 4.3-kb fragment present in pMAR172 (Fig. 2) was cloned into pMAR976 to generate plasmid pMAR188. Using pMAR188 as a template and the appropriate oligonucleotides (pr11-5 and pr11-6; Fig. 1B), inverse PCR was carried out (see below). The oligonucleotides included EcoRI overhangs for further digestion and self-ligation of the amplified fragment to generate plasmid pMAR189. This carries a deletion of orf11, which extends from the ATG start codon to a position located 22 nucleotides upstream of the stop codon.
To introduce the three indicated deletions into M. xanthus, each deleted plasmid was separately transferred (Kmr selection) to the standard strain DK1050. Plasmids pMAR155 and pMAR173 were transferred by P1-mediated transduction, and plasmid pMAR189 was transferred by electroporation. Kmr cells will arise by integration of the incoming plasmid by homologous recombination, so they should carry both the wild-type and the deleted copies of the cloned DNA. Cells carrying only the deleted copy can arise by intramolecular recombination events that remove the plasmid vector and the wild-type version of the corresponding gene (see Fig. 2B for the case of the orf10 deletion). Those cells were selected in two different ways. For the orf9 and orf10 deletions, independent merodiploid transductants were grown for at least 60 generations in the absence of kanamycin, plated for single colonies, and screened for a Car mutant phenotype (see Results). For the orf11 deletion, merodiploid transductants were grown in the absence of kanamycin and plated in CTT supplemented with 10 mg of galactose per ml. Several Galr Kms colonies were then checked (PCR) for the presence of the expected orf11 deletion.
PCR.
PCR was used to check for the presence of the orf10 and orf11 deletions. DNA from the corresponding M. xanthus strain (Promega genomic DNA purification kit) was used as a template. Synthetic primers were as follows (see Fig. 1B): pr8-1 (5′-CGCCGCGAAGGCGCGTCGACGGCTCGG-3′) and pr10-1 (5′-AAACTCGAGCTACGGGCGATTCTGAGCGT-3′) were used to detect the orf10 deletion; and pr10-E (5′-AAAGAATTCATGACGTTGCGCATCCGCAC-3′) and pr11-2 (5′-GTCTTCAATCACTCCGATATCAACACGCCCTGAC-3′) were used to detected the orf11 deletion. Polymerase mix and reaction buffer were from Roche Molecular Biochemicals. The samples were subjected to 30 cycles of denaturation (45 s, 95°C), primer hybridization (60 s, 65°C), and polymerization (2 min, 72°C).
Inverse PCR (18) was used to obtain the orf11 deletion. Plasmid pMAR188 (see above) was used as a template. The following synthetic oligonucleotides were used as primers (see Fig. 1B): pr11-5 (5′-AAAGGAATTCGTCGCTAGCGCAAGACTACG-3′) and pr11-6 (5′-AAAGGAATTCTGGGACCGCCTCGCGGGTAC-3′). The sample was subjected to 18 cycles of denaturation (30 s, 95°C), primer hybridization (60 s, 62°C), and polymerization (9 min, 68°C). The Expand long template PCR system from Roche Molecular Biochemicals was used.
Protein sequence analysis.
For comparison with databases and multiple protein alignments, the Blast and PIMA1.4 programs provided by the BCM search launcher were used (http://dot.imgen.bcm.tmc.edu/1) (34). For protein secondary-structure predictions, the nearest-neighbor prediction method (32) provided by the same launcher was used.
Expression of β-galactosidase.
Rapid determination of β-galactosidase production was carried out by examining colony color on plates containing 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml. Quantitative analysis of β-galactosidase of dark-or light-grown liquid cultures was performed as previously described (5). At least three independent determinations were done in all cases.
RESULTS
A deletion within orf10 but not within orf9 or orf11 causes light-independent expression from the carB promoter.
The carB-carA gene cluster from wild-type M. xanthus as well as the carA locus from the carA1 mutant strain have been cloned and sequenced. The cluster contains 11 different open reading frames (ORFs), named here, for simplicity, orf1 to orf11 (Fig. 1B). The first six ORFs are under the control of the light-inducible promoter PB. The rest are driven by a light-independent promoter located within orf6 (6; our unpublished results). When compared with the wild type, two nucleotide changes, very close to each other, were detected in the carA1 mutant (6) (Fig. 1B). One is a single-nucleotide deletion, located 18 nucleotides upstream of the orf9 stop codon. This mutation should result in the modification of the last six amino acids of Orf9 and in a 76-residue-long extension of its carboxyl end, due to the (frameshifted) reading through of gene orf10.
Genes orf9 and orf10 are translationally coupled, and gene orf11 lacks the consensus sequence for a ribosome-binding site (6), so the orf9 frameshift mutation may be producing a polar effect by affecting the correct translation of both orf10 and orf11. It should be said that the predicted amino acid sequences of proteins Orf10 and Orf11 show an overall identity of 35.2%, with several regions which are particularly well conserved (6) (see below). The second mutation found in the carA1 mutant is a transversion at the fifth codon of orf10, which causes an Ile-Phe substitution. The N terminus of Orf10 contains a helix-turn-helix (HTH) DNA-binding domain similar to that found in the MerR family of transcriptional regulators (6). The aforementioned Ile is one of the conserved residues in the HTH domain. This complex situation made it difficult to assign to a particular gene the effect of the carA1 mutation on the PB promoter (6). To clarify this point, we have now generated M. xanthus strains carrying long, nonpolar deletions within each of the candidate genes (see Materials and Methods).
To generate the orf9-deleted strain, Kmr plasmid pMAR155, which carries an in-frame deletion within orf9 (Δorf9, Fig. 1B), was transduced into wild-type strain DK1050. Transductants should arise by integration of the plasmid into the M. xanthus chromosome by homologous recombination. All the Kmr merodiploid transductants developed an intense orange color in the dark, indicative of light-independent accumulation of carotenoids (CarC phenotype). In the light, the orange colonies turned more reddish. Intramolecular recombination between the directed repeat generated by the integration of pMAR155 would result in loss of plasmid DNA and the normal or deleted copy of orf9, according to whether the recombination event takes place at one or the other side of Δorf9.
Several of the Kmr CarC merodiploids were grown separately in the absence of kanamycin, plated for single colonies, and checked for a change in phenotype. Colonies were found from all cultures which showed either a Car+ Kms or a Car− Kms phenotype. Southern analysis (not shown) demonstrated that the Car+ Kms cells contained a single, wild-type copy of the carA operon, whereas the Car− Kms cells contained a single copy of the carA operon carrying the Δorf9 deletion. This proves that the orf9 gene product is not the negative regulator of PB. Further analyses have shown that the Car− phenotype caused by the Δorf9 deletion is due to a negative effect of this mutation on the activation of the carQRS operon by blue light (to be described elsewhere). As shown in Fig. 1B, the M. xanthus DNA present in pMAR155 is truncated at the orf10 gene. Therefore, all the Kmr merodiploids from the pMAR155 × DK1050 transduction were also truncated at the orf10 gene. The CarC phenotype of all those transductants suggested that orf10 is the negative regulator of the PB promoter.
A procedure similar to that described above was used to introduce the orf10 deletion (Δorf10) in M. xanthus (Fig. 2B). First, plasmid pMAR174 (Kmr) was transferred to wild-type strain DK1050. Two classes of Kmr merodiploids were expected, depending on whether integration of the plasmid had occurred to the left or to the right of the Δorf10 site. In the first case (type I in Fig. 2B), the Δorf10 deletion will replace the endogenous orf10 allele, this being displaced to the truncated copy of the carA operon. This copy lacks a promoter, located within orf6 (Fig. 1B), so the type I merodiploids might be deficient in protein Orf10. The reverse situation is expected in the second class of merodiploids, which should carry a normal version of the entire carA locus (type II in Fig. 2B). Given the length of the DNA stretches available for homologous recombination at the sides of Δorf10, the type II merodiploids were expected to be the most frequent.
More than 100 independent transductant colonies were checked. About 80% of them showed a normal (Car+) phenotype. The remaining 20% showed the CarC phenotype described above for the orf9 merodiploids. Therefore, one might conclude that the class I transductants are those showing a CarC phenotype and that the orf10 gene product is the one responsible for repressing the PB promoter in the dark. As mentioned above, intramolecular recombination events would result in the removal of both the plasmid vector and either the wild-type or the deleted version of the orf10 gene. Four of the Car+ merodiploids were grown in the absence of kanamycin and plated for a CarC phenotype. Colonies showing that phenotype were obtained from all cultures, and all of them proved to be sensitive to kanamycin (Kms).
The presence of a single copy of the carA operon carrying the expected Δorf10 deletion was confirmed by PCR analysis. For this, we used DNA from several independent CarC Kms colonies and the oligonucleotides pr8-1 and pr10-1 (Fig. 1B) (results not shown). The CarC Kms strain was named MR844. The orf10 deletion moves the orf10 stop codon closer to the orf11 start codon (see Materials and Methods). The possibility that the CarC phenotype of MR844 stems from an unusual polar effect of Δorf10 on orf11 was ruled out, as deleting orf11 did not affect the normal response to blue light (see below).
M. xanthus strain MR401 carries a Tn5::lac (Kmr) insertion at the carB operon that generates a lacZ transcriptional fusion to the PB promoter (5) (Table 1). We tested the effect of the Δorf10 mutation on the activity of PB by transducing the indicated insertion (selection for Kmr) into strain MR844. Quantitative analysis of lacZ expression confirmed that Δorf10 causes high expression in the dark from the normally light-inducible promoter PB (Table 2).
TABLE 2.
Effects of deletions Δorf10 and Δorf11 on the light-inducible promoters
| Host strain (genotype) | Transcriptional fusion | Mean β-galactosidase sp acta ± SD
|
||
|---|---|---|---|---|
| Dark
|
Light, 8 h | |||
| 0 h | 8 h | |||
| DK1050 (wild type) | PB::lacZ | 30 ± 4 | 34 ± 3 | 186 ± 16 |
| MR844 (Δorf10) | PB::lacZ | 158 ± 14 | 174 ± 15 | 188 ± 16 |
| MR848 (Δorf11) | PB::lacZ | 11 ± 1 | 12 ± 2 | 144 ± 14 |
| DK1050 (wild type) | PI::lacZ | 14 ± 1 | 15 ± 1 | 439 ± 48 |
| MR844 (Δorf10) | PI::lacZ | 18 ± 2 | 17 ± 2 | 371 ± 39 |
| DK1050 (wild type) | PQRS::lacZ | 8 ± 1 | 6 ± 1 | 218 ± 19 |
| MR844 (Δorf10) | PQRS::lacZ | 9 ± 1 | 9 ± 4 | 23 ± 6 |
| MR856 (Δorf10 crtI) | PQRS::lacZ | 7 ± 1 | 8 ± 1 | 163 ± 17 |
At time zero, cell samples growing exponentially in the dark (CTT medium) were divided in two, one to be kept in the dark and the other to be exposed to light. Strains carrying a PI::lacZ fusion were at the late exponential phase, as entry into the stationary phase is required for activation of the PI promoter by blue light (10). The same was true for the strains carrying a PQRS::lacZ fusion (see the text). Samples for β-galactosidase assay were taken at time zero and after 8 h of additional growth in the indicated conditions. Means and standard deviations are given; units are as defined for Fig. 5.
Plasmid-based lacZ fusions to the PQRS and PI promoters (Fig. 1A) are available. Integration of the plasmids into the M. xanthus chromosome by homologous recombination generates lacZ transcriptional fusions, leaving an intact copy of the corresponding gene or operon. Plasmid pMAR206 (Kmr PI::lacZ; Table 1) was transferred to strain MR844 by P1-mediated transduction and selection for Kmr. On X-Gal plates, all the transductants behaved like a wild-type derivative carrying the same lacZ fusion (a pale blue color in the dark that turned intense blue in the light). The results of β-galactosidase assays confirmed that mutation Δorf10 does not affect the normal regulation of PI (Table 2).
An experiment similar to the one just described was done using plasmid pDAH217 (Kmr PQRS::lacZ; Table 1). In the quantitative analysis of lacZ expression, we noticed that the activation of the PQRS promoter in the MR844-derived transductants was similar to that of the control sample only when the cells were illuminated at the early exponential phase (not shown). When illumination was at the late exponential phase, the activation of PQRS was much lower in the MR844-derived transductants (Table 2). It is known that the activation of PQRS by light decreases with the progressive accumulation of colored carotenoids (16). Since Δorf10 caused light-independent carotenoid synthesis, the reduced activation of PQRS might be an indirect consequence of the progressive accumulation of carotenoids during growth in the dark.
To avoid this interference, a transposon insertion in the crtI gene was used. The crtI gene codes for an enzyme acting at an early step of the carotenoid pathway (10). The inserted transposon, a tetracycline resistance (Tcr) derivative of Tn5, did not affect the light activation of PQRS (not shown). The Tcr transposon was transduced into the strain carrying the Δorf10 deletion and the PQRS::lacZ fusion to obtain the Car− strain MR856. The behavior of all transductants on X-Gal plates and the results of β-galactosidase assays confirmed that mutation Δorf10 by itself does not have a significant effect on the normal regulation of PQRS (Table 2).
An experiment similar to the ones described above was attempted to generate an M. xanthus strain carrying an orf11 deletion. In this case, all the merodiploid transductants were Car+. This indicated that orf11 does not play a critical role in light induction of gene expression in M. xanthus, but it also made it difficult to screen for cells having a single copy of the carA locus mutated at the orf11 gene. We then took advantage of plasmid pMAR976, which carries a positive-negative cassette (see Materials an Methods). As expected, the DK1050-derived strain, which carried a single copy of the carA locus with the orf11 deletion, named MR848, showed a Car+ phenotype. This clearly indicated that orf11 does not play a critical role in the normal regulation of the PB promoter or the other promoters known to participate in the carotenogenic response of M. xanthus. This was confirmed for PB by generating the same Δorf11 deletion in M. xanthus strain MR418, which carries a lacZ fusion to the indicated promoter (Table 1). The β-galactosidase expression data are shown in Table 2.
Cobalamin-binding domain in the orf10 and orf11 gene products.
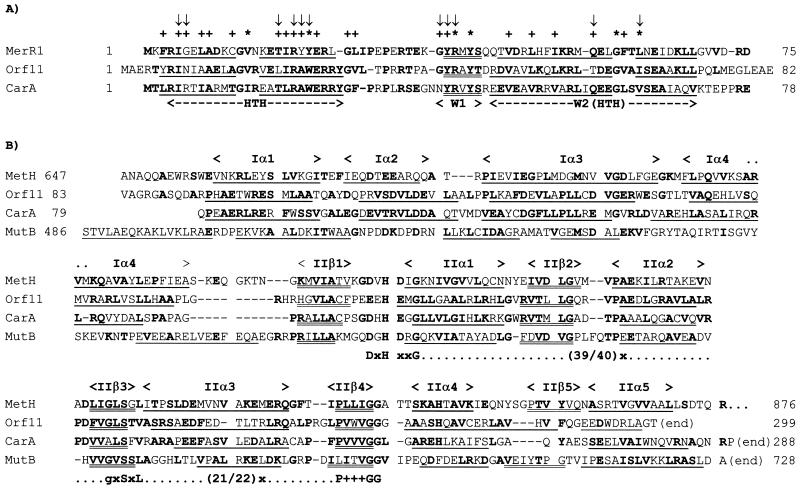
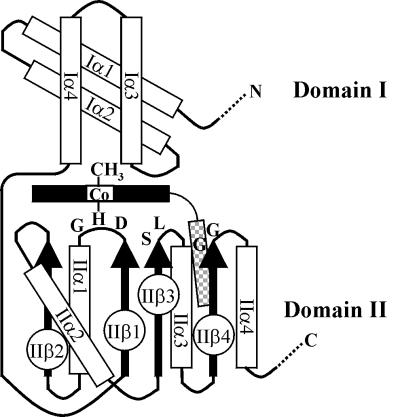
At the time the predicted amino acid sequences of Orf10 and Orf11 were published, we noticed the overall similarity of both sequences and the presence of a helix-turn-helix DNA-binding domain at their N termini (Fig. 3A). No other similarities to the then-known protein domains were noticed (6). In later, computer-aided searches, we found that both Orf10 and Orf11 contained a cobalamin-binding domain. The crystal structure of a methylcobalamin-containing fragment of the E. coli methionine synthase (MetH) has been solved (9) (Fig. 4). The structure revealed an amino-terminal domain, formed by two pairs of antiparallel helices (domain I), and an α/β carboxyl-terminal domain (domain II).
FIG. 3.
(A) Sequence alignment of the N termini of CarA and Orf11 with the DNA-binding domain of protein MerR1 from Bacillus cereus (AF138877), the MerR protein showing the highest sequence similarity to CarA and Orf11. Identical or conserved residues in two or more proteins are in boldface. An HTH motif (α-helices are underlined) and two “wings” (W1 and W2) form the DNA-binding domain. W1 is a β-strand (doubly underlined), and W2 is another HTH motif. The asterisks and the + symbols indicate invariant and conserved residues, respectively, in 11 other members of the MerR family; the arrows point to positions that contact DNA, according to crystallographic data for one member of the family (reference 38 and Fig. 2 of the Supplementary Information in this reference). (B) Sequence alignment of the C termini of CarA and Orf11, the methylcobalamin-binding domain of methionine synthase from E. coli (MetH), and the adenosylcobalamin-binding domain of methylmalonyl-coenzyme A mutase from Propionibacterium shermanii (MutB). Identical or conserved residues in two or more proteins are in boldface. Notice that in a few positions, two proteins share a particular amino acid, and the other two proteins share a different one. The structural elements indicated on top of the alignment correspond to those in Fig. 4. The α-helices (underlined) and β-sheets (doubly underlined) of MetH and MutB are from crystallographic studies (9, 22). Those of CarA and Orf11 are computer-aided predictions (32). The cobalamin-binding sequence fingerprint proposed by Drennan et al. (9) is shown below the alignment (x indicates any amino acid). Added to the fingerprint line are a proline followed by three hydrophobic residues (+ symbols), which are much conserved in methylcobalamin-binding domains (see entry pfam02310 of the Pfam data bank at http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). (Note: amino acid residues A271, A272, and A273 in Orf11 rectify the previously incorrect G271, R272, and R273; M. Elías, personal communication).
FIG. 4.
Topology diagram of the methylcobalamin-binding domain of methionine synthase from E. coli. Domain I, formed by two pairs of antiparallel helices (Iα1 to Iα4), contacts the upper phase of the corrin macrocycle (represented by a black rectangle with a central cobalt atom). The α/β domain II (IIβ1 to IIα4) contacts the lower phase of the corrin macrocycle and accommodates the dimethylbenzimidazole nucleotide tail (shaded rectangle). The invariant residues that form a structure-based sequence fingerprint for cobalamin binding are indicated. Domain II is folded in such a way that the serine and the three glycines line and pack the nucleotide tail. The histidine residue is the cobalt lower ligand, which is adjoined by invariant leucine and aspartate residues. The aspartate is part of a hydrogen-bonded proton pathway from solvent to the buried histidine (adapted from Fig. 3 of reference 9).
Sequence comparison with other methylcobalamin- and adenosylcobalamin-dependent enzymes (9) allowed Drennan et al. to propose a sequence fingerprint from domain II: DxHxxG-(39/40)x-gxSxL-(25/26)x-GG, where x is any amino acid. The conserved residues included in this fingerprint are critical for the binding to or the reactivity of the prosthetic group. As shown in Fig. 3B, the amino acid sequences of both Orf10 and Orf11 are predicted to generate the helical bundle and the α/β region that form, respectively, domains I and II of a cobalamin-binding peptide. The indicated sequence fingerprint is also present, at the right position, in both proteins. Moreover, 51% of the Orf10 residues located between the predicted structural elements IIβ1 and IIβ4 are identical or similar to those found at the same positions in MetH from various bacterial species (45% for Orf11) (entry pfam02310 in the Pfam Data Bank; see the legend to Fig. 3).
Vitamin B12 is required for normal activation of the carB promoter by blue light.
The role of protein Orf10 in the regulation of the PB promoter and the presence in that protein of a cobalamin-binding domain prompted us to test the effect of cobalamin on the activation of PB by light. Three different cobalamin derivatives, cyanocobalamin (vitamin B12), methylcobalamin, and adenosylcobalamin (coenzyme B12), were tested. The data reported are those obtained with vitamin B12, but similar results were observed when either of the other two compounds was used.
In all previous works on light induction of gene expression in M. xanthus, the standard rich medium CTT, which supports fast growth of the cells, had been used. In preliminary experiments using strain MR401 (PB::lacZ in a wild-type background), we checked the effect of adding vitamin B12 to CTT on the activation of PB by light. The vitamin did not affect the low activity of PB in the dark, but it did stimulate the activation of that promoter by light. A reproducible effect was observed at a vitamin concentration as low as 50 ng/ml. The effect reached a plateau, a twofold increase in β-galactosidase specific activity with respect to the control, at about 500 ng of the vitamin per ml (not shown).
CTT medium contains an undetermined amount of vitamins. For better quantification of the effect of vitamin B12, a culture medium based on a vitamin-free mixture of amino acids (CAA) was used. It should be noted that M. xanthus requires vitamin B12 for growth only in the absence of methionine (7). This amino acid is present in the CAA medium, but to avoid a possible nutritional effect of the vitamin, control samples specifically supplemented with methionine were included in the experiments. As shown in Fig. 5A, activation of the PB promoter by light was negligible in the absence of vitamin B12. A very intense activation was observed, however, in the presence of the vitamin. The addition of methionine produced no significant effect.
The product of carS, one of the genes of the light-inducible carQRS operon, acts as a positive element for the activation of PB by light (see the introduction). To test whether the lack of response of PB to illumination in the absence of vitamin B12 was the consequence of a primary effect of the vitamin on the PQRS promoter, an experiment identical to the one described above was done with the wild-type strain carrying the PQRS::lacZ fusion. Vitamin B12 was not required for the activation of PQRS, although it stimulated the response of that promoter to illumination (Fig. 5B).
DISCUSSION
In wild-type M. xanthus, the accumulation of carotenoids is absolutely dependent on the activation of the single gene crtI and the carB operon by blue light. The carA gene of this bacterium was originally defined as the one impaired in a mutant strain that accumulated slightly colored, partially dehydrogenated carotenoids in the dark. In the light, the mutant accumulated the same, fully dehydrogenated carotenoids as the wild type grown in the same conditions. The mutation, named carA1, was found to cause constitutive expression from the normally light-inducible promoter of the carB operon (PB) (5, 24, 25). This explains the phenotype of the carA1 mutant, as the carB operon contains six genes responsible for the synthesis of phytoene, the first carotene precursor, as well as for the partial dehydrogenation of phytoene and other steps downstream in the pathway. The accumulation of fully dehydrogenated carotenoids requires the carA-independent activation of the crtI gene (6, 10) (Fig. 1A).
Sequence analysis had revealed that carA1 is in fact a double mutation that could affect more than one gene. Here we have shown that a deletion affecting one of the candidate genes (orf10 in Fig. 1B) reproduces exactly the known effects of mutation carA1, both on the Car phenotype and on the activity of the PB promoter. Therefore, we retained the original carA designation for orf10. The previous proposal that protein CarA is a negative regulator of PB in the dark (see the introduction) was based on the simple but unproven assumption that carA1 was a lack-of-function mutation. Now, our results with the Δorf10 deletion confirm that CarA normally acts as a negative regulator of the PB promoter in the dark. They also confirm that CarA does not participate directly in the regulation of the other light-inducible promoters known in M. xanthus, PQRS and PI.
Downstream of carA, a gene was found (orf11) which was predicted to generate a protein strikingly similar to CarA (Fig. 3). Thus, carA and orf11 may have originated from the duplication of a common ancestral gene. Deleting the orf11 gene produced no effect on the regulation of the PB promoter, nor did it affect the normal accumulation of carotenoids by M. xanthus cells in the light. The function of the orf11 gene is presently unknown.
The sequence and the structural elements predicted for the N termini of CarA and Orf11 closely resemble the DNA-binding domain found at the N termini of the MerR-like proteins (Fig. 3A). This is a family of bacterial transcriptional activators that regulate the response to stress arising from exposure to toxic compounds or oxygen free radicals. The nonhomologous carboxy-terminal domain of the MerR proteins contains the binding site for the coactivator-toxic compound or a redox [2Fe-2S] center, as in the case of the SoxR protein from E. coli (2, 14, 35). Binding to the coactivator or oxidation of the redox center converts the MerR proteins to active forms.
Many lines of evidence, including data on the crystal structure of a member of the family bound to DNA, indicate that MerR proteins activate transcription initiation by remodeling the unusually long (19 bp) spacer found between the −35 and −10 promoter elements (38 and references therein). CarA is not an activator, but probably acts as a truly classical bacterial repressor. This unusual behavior of CarA as a MerR-like protein might be related to the particular position of its binding site relative to the promoter or to certain structural peculiarities of the protein. For example, the MerR proteins contain an 11-turn-long α-helix that separates the DNA-binding region from the coactivator-binding domain (38). This long linker region is absent in CarA, whose DNA-binding domain is immediately linked to the cofactor-binding domain.
As shown in Fig. 3, CarA is predicted to contain a cobalamin-binding domain that is separated from its DNA-binding region by only a few amino acids. Cobalamin is a complex prosthetic group formed by a heme-like corrin macrocycle linked to the nucleotide dimethylbenzimidazole (15). The crystal structure of the methylcobalamin-binding domain of protein MetH from E. coli reveals the corrin macrocycle lying between an amino-terminal domain (domain I), formed by two pairs of antiparallel helices, and an α/β carboxyl-terminal domain (domain II). A bond to a conserved histidine residue replaces the lower axial bond of dimethylbenzimidazole to the cobalt which is found in free methylcobalamin. As a result, the nucleotide is displaced and extended as a tail that penetrates into a deep pocket of domain II (9) (Fig. 4).
The crystal structure of the adenosylcobalamin-binding domain of methylmalonyl-coenzyme A mutase (MutB) has also been solved (22). Not many residues are conserved, but extensive structural similarities exist between the cobalamin-binding domains of MetH and MutB (Fig. 3B). In particular, MutB contains an α/β domain whose tertiary structure superimposes very well on domain II of MetH (22). In this domain, the best-conserved residues are the aforementioned histidine and several other residues that play critical roles in the cobalamin-protein functional interaction (Fig. 4). CarA contains all but one of the conserved residues, at the right spacing and within the expected structural elements (Fig. 3 and 4). The one exception is a conservative leucine-to-valine substitution. A second conservative substitution, aspartic to glutamic acid, is found in Orf11 (Fig. 3B). The four-helix domain I of MetH interacts with the upper face of the methylcobalamin molecule (9). The motif used by MutB to interact with the upper face of adenosylcobalamin shows no sequence or structural similarity to the MetH domain I (22) (Fig. 3B). CarA is predicted to form a four-helix bundle with a spacing similar to that of the MetH domain I.
Cobalamin is absolutely required for the light activation of PB, the M. xanthus promoter regulated by CarA. That requirement is not observed for the CarA-independent, light-inducible promoter PQRS, although cobalamin increases the effect of light on this promoter (Fig. 5). This increase is likely to be an indirect consequence of the effect of cobalamin on the PB promoter. We have data showing that transcription from PB goes through the carA operon and that the gene orf9 from this operon (Fig. 1B) is positively required for the activation of PQRS by light (unpublished data).
The concurrence of structural and physiological data leads to a model in which cobalamin acts as a prosthetic group of CarA that mediates a light-activated on-off shift of this protein as a repressor of the PB promoter. This process will depend on protein CarS in an as yet unknown way. As already mentioned (see the introduction), the overexpression of CarS counteracts the negative action of CarA on PB in the dark, and the same effect is produced by the gain-of-function mutation carS1, which does not affect the low expression of carS in the dark. The model reinforces the versatility of the MerR-like DNA-binding domain, regarding the cofactor-binding domains with which they have become successfully associated during evolution (38). More interestingly, it points to a rather surprisingly novel capacity of cobalamin, that of acting as a cofactor of regulatory, DNA-binding proteins. To our knowledge, this is the first cobalamin-binding protein known to regulate gene expression. A search of the data banks for a CarA-like product, one having a cobalamin- and a DNA-binding domain in the same molecule, was not fruitful.
The precise mechanism of action of cobalamin in the light-mediated inactivation of CarA is an open question. Three cobalamin derivatives have been effective in our tests. This might suggest that the breaking of a specific cobalt-carbon bond is not involved, contrary to what happens in the well-known cobalamin-dependent enzymatic reactions (9, 22). Very little is known about the metabolism of the cobalamins in M. xanthus, although it is known that it can convert vitamin B12 into methylcobalamin (7). Other interconversions of the different cobalamins within the M. xanthus cells cannot be discarded. A distinctive property of methylcobalamin is particularly propitious for the light-driven inactivation of an associated protein. The cob(I)alamin form of MetH can suffer a photolytic conversion to an oxidized form, cob(II)alamin, that renders the enzyme inactive. Interestingly, the enzyme-bound cob(I)alamin but not other forms of protein-bound cobalamins exhibits strong blue light absorbance, with a maximum near 400 nm (19 and references therein).
Current work in our laboratory is aimed at delimiting the carB promoter and purifying the CarA and CarS proteins. This will allow in vitro studies that should bring forth new insights into the novel action of the cobalamins that we have uncovered here.
ADDENDUM IN PROOF
While this paper was under review, we confirmed DNA binding by CarA to an operator site in PB and that CarS abrogates this by interacting with the CarA DNA-binding domain. We also showed that the cobalamin-binding domain is involved in CarA-CarA interaction (J. J. López-Rubio, M. Elías-Arnanz, S. Padmanabhan, and F. J. Murillo, J. Biol. Chem. 277:7262-7270, 2002). Another recent study has also confirmed CarA-CarS interaction and its effect on CarA-DNA binding, but varies from ours in, among others, the details of the protein-DNA and protein-protein interactions (D. E. Whitworth and D. A. Hodgson, Mol. Microbiol. 42:809-819, 2001).
Acknowledgments
Special thanks are given to Simon P. Gough (Frederiksberg, Denmark), who first pointed out to us that Orf10 contains a cobalamin-binding domain, and to José A. Botella, from our laboratory, who generated the orf9 deletion. We also thank José A. Madrid and Ana C. García for technical assistance.
This work was supported by the Spanish Ministerio de Educación y Cultura (grant PB96-1096 and fellowship to M.C.), Ministerio de Ciencia y Tecnología (grant BMC2000-1006), and Fundación Séneca (fellowship to M.C.).
REFERENCES
- 1.Altschul, S. F., and W. Gish. 1966. Local alignment statistics. Methods Enzymol. 266:460-480. [DOI] [PubMed] [Google Scholar]
- 2.Ansari, A. Z., J. E. Bradner, and T. V. O'Halloran. 1995. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374:371-375. [DOI] [PubMed] [Google Scholar]
- 3.Avery, L., and D. Kaiser. 1983. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol. Gen. Genet. 191:99-109. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre, J. M. 1989. Inducción por la luz de la expresión génica y la carotenogénesis en Myxococcus xanthus. Ph.D. thesis. Universidad de Murcia, Murcia, Spain.
- 5.Balsalobre, J. M., R. M. Ruiz-Vázquez, and F. J. Murillo. 1987. Light induction of gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 84:2359-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botella, J. A., R. M. Ruiz-Vázquez, and F. J. Murillo. 1995. A cluster of structural and regulatory genes for light-induced carotenogenesis in Myxococcus xanthus. Eur. J. Biochem. 223:238-248. [DOI] [PubMed] [Google Scholar]
- 7.Bretscher, A. P., and D. Kaiser. 1978. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 133:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in E. coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 9.Drennan, C. L., S. Huang, J. T. Drummond, R. G. Matthews, and M. L. Ludwig. 1994. How a protein binds B12: a 3.0 A X-ray structure of B12-binding domains of methionine synthase. Science 266:1669-1674. [DOI] [PubMed] [Google Scholar]
- 10.Fontes, M., R. M. Ruiz-Vázquez, and F. J. Murillo. 1993. Growth phase dependence of the activation of a bacterial gene for carotenoid synthesis by blue light. EMBO J. 12:1265-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill, R. E., M. G. Cull, and S. Fly. 1988. Genetic identification and cloning of a gene required for developmental cell interactions in Myxococcus xanthus. J. Bacteriol. 170:5279-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorham, H. C., S. J. McGowan, P. R. H. Robson, and D. A. Hodgson. 1996. Light-induced carotenogenesis in Myxococcus xanthus: light-dependent membrane sequestration of ECF sigma factor CarQ by antisigma factor CarR. Mol. Microbiol. 19:171-186. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1983. Studies of transformation of E. coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 14.Hidalgo, E., H. Ding, and B. Demple. 1997. Redox signal transduction via iron-sulfur clusters in the SoxR transcription activator. Trends Biochem. Sci. 22:207-210. [DOI] [PubMed] [Google Scholar]
- 15.Hodgkin, D. C., J. Kamper, M. Mackay, J. Pickworth, K. Trueblood, and J. White. 1956. Structure of vitamin B12. Nature 178:64-66. [DOI] [PubMed] [Google Scholar]
- 16.Hodgson, D. A. 1993. Light-induced carotenogenesis in Myxococcus xanthus: genetic analysis of the carR region. Mol. Microbiol. 7:471-488. [DOI] [PubMed] [Google Scholar]
- 17.Hodgson, D. A., and A. E. Berry. 1998. Light regulation of carotenoid synthesis in Myxococcus xanthus, p. 186-211. In M. X. Caddick, S. Baumber, D. A. Hodgson, and M. K. Phillips-Jones (ed.), Microbial responses to light and time. Cambridge University Press, Cambridge, United Kingdom.
- 18.Imai, Y., Y. Matsushima, T. Sugimura, and M. Terada. 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarret, J. T., C. W. Goulding, K. Fluhr, S. Huang, and R. G. Mathews. 1997. Purification and assay of cobalamin-dependent methionine synthase from Escherichia coli. Methods Enzymol. 281:196-213. [DOI] [PubMed] [Google Scholar]
- 20.Julien, B., D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashefi, A., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 22.Mancia, F., N. H. Keep, A. Nakagawa, P. L. Leadlay, S. McSweeney, B. Rasmussen, P. Bösecke, O. Diat, and P. R. Evans. 1996. How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 Å resolution. Structure 4:339-350. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Argudo, I., R. M. Ruiz-Vázquez, and F. J. Murillo. 1998. The structure of an ECF-σ-dependent, light-inducible promoter from the bacterium Myxococcus xanthus. Mol. Microbiol. 30:883-893. [DOI] [PubMed] [Google Scholar]
- 24.Martínez-Laborda, A., J. M. Balsalobre, M. Fontes, and F. J. Murillo. 1990. Accumulation of carotenoids in structural and regulatory mutants of the bacterium Myxococcus xanthus. Mol. Gen. Genet. 223:205-210. [DOI] [PubMed] [Google Scholar]
- 25.Martínez-Laborda, A., and F. J. Murillo. 1989. Genic and allelic interactions in the carotenogenic response of Myxococcus xanthus to blue light. Genetics 122:801-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGowan, S. J., H. C. Gorham, and D. A. Hodgson. 1993. Light-induced carotenogenesis in Myxococcus xanthus: DNA sequence analysis of the carR region. Mol. Microbiol. 10:713-735. [DOI] [PubMed] [Google Scholar]
- 27.Moreno, A. J., M. Fontes, and F. J. Murillo. 2001. ihfA gene of the bacterium Myxococcus xanthus and its role in activation of carotenoid genes by blue light. J. Bacteriol. 183:557-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolás, F. J., R. M. Ruiz-Vázquez, and F. J. Murillo. 1994. A genetic link between light response and multicellular development in the bacterium Myxococcus xanthus. Genes Dev. 8:2375-2387. [DOI] [PubMed] [Google Scholar]
- 29.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Vázquez, R. M., M. Fontes, and F. J. Murillo. 1993. Clustering and co-ordinated activation of carotenoid genes in Myxococcus xanthus by blue light. Mol. Microbiol. 10:25-34. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Vázquez, R. M., and F. J. Murillo. 1984. Abnormal motility and fruiting behavior of Myxococcus xanthus bacteriophage-resistant strains induced by a clear plaque mutant of bacteriophage Mx8. J. Bacteriol. 160:818-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salamov, A. A., and V. V. Solovyev. 1995. Prediction of protein secondary structure by combining nearest-neighbour algorithms and multiple sequence alignments. J. Mol. Biol. 247:11-15. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Smith, R. F., and T. F. Smith. 1992. Pattern-induced multi-sequence alignment (PIMA) algorithm employing secondary structure-dependent gap penalties for comparative protein modeling. Protein Eng. 5:35-41. [DOI] [PubMed] [Google Scholar]
- 35.Summers, A. O. 1992. Untwist and shout: a heavy metal-responsive transcriptional regulator. J. Bacteriol. 174:3097-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueki, T., S. Inouye, and M. Inouye. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153-157. [DOI] [PubMed] [Google Scholar]
- 37.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal plasmids. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 38.Zheleznova-Heldwein, E. E., and R. G. Brennan. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409:378-382. [DOI] [PubMed] [Google Scholar]