Abstract
Campylobacter jejuni produces both lipooligosaccharide (LOS) and a higher-molecular-weight polysaccharide that is believed to form a capsule. The role of these surface polysaccharides in C. jejuni-mediated enteric disease is unclear; however, epitopes associated with the LOS are linked to the development of neurological complications. In Escherichia coli and Salmonella enterica serovar Typhimurium the waaF gene encodes a heptosyltransferase, which catalyzes the transfer of the second l-glycero-d-manno-heptose residue to the core oligosaccharide moiety of lipopolysaccharide (LPS), and mutation of waaF results in a truncated core oligosaccharide. In this report we confirm experimentally that C. jejuni gene Cj1148 encodes the heptosyltransferase II enzyme, WaaF. The Campylobacter waaF gene complements an S. enterica serovar Typhimurium waaF mutation and restores the ability to produce full-sized lipopolysaccharide. To examine the role of WaaF in C. jejuni, waaF mutants were constructed in strains NCTC 11168 and NCTC 11828. Loss of heptosyltransferase activity resulted in the production of a truncated core oligosaccharide, failure to bind specific ligands, and loss of serum reactive GM1, asialo-GM1, and GM2 ganglioside epitopes. The mutation of waaF did not affect the higher-molecular-weight polysaccharide supporting the production of a LOS-independent capsular polysaccharide by C. jejuni. The exact structural basis for the truncation of the core oligosaccharide was verified by comparative chemical analysis. The NCTC 11168 core oligosaccharide differs from that known for HS:2 strain CCUG 10936 in possessing an extra terminal disaccharide of galactose-β(1,3) N-acetylgalactosamine. In comparison, the waaF mutant possessed a truncated molecule consistent with that observed with waaF mutants in other bacterial species.
The human pathogen Campylobacter jejuni is the principal cause of bacterial enteric disease in many industrializedcountries. C. jejuni enteritis is normally a self-limiting and uncomplicated illness, with symptoms that range from mild diarrhea to severe inflammatory enteritis (21). In a small proportion of cases the serious neurological complication Guillain-Barré syndrome (GBS) or the related Miller-Fisher syndrome (MFS) can develop (32, 37). The consumption of contaminated food or water is the usual route for bacterial acquisition. Due to a high and increasing frequency of infection, C. jejuni is of major health and economic importance (50). In contrast to those of many other enteric pathogens, the biology of C. jejuni and the pathophysiology of Campylobacter-mediated disease remain poorly understood (50).
Lipopolysaccharide (LPS) and lipooligosaccharide (LOS) constituents of the outer membrane are important virulence factors in many bacterial infections, conferring, for example, serum resistance, antibiotic resistance, and endotoxicity to the pathogen (39). LPS is composed of three regions, lipid A, a core oligosaccharide, and an O-chain consisting of repeating oligosaccharide units; LOS lacks the O-chain (40). Until recently, C. jejuni was believed to produce either LPS or LOS. However, repeating oligosaccharide units once considered to be an LPS-associated O-chain are now believed to be capsular (18, 50), and there is also a considerable level of surface protein glycosylation (47). Thus, although there is a greater capacity for polysaccharide biosynthesis than previously supposed, C. jejuni may only express LOS and not LPS. C. jejuni heat-stable antigens, considered to include a capsule and LOS, form the basis of the Penner serotyping system (18, 29, 38). Certain serotypes, and hence LOS structures, have been linked to the development of C. jejuni-associated GBS or MFS (37). In addition, cell surface polysaccharides may play a role in cell adhesion, as purified C. jejuni “LPS” was shown to adhere to tissue culture cells and intestinal mucus (28), and it has been suggested that sialylation of C. jejuni LOS affects immunogenicity and serum resistance (13).
The genetic basis of LOS biosynthesis in Campylobacter was first addressed in a study describing regions of the genome of Campylobacter coli (Campylobacter hyoilei), which contained open reading frames (ORFs) with similarity to known genes involved in LPS biosynthesis (24). This study was extended to identify a similar region in C. jejuni NCTC 11828 (also known as 81116), which was able to complement LPS biosynthesis in an Escherichia coli mutant background (11). The region was found to contain 13 ORFs, designated galE and wlaB to wlaM, which had similarity to genes involved in LPS biosynthesis. Mutation of some of these genes in strain NCTC 11168 resulted in a phenotype of altered LOS reactivity, with serotyping antisera directly supporting a role for this region in LOS biosynthesis (52). Interestingly, an additional role for genes in the wla region in C. jejuni strain 81-176 was demonstrated (47). In contrast to what was found previously (52), mutation of several wla (alternatively named pgl [47]) genes in 81-176 had no discernible effect on LOS and capsule biosynthesis but affected the glycosylation of several C. jejuni proteins including flagellin (47).
Analysis of the complete genome sequence of C. jejuni NCTC 11168 led to the identification of a large cluster of LOS-related ORFs (35), which includes the previously described wla genes. This cluster contains several additional ORFs with similarity to genes involved in inner core biosynthesis including galE (Cj1131), gmhA (Cj1149), gmhD (Cj1151), gmhE (Cj1150), waaC (Cj1133), and waaF (Cj1148). The putative roles of both the galE and waaC genes of C. jejuni have now been verified by functional studies (10, 23), but the roles of other inner core biosynthesis genes still require experimental confirmation.
In E. coli and Salmonella enterica serovar Typhimurium the waaF gene encodes a heptosyltransferase, which catalyzes the transfer of the second l-glycero-d-manno-heptose (LD-Hep) residue to the core oligosaccharide moiety of LPS (8, 46). Mutation of waaF prevents the incorporation of the heptose residue and subsequently blocks the addition of other sugar moieties. Thus, a greatly altered LPS molecule, consisting of a truncated core oligosaccharide and no attached O-chain polysaccharide, is produced (42). Such so-called deep rough Salmonella mutants have increased sensitivity to antibiotics, detergents, and bile salts (9). In particular, they are highly susceptible to hydrophobic antibiotics such as novobiocin. Functional complementation restores the ability of the mutant to synthesize a complete core oligosaccharide with attached O-chain and consequently also restores antibiotic resistance. Complementation of antibiotic resistance has been used as an approach to isolate several waaF genes from various bacterial species including Pseudomonas aeruginosa (22), Bordetella pertussis (1), Haemophilus influenzae (33), and Haemophilus ducreyi (7).
Here we report experimental confirmation that Cj1148 encodes the heptosyltransferase II enzyme, WaaF, by functional complementation and characterization of the mutant phenotype. Furthermore, analysis of LOS extracts from the waaF mutant showed directly that the previously postulated O-chain polysaccharide observed in electrophoretic gels and immunoblots is not covalently attached to the core oligosaccharide and is an independent extracellular polysaccharide.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
All bacterial strains used in this study are described in Table 1. C. jejuni strains NCTC 11168 and NCTC 11828 used in this study are from Penner serotypes HS:2 and HS:6, respectively. Both strains were cultured on Campylobacter serum-free agar (Oxoid) or in Mueller-Hinton broth at 37°C in microaerophilic conditions (85% N2, 10% CO2, 5% O2). S. enterica serovar Typhimurium SL3770 (wild-type) and SL3789 (waaF511) and E. coli strain DH5α were cultured on Luria broth or agar (41). Antibiotic selection with chloramphenicol (20 μg ml−1), ampicillin (100 μg ml−1), and novobiocin (5 μg ml−1) was used as appropriate. All antibiotics and chemical reagents were obtained from Sigma Chemical Co.
TABLE 1.
Bacterial strains, plasmids, and primers used
| Strain, plasmid, or primer | Relevant genotype, phenotype, characteristics, or nucleotide sequence (5′-3′) | Reference or source |
|---|---|---|
| Strains | ||
| C. jejuni | ||
| NCTC 11168 | Parental | National Collection of Type Cultures |
| NCTC 11828 | Parental; also known as 81116; produces high-molecular-weight polysaccharide | 34 |
| 11168NOL22/23 | NCTC 11168 waaF::cat | This study |
| 11828NOL11/12 | NCTC 11828 waaF::cat | This study |
| S. enterica serovar Typhimurium | ||
| SL3770 | Parental | T. Isobea |
| SL3789 | waaF511 | T. Isobe |
| E. coli DH5α | F− φ80d lacZΔM15 | 14 |
| Plasmids | ||
| pUC19 | Cloning vector, Apr | Stratagene |
| pAV35 | C. coli Cmr in pBluescript | 49 |
| pNOL8 | 2.1-kb fragment containing waaF from NCTC 11828 in pUC19 | This study |
| pNOL11 | Cmr cassette of pAV35 cloned (forward orientation) into unique BamHI site generated in pNOL8 by inverse PCR | This study |
| pNOL12 | Cmr cassette of pAV35 cloned (reverse orientation) into unique BamHI site generated in pNOL8 by inverse PCR | This study |
| pNOL17 | 2.0-kb fragment containing waaF from NCTC 11168 in pUC19 | This study |
| pNOL22 | Cmr cassette of pAV35 cloned (forward orientation) into unique BamHI site generated in pNOL17 by inverse PCR | This study |
| pNOL23 | Cmr cassette of pAV35 cloned (reverse orientation) into unique BamHI site generated in pNOL17 by inverse PCR | This study |
| Primers | ||
| GMHAF3 | CGGGGTACCCAAATCGCTAAAGTAGGTGAGC | This study |
| WLASAR3 | AAACTGCAGCACTTAGCCCAAACCGACCAGC | This study |
| WLAXAF3 | CGGGGTACCGAGCTAAAAGCATAAACAAC | This study |
| WAAFF2 | CGCGGATCCCGCCTAACCAGGTGGGAAGATG | This study |
| WAAFR4 | CGCGGATCCGCCTGCTGAAAAAGTGATAGAACAGGC | This study |
| NOL8F1 | CGCGGATCCGCATGAAAGATCTAAAGCCTG | This study |
Department of Clinical Veterinary Medicine, University of Cambridge, Cambridge, United Kingdom.
Molecular genetics methods.
The Expand High Fidelity PCR system (Boehringer Mannheim) was used for DNA amplification with the primers described in Table 1. C. jejuni-specific primers were designed by using the NCTC 11168 genome sequence (available at http://www.sanger.ac.uk/Projects/C_jejuni/) or sequence data obtained from studies on NCTC 11828 (N. J. Oldfield and J. M. Ketley, unpublished results). DNA modification was carried out by standard methods (41) with DNA isolated and purified by Qiagen purification kit technology. DNA was sequenced with the BigDye terminator cycle sequencing kit (ABI, Applied Biosystems) on an ABI 377 DNA sequencer. Restriction and modifying enzymes were obtained from Gibco-BRL. Molecular genetics methods utilized with C. jejuni are based on those described previously (49).
Cloning and mutation of the putative waaF gene from C. jejuni.
Chromosomal DNA was extracted from C. jejuni by a guanidinium thiocyanate-based method (36). A 2-kb fragment containing the waaF gene and approximately 500 bp of flanking sequence from the neighboring ORFs Cj1147 and Cj1149 was amplified with primers GMHAF3 and WLASAR3 (NCTC 11168) or WLAXAF3 and WLASAR3 (NCTC 11828) (Table 1), cloned into pUC19, and confirmed by PCR and restriction analysis.
A deletion in the waaF gene was constructed by inverse PCR (53) using primers WAAFF2 and WAAFR4 for pNOL17 and primers WAAFF2 and NOL8F1 for pNOL8 (Table 1). A chloramphenicol acetyltransferase cassette (cat) (51) was inserted (49) into the unique restriction site generated during the PCR. The resulting plasmids (Table 1) containing the cat resistance cassette in the forward and reverse orientations were electroporated into C. jejuni (52), and recombinants were selected on medium containing chloramphenicol.
LPS and LOS extracts and SDS-PAGE.
Late-exponential-phase cells (1 ml at an optical density at 600 nm of 0.6) were harvested by centrifugation (3,020 × g, 20 min) and resuspended in 200 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (100 mM Tris-Cl [pH 8.0], 2% β-mercaptoethanol, 4% SDS, 0.2% bromophenol blue, 0.2% xylene cyanol, 20% glycerol). After the cells were boiled for 10 min, proteinase K was added to a final concentration of 0.5 mg ml−1 and the sample was incubated at 65°C for 2 h. LPS preparations were boiled for 10 min before being loaded onto an SDS-PAGE gel (52) (4% [vol/vol]) stacking gel and 15% [vol/vol] separating gel) and electrophoresed at 15 mA per gel for 16 h. Gels were silver stained (48), or the separated LPS and LOS extracts were transferred to nitrocellulose (41). Penner serotyping antisera (HS:2 and HS:6) were used at a dilution of 1/5,000 and detected with the Bio-Rad Immun-Star Western blotting kit.
TLC.
Biomass of C. jejuni NCTC 11168 and mutant strain 11168NOL22, which had been grown as described above, was subjected to hot phenol-water extraction, and the crude LPS and LOS from the water phase of extracts were purified by enzymatic treatments with RNase A, DNase II, and proteinase K and by ultracentrifugation, according to the protocol of Moran et al. (31). The LOSs (1 μg) from C. jejuni NCTC 11168 and C. jejuni 11168NOL22 were analyzed by thin-layer chromatography (TLC) on precoated silica gel 60 glass plates (Merck, Darmstadt, Germany). A solvent system consisting of n-propanol-water-25% NH4OH (60:30:10 [vol/vol/vol]) was used as a developer, and bands were visualized by spraying plates with resorcinol-HCl reagent (45).
TLC with immunostaining was performed by the procedure of Schwerer et al. (44). Briefly, developed TLC plates were dried for 30 min in a vacuum dessiccator, fixed in 0.2% polyisobutylmethacrylate (Aldrich) in n-hexane (Merck) for 1.5 min, and dried as before. Nonspecific binding was reduced by submerging the plates for 1 h in a solution of phosphate-buffered saline (PBS) containing 0.3% gelatin (gelatin-PBS). Subsequently, lanes were overlaid with rabbit antiserum to gangliosides (Matreya Inc.) and diluted 1:100 in gelatin-PBS. Plates were incubated at 4°C overnight, washed three times with cold PBS, overlaid with peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma) diluted 1:500 in gelatin-PBS, and incubated at room temperature for 1 h with gentle rocking. The plates were washed with cold PBS, and the immunoreactants were visualized by use of a horseradish-peroxidase development system (Bio-Rad). Binding studies with cholera toxin (CT)-peroxidase conjugate (Sigma), peanut agglutinin (PNA)-peroxidase conjugate (Kem-En-Tec, Copenhagen, Denmark), and tetanus toxin C (TTC; Calbiochem) were performed under the same conditions as described for immunostaining, but only one overlay step with peroxidase-conjugated CT (1 μg/ml), PNA (20 μg/ml), or TTC (1 μg/ml) was necessary.
Chemical characterization of LOS.
Core oligosaccharide was liberated from LOS by mild acid hydrolysis with 1% acetic acid at 100°C for 1 h and isolated by gel permeation chromatography, and the oligosaccharides (400 to 500 μg) were methylated (5). Subsequently, the fast atom bombardment-mass spectrometry (FAB-MS) spectra of the permethylated sample in methanol (1 to 2 μl) were recorded with an instrument equipped with an Ion Tech saddle field gun under the conditions described previously (5). Interpretations of positive-ion mass spectra of permethylated derivatives were as used in earlier studies (4-6).
Nucleotide sequence accession numbers.
The complete sequence of the C. jejuni NCTC 11168 genome can be obtained from the Sanger Centre (http://www.sanger.ac.uk/Projects/C_jejuni/) or from EMBL with identification no. CJ11168 (accession no. AL11168). The DNA sequence of the NCTC 11828 insert from pNOL8 described here has been deposited in the GenBank database under accession no. AF343914.
RESULTS
Cloning of the waaF gene.
The genome sequence of C. jejuni NCTC 11168 (35) contains an ORF (Cj1148) with similarity to several waaF genes (data not shown). A 2-kb fragment containing the waaF gene was amplified with primers that annealed to the flanking genes, Cj1147 and Cj1149, and cloned into pUC19 (data not shown) to form pNOL17 (Table 1). The corresponding waaF gene of C. jejuni NCTC 11828 on a 2.1-kb fragment was cloned by PCR, but with one different flanking primer due to the presence of a novel gene immediately downstream of waaF in the strain NCTC 11828 wla locus (N. J. Oldfield et al., unpublished data). The resulting plasmid containing the strain NCTC 11828 waaF gene was named pNOL8 (Table 1).
Complementation of S. enterica serovar Typhimurium SL3789 with C. jejuni NCTC 11168 waaF.
The mutant S. enterica serovar Typhimurium SL3789 (waaF511) was transformed with plasmid pNOL17, and transformants were selected with ampicillin and novobiocin. The deep rough S. enterica serovar Typhimurium SL3789 is normally unable to grow in the presence of novobiocin due to the truncated LPS produced by the waaF511 mutation (1). The presence of pNOL17 in novobiocin-resistant transformants was confirmed, indicating that the putative C. jejuni waaF gene was able to complement the waaF511 mutation in S. enterica serovar Typhimurium SL3789. In contrast, plasmids pNOL22 and pNOL23 (Table 1; see below), which contain mutated C. jejuni waaF genes, were unable to complement the waaF511 mutation in SL3789.
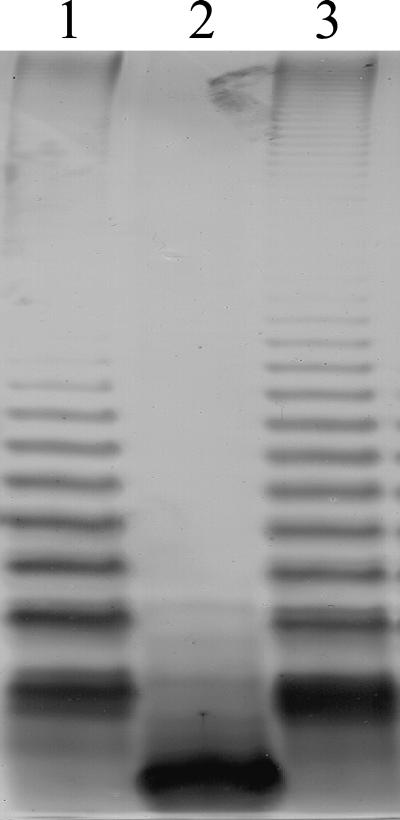
Growth of S. enterica serovar Typhimurium SL3789(pNOL17) on novobiocin implies functional complementation of the waaF mutation due to the production of smooth LPS, which confers resistance to the antibiotic. The production of a complete LPS molecule with an O-chain polysaccharide was confirmed by analysis of the LPS from the complemented Salmonella strain (Fig. 1). In contrast to what is found for S. enterica serovar Typhimurium SL3789, which shows only the presence of a truncated core molecule, the production of attached O-chain is restored in strain SL3789(pNOL17), further supporting the identification of Cj1148 as the C. jejuni waaF gene. Moreover, the truncated core molecule seen in S. enterica serovar Typhimurium SL3789 is not observed in strain SL3789(pNOL17), indicating that complementation is complete. In contrast, the waaF gene of B. pertussis was shown to confer only partial functional complementation in strain SL3789, as the truncated core molecule was still observed in LPS preparations (1).
FIG. 1.
Complementation of the S. enterica serovar Typhimurium waaF511 mutation using the C. jejuni NCTC 11168 waaF gene. S. enterica serovar Typhimurium LPS was isolated from the wild type, waaF mutant, and waaF mutant complemented with C. jejuni waaF, separated electrophoretically on an SDS-PAGE gel, and silver stained. Lane 1, wild-type S. enterica serovar Typhimurium SL3770; lane 2, mutant S. enterica serovar Typhimurium SL3789 (waaF511); lane 3, S. enterica serovar Typhimurium SL3789 complemented by the C. jejuni NCTC 11168 waaF gene.
Construction and characterization of C. jejuni NCTC 11168 waaF mutants.
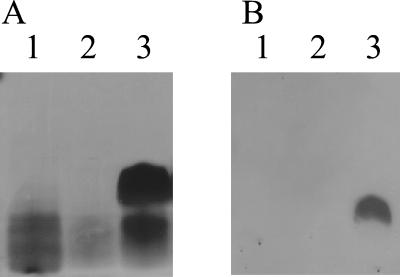
To confirm that Cj1148 encoded a heptosyltransferase II in C. jejuni, the gene was mutated and the effect on LOS biosynthesis was determined. Inverse PCR was used to construct a deletion in pNOL17 that resulted in the removal of approximately 870 bp of waaF coding sequence, and an 850-bp chloramphenicol resistance cassette from pAV35 (49, 51) was inserted in both orientations at the site of the deletion, forming plasmids pNOL22 (cat forward orientation) and pNOL23 (cat reverse orientation) (Table 1). Constructs pNOL22 and pNOL23 were each transformed into C. jejuni NCTC 11168, and putative recombinants were assessed by PCR and Southern blotting to verify that allelic replacement with the mutant waaF allele had occurred (data not shown). The C. jejuni waaF mutant strains were named 11168NOL22 and 11168NOL23 (Table 1). The LOS produced by these mutants was characterized by SDS-PAGE followed by silver staining or Western blotting (immunoblotting) using Penner 2 antisera (Fig. 2). Silver staining revealed that the core molecules produced by the waaF mutant strains were notably truncated compared to that produced by C. jejuni NCTC 11168. Immunoblotting the waaF mutants confirmed truncation of the LOS molecules to such an extent as to render them no longer detectable with Penner serum, and thus the predicted absence of the outer core can be deduced to have removed the majority of HS:2 serum-reactive epitopes in the C. jejuni waaF mutants. Consistent with these findings, in TLC with immunostaining, LOS from C. jejuni 11168NOL22 did not react with antisera against GM1, asialo-GM1, or GM2 gangliosides whereas LOS of the C. jejuni NCTC 11168 serostrain reacted with all three antisera. Similarly, although C. jejuni NCTC 11168 LOS bound PNA, CT, and TTC ligands, LOS of 11168NOL22 did not. Hence, compared to the parental strain, the waaF mutant has lost the outer core, which contains ganglioside-like epitopes, due to the truncation of the core oligosaccharide.
FIG. 2.
Mutation of the waaF gene in C. jejuni NCTC 11168 affects the size and antigenicity of core oligosaccharide. SDS-PAGE gels of LOS extracts of wild-type C. jejuni NCTC 11168 and waaF mutants were silver stained (A) or immunoblotted with HS:2 antisera (B). The truncated LOS is not detectable by Western blotting using HS:2 antisera. Lane 1, waaF mutant 11168NOL22 (cat forward orientation); lane 2, waaF mutant 11168NOL23 (cat reverse orientation); lane 3, NCTC 11168.
Chemical analysis of core oligosaccharide produced by wild-type and waaF mutant strains of C. jejuni NCTC 11168.
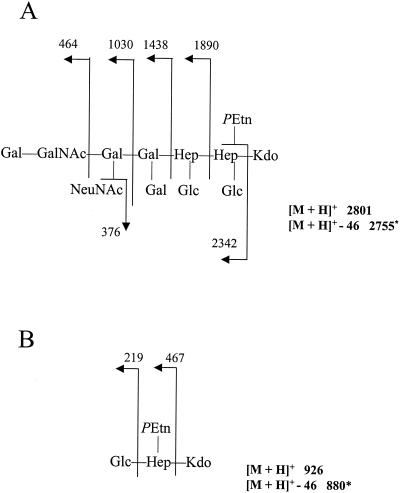
Loss of WaaF, a heptosyltransferase, is predicted to result in a truncation of the core oligosaccharide after the first LD-Hep residue that is added by WaaC. To verify the exact structural basis for the truncation seen in the NCTC 11168 waaF mutant, comparative chemical analysis of the mutant and parental strains was undertaken. Core oligosaccharides were liberated from LOSs of C. jejuni NCTC 11168 and strain 11168NOL22, methylated, and subsequently analyzed by FAB-MS. As shown in Fig. 3A, the permethylated core oligosaccharide of strain NCTC 11168 possessed a pseudomolecular ion (m/z = 2,801 [M+H]+). Consistent with the observed reactivity with antiganglioside antibodies and various ligands in TLC, the sugar sequence of the outer tetrasaccharide is identical to that in the GM1 ganglioside. Moreover, a complete structural analysis of the core oligosaccharide of NCTC 11168 has confirmed this and has shown that the core oligosaccharide differs to that of the previously published core of CCUG 10936 (also HS:2) in possessing an extra disaccharide of Gal-β(1,3)GalNAc (A. P. Moran et al., unpublished results). In contrast, the permethylated oligosaccharide of strain 11168NOL22 (Fig. 3B) possessed a pseudomolecular ion (m/z = 926 [M+H]+) and daughter ions indicative of a truncated molecule composed of glucose, heptose, 3-deoxy-d-manno-2-octulosonic acid (Kdo), and phosphorylethanolamine. Therefore, the phenotypic analysis of LOS produced by the C. jejuni waaF mutant supports the presence of a truncated core oligosaccharide, a phenotype entirely consistent with those of waaF mutants in other species.
FIG. 3.
Analysis of positive-ion FAB-MS of permethylated core oligosaccharide from LOS of wild-type C. jejuni NCTC 11168 (A) and waaF mutant 11168NOL22 (B). Numbers refer to m/z values for ions. Abbreviations: GalNAc, N-acetyl-d-galactosamine; Gal, d-galactose; Glc, d-glucose; Hep, heptose; PEtn, phosphorylethanolamine. ∗, derivatization results in partial degradation of the Kdo with loss of 46 mass units.
Construction and characterization of C. jejuni NCTC 11828 waaF mutants.
Until recently the high-molecular-weight polysaccharides produced by some strains of C. jejuni were proposed to be O-chain polysaccharides attached to core oligosaccharides (38). However, the presence of genes possibly involved in capsular biosynthesis in the C. jejuni genome (35) was reported, and the mutation of several of these kps genes leads to loss of high-molecular-weight polysaccharide biosynthesis (18). Thus, C. jejuni produces an extracellular polysaccharide encoded by the locus containing the kps genes, but it remained to be shown that this polysaccharide is not attached to the core oligosaccharide, and thus is not an O-chain polysaccharide.
C. jejuni NCTC 11828 (HS:6) produces a high-molecular-weight polysaccharide that can be visualized by immunoblotting on a nitrocellulose membrane (52). The putative waaF gene from NCTC 11828 cloned into plasmid pNOL8 (see above) shows 94% identity at the DNA level to the NCTC 11168 waaF gene (data not shown). A deletion mutation in the C. jejuni NCTC 11828 waaF gene of pNOL8 was obtained by inverse PCR, and the approximately 820 bp of deleted sequence was replaced with a cat resistance cassette to form plasmids pNOL11 (cat in the forward orientation) and pNOL12 (cat in the reverse orientation). These two constructs were transformed into C. jejuni NCTC 11828, the resultant recombinant strains were selected on chloramphenicol, and the expected allelic replacement in each strain was verified by PCR and Southern blotting (data not shown).
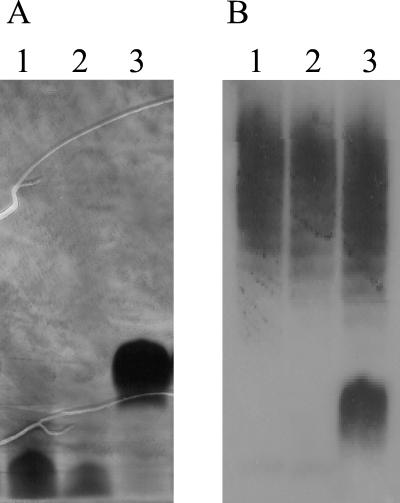
Extracts of the NCTC 11828 waaF mutants (11828NOL11 and 11828NOL12) were analyzed electrophoretically (Fig. 4). Silver staining and Western blotting with HS:6 antisera showed that, as with 11168NOL22 and 11828NOL23, the core molecules produced by 11828NOL11 and 11828NOL12 were significantly truncated. However, significantly, the mobility and intensity of the high-molecular-weight polysaccharides in both 11828NOL11 and 11828NOL12 were unaffected by mutation of waaF. As would be predicted for an LOS-independent polysaccharide, the high-molecular-weight polysaccharide does not appear to be covalently attached to the lipid A core oligosaccharide molecule.
FIG. 4.
Mutation of the waaF gene in C. jejuni NCTC 11828 affects the size and antigenicity of the core oligosaccharide but not of the high-molecular-weight polysaccharide. SDS-PAGE gels of polysaccharide extracts from parental C. jejuni NCTC 11828 and waaF mutants were stained with silver (A) and immunoblotted (B) by using HS:6 antisera. Lane 1, waaF mutant 11828NOL11 (cat forward orientation); lane 2, waaF mutant 11828NOL12 (cat reverse orientation); lane 3, parental C. jejuni NCTC 11828.
DISCUSSION
Before the completion of the C. jejuni genome sequencing project (35) little was known about the genetic basis of cell surface-associated oligosaccharide production. Functional analysis of genes present in two of the three chromosomal regions containing polysaccharide biosynthesis genes (13, 18, 25, 26, 52) has revealed that C. jejuni may produce LOS and extracellular capsule-related polysaccharide(s) rather than rough and smooth LPS. However, the exact structure of the LOS molecule of C. jejuni NCTC 11168 was not known, and although genes that are involved in the biosynthesis of a putative capsule were identified (18), there was no genetic evidence to support the suggestion from previous structural studies (2, 15) of the occurrence of an extracellular polysaccharide not covalently linked to the core oligosaccharide.
The core oligosaccharide structures of several C. jejuni serotypes have been determined, and all contain two LD-Hep residues attached to Kdo, forming a conserved trisaccharide of LD-Hep-α(1,3)-LD-Hep-α(1,5)-Kdo (30, 37). These core structures include that of an HS:2 strain (CCUG 10936) (5). Moreover, the FAB-MS data presented in this study are consistent with the occurrence of such an inner core in C. jejuni NCTC 11168. Furthermore, recent detailed chemical analysis of the latter core oligosaccharide has confirmed the occurrence of this same inner core (A. P. Moran et al., unpublished results).
Analysis of the complete genome sequence of C. jejuni NCTC 11168 identified Cj1148 as a candidate for the waaF gene. Complementation of an S. enterica serovar Typhimurium waaF mutant showed that Cj1148 can restore the production of a complete core oligosaccharide and, thus, attachment of the O-chain. Mutation of Cj1148 in C. jejuni NCTC 11168 resulted in the production of a truncated LOS molecule. This mutant phenotype is consistent with that reported for waaF mutants of other gram-negative bacteria including E. coli (8), B. pertussis (1), H. influenzae (16, 33), H. ducreyi (7), Neisseria gonorrhoeae (43), and Neisseria meningitidis (17). WaaF is a heptosyltransferase which catalyzes the transfer of the second LD-Hep residue to the core oligosaccharide moiety (8, 46), the first heptose being added to Kdo by the heptosyltransferase I enzyme, WaaC. Mutation of waaC has been reported not to be possible in C. jejuni (23; M. Konkel, personal communication). Our C. jejuni waaF mutant is viable, and the loss of WaaF activity resulted in the truncation of the core oligosaccharide of the LOS as determined by SDS-PAGE. Also, this was reflected by the lack of reactivity of this LOS with all the ganglioside and ligand reagents tested, due to the loss of ganglioside mimicry in the outer core oligosaccharide. Furthermore, comparison with the core oligosaccharide of the parental strain, NCTC 11168, by FAB-MS analyses confirmed that truncation was due to the lack of incorporation of the second heptose residue.
Some strains of C. jejuni produce low-molecular-weight glycolipids similar to the LOS molecules produced by Haemophilus and Neisseria spp. (29); however, the additional presence in some strains of ladder-like high-molecular-weight molecules was at first thought to suggest that such strains also produced high-molecular-weight O-chain-bearing LPS (38). The structural similarity of some of the polysaccharide repeat units to those found in other gram-negative O-chains supported the assumption that the ladder-like polysaccharide was the O-chain and, thus, was linked to the core oligosaccharide (29). Although there were limited structural data to support the covalent attachment of an O-chain polysaccharide to a lipid A core molecule (3), there were data from structural studies to suggest the production of other types of extracellular polysaccharide molecules (2, 15). Furthermore, Karlyshev et al. (18) identified genes involved in the production of the high-molecular-weight polysaccharide molecules. Given the deduced amino acid sequence similarity to enzymes associated with capsular biosynthesis, the latter investigators proposed the production of a polysaccharide capsule in C. jejuni; capsular material in NCTC 11828 has been demonstrated by electrophoretic analysis (20) and has been demonstrated in another strain by electron microscopy (19). We established that Cj1148 encodes WaaF, a heptosyltransferase II enzyme, and that a waaF mutant contains a substantially truncated core oligosaccharide. Given that it has not proved possible to produce a waaC mutant, the waaF mutant is the deepest rough strain of C. jejuni available. The waaF mutation has enabled us to clearly establish that, as expected for LPS-independent material, the high-molecular-weight molecules are not attached in significant amounts to the lipid A core oligosaccharide. Moreover, preliminary chemical studies have isolated and identified extracellular polysaccharide molecules produced by strain NCTC 11828 (81116) in which core sugars such as heptose and Kdo are not detectable (A. P. Moran and A. V. Savage, unpublished results). Thus, by complementary study of the core oligosaccharide structure, our data extend the study of Karlyshev et al. (18) and also support the deductions of previous structural studies (2, 15) that C. jejuni strains produce LOS and elaborate a non-LPS-related polysaccharide.
The confirmation of the function of WaaF adds it to a list of only a few enzymes involved in the biosynthesis of the C. jejuni LOS molecule whose functions have been verified experimentally. These include heptosyltransferase I (waaC) (23), UDP-galactose-4-epimerase (galE) (10), galactosyltransferases (wlaN and cgtB) (12, 25), sialyltransferases (cstI and cstII) (12), an N-acetylgalactosaminyltransferase (cgtA) (12), and N-acetylneuraminic acid (NeuNAc) synthetase (neuB1) (26). Only waaC, wlaN, galE, and neuB1 are found in NCTC 11168, and there is a fusion of cgtA and a CMP-NeuNAc synthetase gene (designated neuA1 in NCTC 11168 [35]) (13, 26). Most of the genes involved in LOS biosynthesis in C. jejuni are associated with the formation of the LOS-associated GM1-like epitope found in some strains. The heptosyltransferases WaaC and WaaF attach the first two heptoses to the Kdo of the growing inner core lipid A molecule. GalE is required for the production of the galactose residues found in the outer core molecule and β(1,3)-galactosyltransferases WlaN and CgtB have been shown to be required for the attachment of the galactose residue in the outer core that forms part of the GM1-like epitope. This GM1-like epitope also contains NeuNAc, the biosynthesis of which has been shown to involve NeuB1 (the product of one of three NeuNAc synthetase genes in the C. jejuni NCTC 11168 genome [35]) and which is attached by a sialyltransferase, for example, α(2,3)-sialyltransferase (CstI) or α(2,3)- and α(2,8)-sialyltransferase (CstII). The attachment of the terminal N-acetylgalactosamine of the GM1 epitope requires CgtA or NeuA1. Having the C. jejuni NCTC 11168 genome sequence combined with the exact structure of the LOS of the strain will facilitate the confirmation of proposed models of LOS biosynthesis (10, 27) and will prove useful in the determination of the functions of new LOS biosynthesis genes found in other strains (for example, see reference 12).
Moreover, the waaF mutation described in this study should prove a useful tool for the evaluation of the contribution of C. jejuni LOS structures to pathogenesis. The waaF mutant strains could be used in both in vivo and in vitro studies to study the effects of core truncation on processes such as colonization, adherence, invasion, and the ability to stimulate the host inflammatory response. However, such studies are hampered by the lack of a suitable animal model to closely mimic the clinical manifestations of Campylobacter enteritis (54). Also, the waaF mutant can be used to investigate the contribution of the LOS core oligosaccharide to the development of GBS. However, again, no suitable animal model is currently available to determine the ability of such mutant strains to induce GBS (37).
Acknowledgments
This study was supported by studentships to N.O. and L.M. by the Biotechnology and Biological Sciences Research Council and by a grant to A.M. from the Irish Health Research Board.
The S. enterica serovar Typhimurium strains were a kind gift from T. Isobe (Department of Clinical Veterinary Medicine, University of Cambridge, United Kingdom).
REFERENCES
- 1.Allen, A. G., T. Isobe, and D. J. Maskell. 1998. Identification and cloning of waaF (rfaF) from Bordetella pertussis and use to generate mutants of Bordetella spp. with deep rough lipopolysaccharide. J. Bacteriol. 180:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspinall, G. O., C. M. Lynch, H. Pang, R. T. Shaver, and A. P. Moran. 1995. Chemical structures of the core region of Campylobacter jejuni O:3 lipopolysaccharide and an associated polysaccharide. Eur. J. Biochem. 231:570-578. [PubMed] [Google Scholar]
- 3.Aspinall, G. O., A. G. McDonald, and H. Pang. 1992. Structures of the O chains from lipopolysaccharides of Campylobacter jejuni serotypes O:23 and O:36. Carbohydr. Res. 231:13-30. [DOI] [PubMed] [Google Scholar]
- 4.Aspinall, G. O., A. G. McDonald, H. Pang, L. A. Kurjanczyk, and J. L. Penner. 1994. Lipopolysaccharides of Campylobacter jejuni serotype O:19—structure of core oligosaccharide regions from the serostrain and two bacterial isolates from patients with the Guillain-Barre syndrome. Biochemistry 33:241-249. [DOI] [PubMed] [Google Scholar]
- 5.Aspinall, G. O., A. G. McDonald, T. S. Raju, H. Pang, L. A. Kurjanczyk, J. L. Penner, and A. P. Moran. 1993. Chemical structure of the core region of Campylobacter jejuni serotype O:2 lipopolysaccharide. Eur. J. Biochem. 213:1029-1037. [DOI] [PubMed] [Google Scholar]
- 6.Aspinall, G. O., A. G. McDonald, T. S. Raju, H. Pang, A. P. Moran, and J. L. Penner. 1993. Chemical structures of the core regions of Campylobacter jejuni serotypes O:1, O:4, O:23, and O:36 lipopolysaccharides. Eur. J. Biochem. 213:1017-1027. [DOI] [PubMed] [Google Scholar]
- 7.Bauer, B. A., S. R. Lumbley, and E. J. Hansen. 1999. Characterization of a WaaF (RfaF) homolog expressed by Haemophilus ducreyi. Infect. Immun. 67:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brabetz, W., S. Muller-Loennies, O. Holst, and H. Brade. 1997. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in an Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur. J. Biochem 247:716-724. [DOI] [PubMed] [Google Scholar]
- 9.Brooke, J. S., and M. A. Valvano. 1996. Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. J. Biol. Chem. 271:3608-3614. [DOI] [PubMed] [Google Scholar]
- 10.Fry, B. N., S. Feng, Y.-Y. Chen, D. G. Newell, P. J. Coloe, and V. Korolik. 2000. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 68:2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fry, B. N., V. Korolik, J. A. ten Brinke, M. T. T. Pennings, R. Zalm, B. J. J. Teunis, P. J. Coloe, and B. A. M. van der Zeijst. 1998. The lipopolysaccharide biosynthesis locus of Campylobacter jejuni 81116. Microbiology 144:2049-2061. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, M., J. R. Brisson, M. F. Karwaski, J. Michniewicz, A. M. Cunningham, Y. Wu, N. M. Young, and W. W. Wakarchuk. 2000. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. J. Biol. Chem. 275:3896-3906. [DOI] [PubMed] [Google Scholar]
- 13.Guerry, P., C. P. Ewing, T. E. Hickey, M. M. Prendergast, and A. P. Moran. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68:6656-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of E. coli with plasmids. J. Mol. Biol. 166:557-579. [DOI] [PubMed] [Google Scholar]
- 15.Hanniffy, O. M., A. S. Shashkov, A. P. Moran, M. M. Prendergast, S. N. Senchenkova, Y. A. Knirel, and A. V. Savage. 1999. Chemical structure of a polysaccharide from Campylobacter jejuni 176.83 (serotype O:41) containing only furanose sugars. Carbohydr. Res. 319:124-132. [DOI] [PubMed] [Google Scholar]
- 16.Hood, D., M. Deadman, T. Allen, H. Masoud, A. Martin, J. Brisson, R. Fleischmann, J. Venter, J. Richards, and E. Moxon. 1996. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol. Microbiol. 22:951-965. [DOI] [PubMed] [Google Scholar]
- 17.Jennings, M. P., M. Bisercic, K. L. Dunn, M. Virji, A. Martin, K. E. Wilks, J. C. Richards, and E. R. Moxon. 1995. Cloning and molecular analysis of the Isi1 (rfaF) gene of Neisseria meningitidis which encodes a heptosyl-2-transferase involved in LPS biosynthesis: evaluation of surface exposed carbohydrates in LPS mediated toxicity for human endothelial cells. Microb. Pathog. 19:391-407. [DOI] [PubMed] [Google Scholar]
- 18.Karlyshev, A. V., D. Linton, N. A. Gregson, A. J. Lastovica, and B. W. Wren. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529-541. [DOI] [PubMed] [Google Scholar]
- 19.Karlyshev, A. V., M. V. McCrossan, and B. W. Wren. 2001. Demonstration of polysaccharide capsule in Campylobacter using electron microscopy. Infect. Immun. 69:5921-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlyshev, A. V., and B. W. Wren. 2001. Detection and initial characterization of novel capsular polysaccharide among diverse Campylobacter jejuni strains using Alcian blue dye. J. Clin. Microbiol. 39:279-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 22.Kievit, T. R., and J. S. Lam. 1997. Isolation and characterization of two genes, waaC (rfaC) and waaF (rfaF), involved in Pseudomonas aeruginosa serotype 5 inner core biosynthesis. J. Bacteriol. 179:3451-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klena, J. D., S. A. Gray, and M. E. Konkel. 1998. Cloning, sequencing, and characterization of the lipopolysaccharide biosynthetic enzyme heptosyltransferase I gene (waaC) from Campylobacter jejuni and Campylobacter coli. Gene 222:177-185. [DOI] [PubMed] [Google Scholar]
- 24.Korolik, V., B. N. Fry, M. R. Alderton, B. A. M. van der Zeijst, and P. J. Coloe. 1997. Expression of Campylobacter hyoilei lipo-oligosaccharide (LOS) antigens in Escherichia coli. Microbiology 143:3481-3489. [DOI] [PubMed] [Google Scholar]
- 25.Linton, D., M. Gilbert, P. G. Hitchen, A. Dell, H. R. Morris, W. W. Wakarchuk, N. A. Gregson, and B. W. Wren. 2000. Phase variation of a beta-1,3-galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 37:501-514. [DOI] [PubMed] [Google Scholar]
- 26.Linton, D., A. V. Karlyshev, P. G. Hitchen, H. R. Morris, A. Dell, N. A. Gregson, and B. W. Wren. 2000. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol. Microbiol. 35:1120-1134. [DOI] [PubMed] [Google Scholar]
- 27.Linton, D., A. V. Karlyshev, and B. W. Wren. 2001. Deciphering Campylobacter jejuni cell surface interactions from the genome sequence. Curr. Opin. Microbiol. 4:35-40. [DOI] [PubMed] [Google Scholar]
- 28.McSweegan, E., and R. I. Walker. 1986. Identification and characterization of two Campylobacter jejuni adhesins for cellular and mucous substrates. Infect. Immun. 53:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran, A. P., and J. L. Penner. 1999. Serotyping of Campylobacter jejuni based on heat-stable antigens: relevance, molecular basis and implications in pathogenesis. J. Appl. Microbiol. 86:361-377. [DOI] [PubMed] [Google Scholar]
- 30.Moran, A. P., J. L. Penner, and G. O. Aspinall. 2000. Campylobacter lipopolysaccharides, p. 241-257. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 31.Moran, A. P., E. T. Rietschel, T. U. Kosunen, and U. Zahringer. 1991. Chemical characterization of Campylobacter jejuni lipopolysaccharides containing N-acetylneuraminic acid and 2,3-diamino-2,3-dideoxy-d-glucose. J. Bacteriol. 173:618-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nachamkin, I., B. M. Allos, and T. W. Ho. 2000. Campylobacter jejuni infection and the association with Guillain-Barré syndrome, p. 155-175. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 33.Nichols, W. A., B. W. Gibson, W. Melaugh, N. G. Lee, M. Sunshine, and M. A. Apicella. 1997. Identification of the ADP-l-glycero-d-manno-heptose-6-epimerase (rfaD) and heptosyltransferase II (rfaF) biosynthesis genes from nontypeable Haemophilus influenzae 2019. Infect. Immun. 65:1377-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer, S. R., P. R. Gully, J. M. White, A. D. Pearson, W. G. Suckling, D. M. Jones, J. C. L. Rawes, and J. L. Penner. 1983. Water-borne outbreak of Campylobacter gastroenteritis. Lancet i:287-290. [DOI] [PubMed] [Google Scholar]
- 35.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 36.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial chrmosomal DNA with guanidinium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 37.Prendergast, M. M., and A. P. Moran. 2001. Lipopolysaccharides in the development of the Guillain-Barré syndrome and Miller Fisher syndrome forms of acute inflammatory peripheral neuropathies. J. Endotoxin Res. 6:341-359. [PubMed] [Google Scholar]
- 38.Preston, M. A., and J. L. Penner. 1987. Structural and antigenic properties of lipopolysaccharides from serotype reference strains of Campylobacter jejuni. Infect. Immun. 55:1806-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rietschel, E. T., L. Brade, O. Holst, V. A. Kulschin, B. Lindner, A. P. Moran, U. F. Schade, U. Zähringer, and H. Brade. 1990. Molecular structure of bacterial endotoxin in relation to bioactivity, p. 15-32. In A. Nowotny, J. J. Spitzer, and E. J. Ziegler (ed.), Cellular and molecular aspects of endotoxin reactions, vol. I. Elsevier Science Publishers, Amsterdam, The Netherlands. [Google Scholar]
- 40.Rietschel, E. T., T. Kirikae, F. U. Schade, U. Mamat, G. Schmidt, H. Loppnow, A. J. Ulmer, U. Zähringer, U. Seydel, F. Di Padova, M. Schreier, and H. Brade. 1994. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 8:217-225. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schnaitman, C. A., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwan, E. T., B. D. Robertson, H. Brade, and J. P. van Putten. 1995. Gonococcal rfaF mutants express Rd2 chemotype LPS and do not enter epithelial host cells. Mol. Microbiol. 15:267-275. [DOI] [PubMed] [Google Scholar]
- 44.Schwerer, B., A. Neisser, R. J. Polt, H. Bernheimer, and A. P. Moran. 1995. Antibody crossreactivities between gangliosides and lipopolysaccharides of Campylobacter jejuni serotypes associated with Guillain-Barré syndrome. J. Endotoxin Res. 2:395-403. [Google Scholar]
- 45.Schwimmer, S., and A. Bevenue. 1956. Reagent for differentiation of 1,4- and 1,6-linked glucosaccharides. Science 123:543-544. [DOI] [PubMed] [Google Scholar]
- 46.Sirisena, D. M., P. R. Maclachlan, S. L. Liu, A. Hessel, and K. E. Sanderson. 1994. Molecular analysis of the rfaD gene, for heptose synthesis, and the rfaF gene, for heptose transfer, in lipopolysaccharide synthesis in Salmonella typhimurium. J. Bacteriol. 176:2379-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szymanski, C. M., R. J. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 48.Tsai, C.-M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 49.van Vliet, A., A. Wood, J. Henderson, K. Wooldridge, and J. Ketley. 1998. Genetic manipulation of enteric Campylobacter species, p. 407-419. In P. Williams, J. Ketley, and G. Salmond (ed.), Techniques for bacterial pathogenesis. Academic Press, London, United Kingdom.
- 50.van Vliet, A. H. M., and J. M. Ketley. 2001. Pathogenesis of enteric Campylobacter infection. J. Appl. Microbiol. 90:45S-56S. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 52.Wood, A. C., N. J. Oldfield, C. A. O'Dwyer, and J. M. Ketley. 1999. Cloning, mutation and distribution of a putative lipopolysaccharide biosynthesis locus in Campylobacter jejuni. Microbiology 145:379-388. [DOI] [PubMed] [Google Scholar]
- 53.Wren, B. W., J. Henderson, and J. M. Ketley. 1994. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques 16:994-996. [PubMed] [Google Scholar]
- 54.Young, V. B., D. B. Schauer, and J. G. Fox. 2000. Animal models of Campylobacter infection, p. 287-301. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM, Washington, DC.