Abstract
ChiA, an 88-kDa endochitinase encoded by the chiA gene of the gram-negative enteropathogen Vibrio cholerae, is secreted via the eps-encoded main terminal branch of the general secretory pathway (GSP), a mechanism which also transports cholera toxin. To localize the extracellular transport signal of ChiA that initiates transport of the protein through the GSP, a chimera comprised of ChiA fused at the N terminus with the maltose-binding protein (MalE) of Escherichia coli and fused at the C terminus with a 13-amino-acid epitope tag (E-tag) was expressed in strain 569B(chiA::Kanr), a chiA-deficient but secretion-competent mutant of V. cholerae. Fractionation studies revealed that blockage of the natural N terminus and C terminus of ChiA did not prevent secretion of the MalE-ChiA-E-tag chimera. To locate the amino acid sequences which encoded the transport signal, a series of truncations of ChiA were engineered. Secretion of the mutant polypeptides was curtailed only when ChiA was deleted from the N terminus beyond amino acid position 75 or from the C terminus beyond amino acid 555. A mutant ChiA comprised of only those amino acids was secreted by wild-type V. cholerae but not by an epsD mutant, establishing that amino acids 75 to 555 independently harbored sufficient structural information to promote secretion by the GSP of V. cholerae. Cys77 and Cys537, two cysteines located just within the termini of ChiA(75-555), were not required for secretion, indicating that those residues were not essential for maintaining the functional activity of the ChiA extracellular transport signal.
In gram-negative bacteria, extracellular secretion is a complex process which requires translocation of proteins across both the cytoplasmic membrane and the outer membrane. In the first stage of secretion, newly synthesized proteins are routed to the cytoplasmic membrane via signal peptides which interact with the sec-dependent transport system (for a review, see Pugsley et al. [25] and Sandkvist [26]). Transit of the protein across the cytoplasmic membrane is followed by cleavage of the signal peptide, which releases the protein into the periplasmic space. After crossing the cytoplasmic membrane, the transported proteins fold into their mature configuration. At this intermediate stage of secretion, the prospective extracellular protein is sandwiched between the hydrophobic interfaces of the cytoplasmic membrane and the outer membrane.
Although a variety of mechanisms have evolved to convey the mature protein across the outer membrane, for many bacteria, translocation is accomplished by the main terminal branch (MTB) of the general secretory pathway (10), otherwise known as the type II system for extracellular transport (25). The MTB commonly comprises a minimum of 11 proteins which are believed to form a large multimeric complex that extends from the cytoplasmic membrane to the outer membrane, thereby spanning the periplasmic space (25). MTB systems have been shown to secrete several important virulence factors, including exotoxin A of Pseudomonas aeruginosa (2), aerolysin of Aeromonas salmonicida (36), and pectate lyase of Erwinia chrysanthemi and Erwinia carotovora (20).
In the enteropathogen Vibrio cholerae, the extracellular transport of cholera toxin (CT), an oligomeric heat-labile enterotoxin, is essential for the pathogenesis of the bacterium. Several studies confirmed that secretion of CT is accomplished by the MTB secretory system (22, 28), which is encoded by at least 13 genes, including vcpD in association with the eps cluster of genes (epsCDEFGHIJKLMN) (18, 22, 27, 28).
The MTB of V. cholerae is one of the more promiscuous type II secretory systems in regard to the types of proteins which are secreted by the bacterium (5). Unlike many of the type II secretory systems, which transport one or at most two proteins to the extracellular milieu, the MTB of V. cholerae has the capacity to transport at least six proteins, including CT (22, 28), the type I heat-labile enterotoxin (LT-I) of Escherichia coli (22), the type II heat-labile enterotoxins (LT-IIa and LT-IIb) of E. coli (4, 6), one or more proteases (22), and an endochitinase (5). Polysaccharide transport, which is required for establishment of the rugose phenotype in V. cholerae (1), is also believed to require the eps secretory system. Secretion by the MTB of V. cholerae may not be limited to protein translocation but may extend to the transport of larger multicomponent particles. Release of vibriophage CTXφ from V. cholerae was shown to require epsD, which encodes the outer membrane pore of the MTB machinery (8).
Numerous experiments have established that the MTB of V. cholerae and the MTBs of other bacteria discriminate between secreted and nonsecreted proteins (15, 23, 29). While the periplasmic space of V. cholerae contains numerous types of proteins, only a few of those are transported across the outer membrane into the extracellular milieu. These data indicated that prospective extracellular proteins of V. cholerae contain an extracellular transport signal (ETS) which directs proteins fated for extracellular transport to the eps-encoded transport machinery.
In this study, our efforts were focused on identifying the ETS of ChiA, an 88-kDa extracellular endochitinase which is transported by the eps-encoded MTB of V. cholerae (5). As ChiA has little or no amino acid homology to either CT or the other secreted proteins of V. cholerae (i.e., LT-I, LT-IIa, or LT-IIb), the ETS of ChiA was not identifiable from comparisons of primary amino acid sequences. Rather, a genetic approach was used to delimit the region(s) of ChiA which was essential for secretion. From these studies, it was revealed that a region of ChiA located between amino acids 75 and 555 was necessary and sufficient to promote secretion by the eps-encoded secretory machinery of V. cholerae. Furthermore, these studies lent further credence to the hypothesis that the ETS of ChiA is likely a structural motif whose conformation requires the interaction of distally located regions of the protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
Plasmids and strains used are listed in Table 1. Escherichia coli DH5αF′tet (Life Technologies, Inc., Gaithersburg, Md.) was used as the host in cloning experiments. Unless otherwise noted, all secretion experiments were performed in strain 569B(chiA::Kanr), a chiA-deficient strain of V. cholerae 569B (5). E. coli and V. cholerae strains were cultured in Luria-Bertani (LB) broth and maintained on LB agar. Chemical reagents were obtained from Sigma Biochemicals (St. Louis, Mo.), Fisher Scientific (Springfield, N.J.), and Life Technologies, Inc. Antibiotics were purchased from Sigma Biochemicals. Ampicillin was used at 200 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| V. cholerae | ||
| 569B(chiA::Kanr) | Derivative of 569B, O1 classical biotype, with Kanr inserted into chiA | 5 |
| N16961 | Wild-type, serogroup O1 El Tor biotype | 1 |
| NS1 | Derivative of N16961, epsD::TnphoA, Kanr | 1 |
| E. coli DH5αF′ tet | DH5α transconjugate with (F′ proAB lacIqZΔM15 Tn10(Tetr)] | Life Technologies |
| Plasmids | ||
| pBluescript KS+ | Phagemid cloning vector, Ampr | Stratagene |
| pBluescript SKII+ | Phagemid cloning vector, Ampr | Stratagene |
| pmal-p2 | Expression vector for engineering translational fusions to MalE, Ampr | New England Biolabs |
| pTDCC2 | 2.9-kbp Sau3AI fragment from pTDCC1 into the BamHI site of pBluescript KS− | 5 |
| pJPF2 | Wild-type ChiA; 2.9-kbp SstI/HindIII fragment from pTDCC2 into XbaI and HindIII sites of pBluescript SKII+ | 5 |
| pJL1 | MalE-ChiA-E-tag; chiA/E-tag hybrid gene of pTDCC2.6 fused to 3′ end of malE in pmal-p2 | 5 |
| pchiAC756 | C756; mutant chiA encoding amino acids 1-756 | This study |
| pchiAC608 | C608; mutant chiA encoding amino acids 1-608 | This study |
| pchiAC555 | C555; mutant chiA encoding amino acids 1-555 | This study |
| pchiAC478 | C478; mutant chiA encoding amino acids 1-478 | This study |
| pchiAC378 | C378; mutant chiA encoding amino acids 1-378 | This study |
| pchiAC179 | C179; mutant chiA encoding amino acids 1-179 | This study |
| pchiA(1-28) | ChiA(1-28); mutant chiA encoding amino acids 1-28 | This study |
| pchiAN66 | N66; mutant chiA encoding amino acids 1-28 fused to amino acids 66-846 | This study |
| pchiAN75 | N75; mutant chiA encoding amino acids 1-28 fused to amino acids 75-846 | This study |
| pchiAN125 | N125; mutant chiA encoding amino acids 1-28 fused to amino acids 125-846 | This study |
| pchiAN180 | N180; mutant chiA encoding amino acids 1-28 fused to amino acids 180-846 | This study |
| pchiAN225 | N225; mutant chiA encoding amino acids 1-28 fused to amino acids 225-846 | This study |
| pchiAN276 | N276; mutant chiA encoding amino acids 1-28 fused to amino acids 276-846 | This study |
| pchiAN429 | N429; mutant chiA encoding amino acids 1-28 fused to amino acids 429-846 | This study |
| pchiA(75-555) | ChiA(75-555); mutant chiA encoding amino acids 1-28 fused to chiA sequences encoding amino acids 75-555 | This study |
| pchiA(C77A) | ChiA(C77A); mutant chiA encoding amino acids 1-28 fused to chiA sequences encoding amino acids 75-846 with a cysteine-to-alanine substitution at amino acid position 77 | This study |
| pchiA(C537A) | ChiA(C537A); mutant chiA encoding amino acids 1-540, with a cysteine-to-alanine mutation at amino acid position 537 | This study |
Isolation and manipulation of plasmids.
Plasmid DNA was obtained from E. coli DH5αF′tet by use of a modified alkaline lysis protocol (12) or by a QIAprep spin plasmid miniprep kit (Qiagen, Inc., Valencia, Calif.). Restriction enzymes and DNA-modifying enzymes were purchased from Life Technologies, Inc., or from MBI Fermentas Inc. (Amherst, N.Y.). Agarose was purchased from J.T. Baker (Phillipsburg, N.J.). Plasmids were transformed into E. coli by osmotic shock (16) and into V. cholerae by electroporation (17). Transformants were selected on LB agar containing appropriate antibiotics.
Engineering of ChiA truncations.
pTDCC2 was used as the source of the chiA gene for engineering pJPF2, N-terminal truncations, and C-terminal truncations of chiA.
To construct pJPF2, pTDCC2 was digested with SstI and HindIII to produce a 2.9-kbp fragment encoding chiA. The 2.9-kbp fragment was directionally ligated into pBluescript SKII+ (Stratagene, La Jolla, Calif.). The fragment was orientated in pJPF2 so that expression of chiA was under control of the vector's lac promoter (Plac).
Synthetic oligonucleotides and PCR were used to engineer all truncation mutants of chiA. pTDCC2 which had been linearized by digestion with KpnI was used as a template. The upstream oligonucleotide primer used to engineer all six C-terminal truncations was Blue-619 (5′-GTAAAACGACGGCCAGTGA-3′), which was homologous to sequences located in the multicloning region of the vector. The downstream oligonucleotide for each truncation was synthesized with an in-frame TAA translational stop codon (bold) and an EcoRI restriction site (underlined). The nucleotide sequences of the downstream oligonucleotides were as follows: chiA756, 5′-GGAATTCTTACCCGTTGGCTAAAGT-3′; chiA608, 5′-GGAATTCTTAAAGCGCCACCTAAGGAAA-3′; chiA555, 5′-GGAATTCTTAAAAGGCAATATCAATCAC-3′; chiA478, 5′-GGAATTCTTAAGGCTCTGTTGTATCC-3′; chiA378, 5′-GGAATTCTTAGTTGTTGCCCGCAAAA-3′; and chiA179, 5′-GGAATTCTTATGGCGGAACTGGGTTA-3′.
DNA amplification reactions were performed with a DNA thermal cycler model 480 (Perkin-Elmer, Norwalk, Conn.). Conditions for the reactions were: denaturation at 92°C for 30 s, annealing at 54°C for 45 s, and extension at 72°C for 3 min. Amplified DNA fragments were resolved by electrophoresis on 0.8% agarose gels and isolated from gel slices using a GeneClean II kit (Bio 101, Inc., Vista, Calif.). DNA fragments digested with XbaI and EcoRI were directionally ligated into pBluescript SKII+ so that expression of the engineered genes was under control of Plac. The 3′ ends of the truncated genes were confirmed by nucleotide sequencing.
To engineer the N-terminal truncations, an intermediate plasmid containing the signal sequence of ChiA was constructed. Primers Chi-15 (5′-GGAATTCCGAAAAAGAATTGAAAGC-3′, EcoRI site underlined) and chi3′V28 (5′-CATCTAGACACTCCGGCACAGTTAT-3′, XbaI site underlined) were used to amplify from pJPF2 the DNA encoding the first 28 amino acids of ChiA. DNA amplification and fragment isolation were performed as described above. The DNA fragment was ligated into pGEM-T (Promega, Madison, Wis.). pGEM-chiA(1-28) was digested with SacI and XbaI, and the fragment encoding the ChiA signal peptide was subsequently ligated into pBluescript SKII+. The chiA sequence of pchiA(1-28) was confirmed by nucleotide sequencing. Blue-619 was used as the downstream oligonucleotide primer to engineer all seven N-terminal truncations. The upstream oligonucleotide for each truncation was synthesized with an in-frame XbaI restriction site (underlined). The nucleotide sequences of the upstream oligonucleotides were as follows: chiAN66, 5′-CCTCTAGAGGTCAATGGGACGCA-3′; chiAN75, 5′-AATCTAGAGGACAGTGTGACGG-3′; chiAN125, 5′-CCTCTAGAGGATTACAAGCCAATGC-3′; chiAN180, 5′-CGTCTAGAGTGACGTTGACTAGC-3′; chiAN225, 5′-TTTCTAGAATCGATAGCTCTGAGCCT-3′; chiAN276, 5′-GCTCTAGATTACCTGTGTATTCGGTG-3′; and chiAN429, 5′-ATTCTAGAGTTTCACTGACTTCACCGA3′.
DNA amplifications and fragment isolation were performed as described earlier. Amplified DNA fragments were digested with XbaI and HindIII and ligated into pchiA(1-28) at equivalent sites. The junction between chiA(1-28) and each N-terminal truncation was confirmed by nucleotide sequencing. Fragments were oriented so that expression of the mutant genes was under control of Plac.
After transformation of the plasmids into E. coli DH5αF′tet, transformants were initially screened for expression of recombinant proteins by immunoblotting (16) using a rabbit hyperimmune anti-MalE-ChiA-E-tag antiserum for detection (5). Plasmids were subsequently transformed by electroporation (17) into V. cholerae 569B(chiA::Kanr). Transformants were analyzed for ChiA secretion by immunoblotting culture supernatant and periplasmic extract using the anti-MalE-ChiA-E-tag antiserum (5).
Engineering of MalE-ChiA-E-tag fusion protein, ChiA(75-555), and other ChiA mutants.
Construction of pJL1, the plasmid encoding the MalE-ChiA-E-tag chimera, has been described (5). Plasmids encoding ChiA(75-555), ChiA(C77A), and ChiA(C537A) were engineered by PCR.
To engineer ChiA(75-555), the two synthetic oligonucleotides used for the N- and C-terminal truncation studies, ChiAN75 and ChiAC555, were used as primers to amplify the specific DNA fragment from pTDCC2. Conditions for PCR were denaturation at 92°C for 30 s, annealing at 54°C for 45 s, and extension at 72°C for 2 min. Amplified DNA fragments were resolved by electrophoresis on 0.8% agarose gels, isolated from gel slices using a GeneClean kit, and digested with XbaI and HindIII for subsequent ligation into pchiA(1-28). The DNA fragment was oriented in-frame with respect to the natural ChiA signal peptide encoded by pchiA(1-28) and downstream of the vector's Plac.
To engineer ChiA(C77A), primer Blue-619 and primer chiA(C77A) (5′-AATCTAGAGGACAGGCGGACGGAG-3′, XbaI site underlined, nucleotide change in italic; identical in sequence to primer chiAN75 with the exception of a three-nucleotide change at amino acid position 77 of wild-type ChiA) were used to amplify an internal region of chiA from pJPF2 encoding amino acids 75 to 846 of ChiA with an alanine for cysteine substitution at amino acid position 77 (C77A). The DNA fragment encoding the C77A mutation was digested with XbaI and HindIII and ligated into pchiA(1-28) to fuse the gene sequences in-frame to the chiA signal sequence.
To engineer ChiA(C537A), primer Blue-619 and ChiA(C537A) (5′-TCAAGCTTTTAACGAATTGGCGCACCCGCA-3′, HindIII site underlined, stop codon in bold, and nucleotide change in italic) were used to amplify an internal region of chiA from pTDCC2, which encodes amino acids 1 to 540, but substituted with an alanine for cysteine at amino acid position 537 (C537A). The amplification reaction also incorporated an in-frame TAA translational stop codon at the 3′ end of the sequence. The DNA fragment encoding the C537A mutation was digested with XbaI and HindIII and ligated into pBluescript SKII+. Nucleotide sequencing confirmed the sequence of the entire coding region of pchiA(75-555) and the cysteine to alanine mutation of both mutants, pchiA(C77A) and pchiA(C537A).
All three recombinant plasmids [pchiA(75-555), pchiA(C77A), and pchiA(C537A)] were transformed into E. coli DH5αF′tet for immunoblotting using rabbit anti-MalE-ChiA-E-tag antiserum to confirm that the plasmids expressed the appropriate immunoreactive mutant ChiA proteins. After confirmation, plasmids were transformed into 569B(chiA::Kanr), V. cholerae N16961, and V. cholerae NS1(epsD). Clones were analyzed for secretion of the chimeric proteins by immunoblotting using rabbit anti-MalE-ChiA-E-tag antiserum (5).
Preparation of culture supernatant and periplasmic extract.
Culture supernatant and periplasmic extract were obtained from bacteria grown to an optical density at 600 nm (OD600) of 0.9 to 1.4 (6). In brief, bacterial cultures were centrifuged to separate cells and culture supernatant. Sodium azide was added to the culture supernatant at a final concentration of 0.05% (wt/vol) to inhibit continuing growth of residual viable cells. Pelleted cells, resuspended in phosphate-buffered saline (PBS) containing 1 mM EDTA and 1 mg of polymyxin B sulfate per ml, were incubated at 37°C for 30 min to release periplasmic contents. The cell suspensions were centrifuged to separate the periplasmic extract from the extracted cells.
SDS-PAGE and Western analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were performed as previously described (5). Isoelectric focusing grade acrylamide and bisacrylamide were purchased from Pharmacia Biotech (Uppsala, Sweden). Proteins were resolved by SDS-PAGE in 8.75% gels and electrophoretically transferred to nitrocellulose filters, and the filters were immunoblotted with rabbit anti-MalE-ChiA-E-tag antiserum (1:15,000) and horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (1:10,000; Sigma Biochemicals) using previously established procedures (5). Immunoblots were developed using a Renaissance Western blot chemiluminescence kit (Dupont/New England Nuclear, Wilmington, Del.). Fluorescent signals were detected by exposure of the immunoblots on Biomare Blue-sensitive autoradiographic film (Marsh Biomedical Products, Inc., Rochester, N.Y.).
DNA sequencing.
Double-stranded DNA sequencing was performed by the Sequencing Facility of the Center for Applied Molecular Biology and Immunology at the State University of New York at Buffalo or by the Biopolymer Facility at the Roswell Park Cancer Institute (Buffalo, N.Y.). The synthetic, single-stranded oligonucleotide Blue-619 was used to prime the sequencing reactions.
Measurement of β-lactamase activity.
When appropriate, β-lactamase activity was used to confirm that the cells in cultures were intact and that polypeptides found in the culture supernatant were not derived from cell lysis. Culture supernatant and periplasmic extract were prepared as previously described (6). Extract was added to a solution of nitrocefin (Becton Dickinson Microbiology Systems, Cockeysville, Md.) (51.6 μg/ml in 0.05 M phosphate buffer at pH 7.0), and the mixture was measured for increasing absorbency over a 5-min interval at OD482 using a Beckman DU series 600 spectrophotometer (Fullerton, Calif.). β-Lactamase activity was calculated by the method of O'Callaghan et al. (21).
RESULTS
Secretion of ChiA does not require the natural N or C terminus.
While prior studies of ChiA secretion provided strong evidence that the protein contained an extracellular transport signal that interacted with one or more components of the eps-encoded MTB of V. cholerae, the precise location of that signal within ChiA had yet to be identified. Since secretion of polygalacturonase by the MTB of E. carotovora required an intact C terminus (23), it was initially hypothesized that the ETS of ChiA was located at the N terminus or at the C terminus of the protein.
To investigate whether either terminus of ChiA was essential for secretion, a chimeric protein was engineered that fused MalE of E. coli to the N terminus of the protein and a 13-amino-acid epitope tag (E-tag) (Pharmacia Biotech) to the C terminus of the protein (Fig. 1). When the plasmid (pJPL1) encoding the chimera was transformed into E. coli DH5αF′tet and cultivated on ethylene glycol-chitin (EGC) agar, a substrate for ChiA, isopropylthiogalactopyranoside (IPTG)-inducible expression of chitinase activity was observed (5), which was associated with expression of a 128-kDa immunoreactive polypeptide corresponding to the molecular mass of the predicted MalE-ChiA-E-tag chimera (data not shown).
FIG. 1.
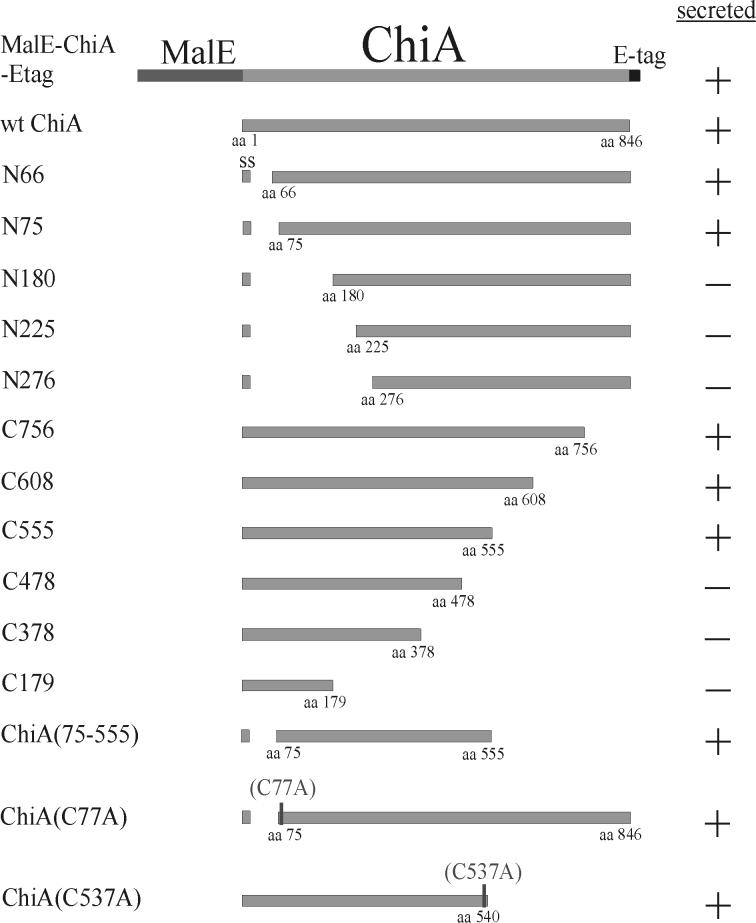
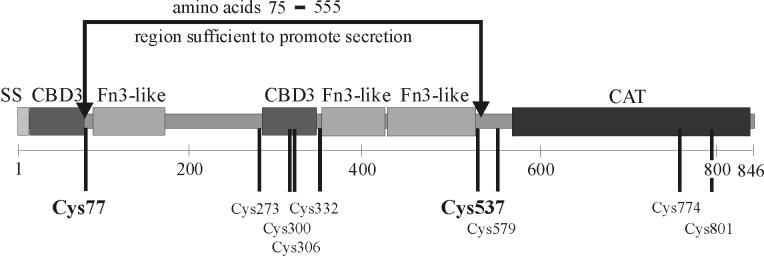
Summary of the structure and secretion phenotypes of the mutant ChiA proteins. Amino acid positions are numbered beginning with the N-terminal methionine of immature ChiA. Truncated mutant ChiA polypeptides were designated by the letter N to denote an N-terminal deletion and by a letter C to denote a C-terminal deletion, while the numbers indicate the N-terminal or C-terminal boundary of the truncated mutant polypeptide (i.e., N66 is a mutant polypeptide comprised of amino acids 66 to 846 of ChiA, while C756 is a mutant polypeptide comprised of amino acids 1 to 756.). The positions of the Cys to Ala substitutions in ChiA(C77A) and ChiA(C537A) are denoted (C77A and C537A). +, secreted into the culture supernatant; −, not secreted into the culture supernatant; SS, signal peptide (amino acids 1 to 28); wt, wild type; aa, amino acid.
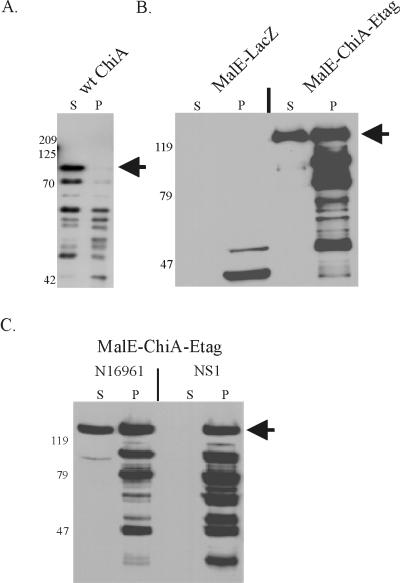
To assess whether the chimera would be secreted by V. cholerae, pJL1 was subsequently introduced into 569B(chiA::Kanr), a secretion-competent but chitinase-negative strain of 569B (5). pmal-p2, which encoded MalE, and pJPF2, encoding wild-type ChiA, were also introduced into the strain as a negative and positive control, respectively. Supernatant and periplasmic extract were obtained from mid-log-phase, IPTG-induced cells, and the presence of anti-ChiA immunoreactive polypeptides in each fraction was determined by immunoblotting (Fig. 2). As expected, MalE was evident only in the periplasmic extract of 569B(chiA::Kanr)(pmal-p2) (Fig. 2B), indicating that this periplasmic protein of E. coli was not secreted by V. cholerae. In contrast, the majority of wild-type ChiA was secreted into the culture supernatant by 569B(chiA::Kanr)(pJPF2) (Fig. 2A). Similarly, an immunoreactive polypeptide of 128 kDa, consistent with the size of the MalE-ChiA-E-tag chimera, was detected in the culture supernatant of 569B(chiA::Kanr)(pJL1) (Fig. 2B). Unlike wild-type ChiA (Fig. 2A), however, considerable amounts of the MalE-ChiA-E-tag chimera were also observed in the periplasmic extract of 569B(chiA::Kanr)(pJL1) (Fig. 2B). These data were suggestive of an inhibitory effect on secretion by the MalE or E-tag portion of the chimera.
FIG. 2.
(A) Secretion of recombinant wild-type (wt) ChiA protein by 569B(chiA:: Kanr)(pJPF2). The position of the mature, secreted ChiA (88 kDa) is denoted by an arrow. (B) Secretion of MalE-ChiA-E-tag chimeric protein by 569B(chiA::Kanr). MalE-LacZ was encoded by pmal-p2; MalE-ChiA-E-tag was encoded by pJL1. (C) Secretion of the MalE-ChiA-E-tag chimera is dependent on the eps-encoded MTB of V. cholerae. MalE-ChiA-E-tag was expressed in N16961 and in NS1, a derivative of N16961 which harbors an epsD mutation (1). The position of the mature, secreted MalE-ChiA-E-tag (128 kDa) is marked with an arrow. Proteins in the immunoblots were detected with a rabbit anti-MalE-ChiA-E-tag antiserum. Cells, with the exception of 569B(chiA::Kanr)(pJPF2), were induced by the addition of IPTG. Culture supernatant (S) and periplasmic extract (P) were harvested at the mid-logarithmic stage of growth. The positions of the molecular mass standards are shown (in kilodaltons).
Several smaller immunoreactive products, probably derived from proteolytic degradation, were observed in the periplasmic extracts of cells expressing both wild-type ChiA and the chimera. These smaller forms of ChiA and the chimera were likely an artifact of overexpression, as similar patterns of degradation have been observed when other recombinant proteins were overexpressed in the periplasm of V. cholerae (T. D. Connell, data not shown).
To investigate whether the presence of extracellular chimera was derived from cell lysis, culture supernatant and periplasmic extract of 569B(chiA::Kanr)(pJL1) and 569B(chiA::Kanr)(pmal-p2) were measured for β-lactamase, a periplasmic enzyme produced by the plasmid-encoded blaM gene. Less than 3% of the total β-lactamase activity was detected in the culture supernatant, confirming that cellular lysis was only a minor contributor to the presence of extracellular polypeptide.
Secretion of the chimera depended on an intact eps-encoded transport system.
Although it had been shown previously (5) that ChiA was secreted by the eps-encoded MTB, it was certainly feasible that the MalE-ChiA-E-tag chimera had been transported via an alternative secretory system which had been coopted by the cell. To confirm that secretion of the chimeric ChiA polypeptide by the cell was eps dependent and to provide further evidence that cell lysis was not a contributing factor, pJL1 was transformed into V. cholerae N16961 and into its epsD (secretion deficient) mutant NS1 (1). When the transformants were analyzed for secretion of the chimera, the MalE-ChiA-E-tag chimera was observed in the culture supernatant of N16961(pJL1), but not in the culture supernatant of NS1(pJL1) (Fig. 2C). As observed for 569B(chiA::Kanr)(pJL1), both full-size and smaller degraded forms of the chimera were detected in the periplasmic extracts of both strains, indicating that the lower efficiency of secretion of the chimera was not strain dependent. Nonetheless, it was concluded from these data that, as observed for secretion of wild-type ChiA, secretion of the chimeric MalE-ChiA-E-tag protein by V. cholerae required an intact eps-encoded MTB.
Secretion of N- and C-terminally truncated mutants of ChiA.
Since previous experiments confirmed that neither the natural N terminus nor C terminus of ChiA was essential for secretion of ChiA, it was reasonable to hypothesize that amino acid sequences located internal to the termini were important for secretion recognition. To delimit the regions of ChiA which were involved in secretion recognition, a nested set of N-terminal and C-terminal truncations of ChiA was engineered (Fig. 1). Mutant ChiA polypeptides truncated from the amino-terminal end were designated by the letter N; mutant ChiA polypeptides truncated from the carboxyl-terminal end were denoted by the letter C (e.g., N66 is a mutant ChiA comprised of amino acids 66 to 846, while C756 is a mutant polypeptide comprised of amino acids 1 to 756.) Since the N-terminal mutants required a signal peptide to direct translocation into the periplasmic space of the cell, all genes encoding N-terminal truncations were fused to nucleotide sequences encoding the native sec-dependent signal peptide of ChiA, including its natural signal peptidase cleavage site (amino acids 1 to 28). Seven N-terminal truncations of ChiA (N66, N75, N125, N180, N225, N276, and N429) and six C-terminal truncations of ChiA (C756, C608, C555, C478, C378, and C179) were engineered.
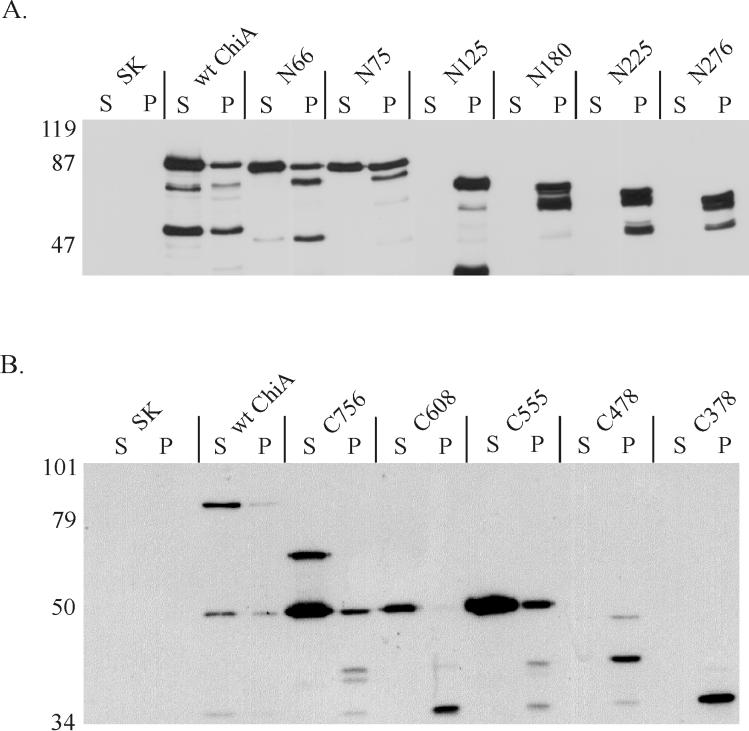
Plasmids encoding the N-terminally truncated mutant ChiA polypeptides were transformed into 569B(chiA::Kanr), and culture supernatant and periplasmic extract were analyzed by immunoblotting for ChiA immunoreactivity. As expected, wild-type ChiA was efficiently secreted by 569B(chiA::Kanr)(pJPF2) (Fig. 3A and 3B). Of the seven N-terminal mutants, however, only mutant ChiA polypeptides N66 and N75 were observed in the culture supernatant (Fig. 3A). For mutant ChiA polypeptides truncated to amino acid 125 (N125) and beyond (N180, N225, and N276), all immunoreactive protein was confined to the periplasmic extract (Fig. 3A). The smallest N-terminal truncation, N429, was also restricted to the periplasmic fraction, although the polypeptide was expressed at exceptionally low levels (data not shown).
FIG. 3.
(A) Secretion of N-terminal truncations of ChiA by V. cholerae. (B) Secretion of C-terminal truncations of ChiA. All proteins were expressed in 569B(chiA::Kanr). N-terminal truncations are designated by N followed by the position of the N-terminal amino acid (not including the signal peptide); C-terminal truncations of ChiA are designated by C followed by the position of the C-terminal amino acid. Numbering of the amino acids begins with the initial methionine at the N terminus of immature ChiA. Extracts were harvested at the mid-logarithmic phase of growth. Proteins in the immunoblots were detected with a rabbit anti-MalE-ChiA-E-tag antiserum. S, culture supernatant; P, periplasmic extract; SK, pBluescript SKII+. The positions of the molecular mass standards are shown (in kilodaltons).
The same protocol was used to investigate secretion of the C-terminal truncated mutant ChiA polypeptides by 569B(chiA::Kanr). Of the six C-terminal truncations, only mutant ChiA polypeptides C756, C608, and C555 were detected in the culture supernatant (Fig. 3B). Neither C478 nor C378 was secreted by 569B(chiA::Kanr), although appreciable amounts of immunoreactive mutant polypeptide was observed in the periplasmic extract of those cells. C478 and C378 were found only in the periplasmic fraction (Fig. 3B).
Initially, the secretion characteristics of C608 were difficult to interpret, as the major immunoreactive polypeptide (≈50 kDa) expressed by the cell did not correspond to its predicted molecular mass (≈63 kDa) (Fig. 3B). However, when significantly large amounts of the culture supernatant from cells expressing this mutant polypeptide were measured for immunoreactive ChiA polypeptides, a faint band of the appropriate molecular mass for C608 was detected (data not shown). The C-terminal truncation with the largest deletion (C179) was expressed at exceptionally low levels but shown by similar methods to be restricted to the periplasmic fraction (data not shown).
Taken together, the results for the N-terminal and the C-terminal mutants established that the essential information required to promote extracellular transport of ChiA by V. cholerae was located between amino acids 75 and 555 (schematically represented in Fig. 1).
Secretion of mutant ChiA(75-555).
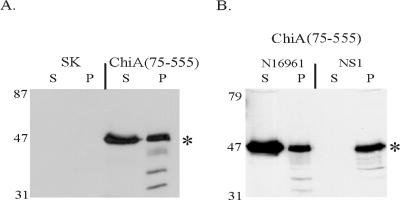
To confirm that the region bounded by amino acids 75 to 555 was sufficient to promote extracellular secretion of ChiA, an N- and C-terminally truncated mutant ChiA polypeptide was engineered that was comprised of only amino acids 75 to 555 fused to the endogenous ChiA signal peptide (Fig. 1). Culture supernatant of 569B(chiA::Kanr)[pchiA(75-555)] was found to contain a 53-kDa immunoreactive polypeptide, which was consistent with the predicted molecular mass of the mutant polypeptide (Fig. 4A). Less than 6% of the total β-lactamase activity was located in the culture supernatant, confirming that the presence of immunoreactive ChiA(75-555) in the culture supernatant was the result of active secretion by the cell and not a result of autolysis.
FIG. 4.
(A) Mutant polypeptide containing amino acids 75 to 555 of ChiA is secreted by 569B(chiA::Kanr). (B) Secretion of the mutant polypeptide ChiA(75-555) is dependent on the eps-encoded MTB of V. cholerae. ChiA(75-555) was expressed in N16961 and in NS1, a derivative of N16961 which harbors an epsD mutation (1). An asterisk denotes the position of the predicted 53-kDa ChiA(75-555) mutant polypeptide. Extracts were harvested at mid-logarithmic growth. Proteins in the immunoblots were detected with a rabbit anti-MalE-ChiA-E-tag antiserum. S, culture supernatant; P, periplasmic extract; SK, pBluescript SKII+. The positions of the molecular mass standards are shown (in kilodaltons).
To confirm that secretion of ChiA(75-555) was eps dependent, the plasmid encoding the mutant polypeptide was transformed into N16961 and its epsD mutant derivative, NS1. ChiA(75-555) was evident in the supernatant of N16961 but not in the culture supernatant of NS1 (Fig. 4B). An accumulation of ChiA(75-555) in the periplasm of NS1 was not observed, probably due to degradation of the mutant polypeptide by proteases located in the periplasm of V. cholerae. These results confirmed that the segment bound by amino acids 75 and 555 contained sufficient information to promote secretion of the polypeptide and that the secretion was dependent upon an intact eps-encoded MTB.
Secretion competence of chitinase A mutants C77A and C537A.
Mature ChiA contains nine cysteines, some of which are likely involved in forming disulfide bonds (Fig. 5). Disulfide bonds are commonly crucial for maintaining the structural integrity of exported proteins (19). It was concluded from the results of the truncated ChiA polypeptides that three of the cysteines (Cys579, Cys774, and Cys801) were not essential for secretion, as these residues are located outside of the 75 to 555 region of ChiA. In contrast, Cys77 and Cys537 are located immediately within the termini of ChiA(75-555). It was deemed feasible that Cys77 and Cys537 participated in disulfide bond formation either with each other or with one of the four other cysteines (Cys273, Cys300, Cys306, and Cys332) and that those disulfide bonds in ChiA were essential for secretion activity. This model was consistent with our results, in which all of the ChiA mutants which were truncated beyond amino acids 75 and 555 were secretion deficient.
FIG. 5.
Predicted domain structure of ChiA, showing the amino acid positions of the nine cysteines. Numbering of the amino acids begins with the initial methionine at the N terminus of immature ChiA. Cysteines are denoted by their position in the amino acid sequence. The region (amino acids 75 to 555) containing the structural information for extracellular secretion is shown in brackets. SS, signal sequence; CBD3, type III chitin-binding domain; Fn3-like, a domain similar to a type III fibronectin domain; CAT, catalytic domain of the chitinase 18 family.
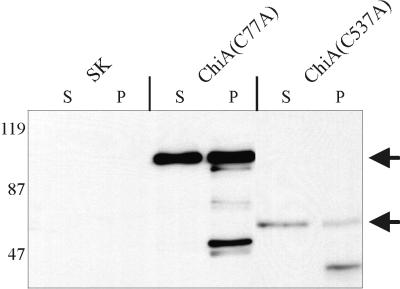
To determine whether Cys77 or Cys537 was essential for secretion of ChiA, mutant polypeptides lacking Cys77 or Cys537 were engineered (Fig. 1). ChiA(C77A) was identical in amino acid sequence to the N-terminally truncated mutant C75 with the exception of a cysteine to alanine mutation at amino acid position 77. ChiA(C537A) was engineered as a mutant ChiA which was C-terminally truncated to amino acid 540, but with a cysteine to alanine mutation at amino acid position 537. Although the expression level of ChiA(C537A) was low, sufficient amounts of mutant polypeptide were produced to demonstrate that 569B(chiA::Kanr) transported both ChiA(C77A) and ChiA(C537A) to the culture supernatant (Fig. 6). Less than 7% of the total β-lactamase activity was located in the culture supernatant of ChiA(C77A) and ChiA(C537A), confirming that the presence of the mutant polypeptides in the culture supernatant was due to active secretion by the cell and not a result of release by passive cell lysis. From these results, it was concluded that neither Cys77 nor Cys537 was essential for secretion of ChiA.
FIG. 6.
Secretion of ChiA mutant polypeptides in 569B(chiA:: Kanr) is unaffected by substitution mutations at Cys77 and Cys537. The positions of the mature ChiA(C77A) and ChiA(C537A) mutant polypeptides are indicated by arrows. Extracts were harvested at mid-logarithmic growth. Proteins in the immunoblots were detected with a rabbit anti-MalE-ChiA-E-tag antiserum. ChiA(C77A), mutant ChiA encoding amino acids 1 to 28 fused to amino acids 77 to 846, with a Cys to Ala substitution at position 77; ChiA(C537A), mutant ChiA encoding amino acids 1 to 540 and having a Cys to Ala substitution at position 537; SK, pBluescript SKII+; S, culture supernatant; P, periplasmic extract. The positions of the molecular mass standards are shown (in kilodaltons).
DISCUSSION
In this study, we focused on identifying the ETS of ChiA, the extracellular endochitinase of V. cholerae. In contrast to cholera toxin (CT), the heat-labile enterotoxin of V. cholerae, which has proven less amenable to genetic analysis of its ETS, ChiA exhibits several features which simplify experimental investigation of the structurally dependent ETS. ChiA is a relatively large (88 kDa) protein which is predicted to be composed of several independent domains (Fig. 5), each of which can be altered without affecting the conformation of the remaining domains of the protein. It is also feasible that ETS formation in CT requires oligomerization of the CT-B subunits (11, 33). Mutations in CT which affect oligomerization therefore may indirectly impact ETS formation. In contrast, ChiA is a monomeric protein, which obviates that particular concern.
As an initial approach to identifying the ETS of ChiA, a fusion strategy was used to determine if the N or C terminus of the protein was essential for secretion (Fig. 1). Since MalE is not secreted by V. cholerae, this protein was chosen as the N-terminal fusion partner for ChiA, and a small E-tag epitope was chosen as the initial C-terminal fusion partner. Secretion studies using this chimera provided strong evidence that, unlike secretion of polygalacturonase by E. carotovora (23), secretion of ChiA did not require the exposure of either of the natural termini (Fig. 2B). The MalE-ChiA-E-tag chimera and wild-type ChiA, however, were not secreted with equivalent efficiency by V. cholerae (Fig. 2A). Considerable amounts of immunoreactive chimera were retained in the periplasmic space.
To address the issue of whether the large amount of chimera in the periplasm was due simply to overloading of the eps-encoded secretory machinery, we decreased the expression levels of wild-type ChiA and the MalE-ChiA-E-tag chimera by use of lower-copy-number vectors and by altering the induction conditions, respectively. Reducing the expression of the recombinant genes had no detectable effect on the relative distribution of these polypeptides (data not shown).
Alternatively, several other factors could be responsible for the less efficient extracellular transport of the chimera from the periplasm. An improperly folded ChiA would exhibit an improperly folded ETS, which would likely be recognized with much lower efficiency by the eps-encoded secretion machinery. In that regard, it is possible that the MalE portion of the MalE-ChiA-E-tag polypeptide interferes with efficient folding of the ChiA portion of the chimera. Second, it cannot be discounted that the size of the chimera (128 kDa) relative to the size of wild-type ChiA (88 kDa ) may have an adverse affect on the ability of the MTB of V. cholerae to transport the protein. In support of this hypothesis is the observation that 569B(chiA::Kanr) was incapable of secreting either a PhoA-ChiA chimera or a BlaM-ChiA chimera (data not shown). The failure of Klebsiella oxytoca to secrete a PhoA-pullulanase chimera was also attributed to size incompatibility (13).
Since the experiments confirmed that neither the N nor the C terminus of ChiA was required to promote secretion, we addressed the alternative hypothesis that the ETS activity was encoded by amino acids located internal to the termini. To locate these amino acids, a series of ChiA mutants with increasing N-terminal and C-terminal truncations were engineered (Fig. 1). These experiments were complicated, however, by the sensitivity of the mutant ChiA polypeptides to in vivo degradation. Degradation was particularly evident for the C-terminal truncations, for which a major 53-kDa degradation product was observed in the culture supernatants of cells expressing mutant C756 and mutant C608 (Fig. 3). In contrast, this 53-kDa derivative polypeptide was not observed in the culture supernatants of any of the N-terminal truncation mutants (Fig. 3). This pattern of degradation suggested to us that the proteolytic cleavage that produces the 53-kDa polypeptide occurs at a site near the C terminus of ChiA. We are currently investigating whether this common degradation product is, itself, proficient for secretion, and thus exhibits an active ETS.
Alternatively, the 53-kDa polypeptide could be produced in the culture supernatant by extracellular proteolysis of a larger, secreted progenitor. Although smaller degradation products were often the major immunoreactive forms in the cells, prospective mutant ChiA polypeptides having the proper predicted molecular masses were always evident in the immunoblots of culture supernatants and/or periplasmic extracts. In some cases, however, large amounts of sample had to be loaded onto the SDS-polyacrylamide gels used for the immunoblots to detect the properly sized mutant ChiA polypeptide (data not shown).
The N- and C-terminal truncation studies revealed that the functional ETS of ChiA was likely located between amino acids 75 and 555. To confirm that this region was independently sufficient to promote secretion, a mutant ChiA polypeptide comprising only amino acids 75 to 555 was engineered and shown to be secreted by V. cholerae (Fig. 1), thus demonstrating that the structural information for secretion competence of ChiA was located solely within the region bounded by amino acids 75 and 555 (Fig. 4A). Expression of the ChiA(75-555) mutant polypeptide in NS1, a secretion-deficient, epsD mutant strain of V. cholerae (Fig. 4B) established that, as is observed for wild-type ChiA, secretion of the ChiA(75-555) mutant polypeptide was dependent upon the eps-dependent extracellular transport system.
Disulfide bonds are often an important contributor to the proper folding and stability of extracellular proteins. The mutant ChiA(75-555) polypeptide contained two cysteines which were located immediately within the terminal boundaries (Cys77 and Cys537). To test whether Cys77 and/or Cys537 contributed to ETS activity, mutant ChiA polypeptides were engineered in which the cysteines were independently replaced with alanines (Fig. 1). Fractionation experiments showed that secretion of the ChiA polypeptides was unaffected by substitution of either amino acid (Fig. 6). The constraints, if any, that the other four cysteines (Cys273, Cys,300, Cys306, and Cys332) located within the 75 to 555 region have on ETS activity has yet to be elucidated.
To investigate the predicted secondary and tertiary structure of ChiA, the amino acid sequence of ChiA was analyzed using the ProDom (7) and SMART (31, 32) databases. ChiA was predicted to contain three types of structural domains: (i) two type III chitin-binding domains (CBD3), (ii) three type III fibronectin-like (Fn3-like) domains, and (iii) a catalytic domain (Fig. 5). Our experiments have confirmed that the catalytic domain of ChiA is localized proximal to the C terminus and downstream of the amino acid position 555 boundary (Fig. 5) (unpublished data). These data provided strong evidence that the catalytic domain of ChiA is not involved in ETS activity.
A role of the Fn3-like domains of ChiA in ETS activity, however, is more likely. Fn3-like domains are hypothesized to encompass strong and stable secondary protein structures that interact with other Fn3-like domains or with other structural domains located elsewhere in a polypeptide. These interactions are believed to promote and maintain the proper conformation of the protein (35). Interestingly, all three Fn3-like domains of ChiA (85 to 180, 336 to 423, and 429 to 528) are located within the 75 to 555 region of the protein (Fig. 5). Two of the three Fn3-like domains (85 to 180 and 429 to 528) of ChiA occur immediately within the boundaries of amino acids 75 to 555. Based on our observations and the pattern of Fn3-like domain distribution, one could hypothesize that the ETS of ChiA is encoded entirely by one or both of these terminally located Fn3-like domains. A more plausible hypothesis, however, is that the two Fn3-like domains (85 to 180 and 429 to 528) promote the proper folding and spacing of the ETS, which is located elsewhere in the protein. Mutant ChiA polypeptides which are deleted for one or more of the Fn3-like domains are currently being engineered to investigate the role of these regions in extracellular secretion of the protein.
In E. chrysanthemi, two nonadjacent regions of pectate lyase C are required for its extracellular transport (14). Similarly, secretion of a β-lactamase-pullulanase fusion protein by K. oxytoca was shown to require the expression of two nonadjacent 80-amino-acid regions of the pullulanase polypeptide (30). While it is possible that the ETS of ChiA is composed entirely of the 480-amino-acid region encompassed by amino acids 75 and 555, an equally plausible hypothesis is that, as for the ETS of pectate lyase and pullulanase, the ETS of ChiA consists of two nonadjacent regions of ChiA, one of which is located immediately beyond amino acid 75 and the other of which is located immediately adjacent to amino acid 555.
To investigate the latter model, we are currently engineering mutant ChiA polypeptides which are deleted for a significant region of the protein located between amino acids 75 and 555. To complement these continuing genetic studies, the three-dimensional structure of wild-type ChiA is being resolved. Upon completion of these genetic and structural investigations, the precise nature of the structural motif comprising the ETS of ChiA will finally be revealed.
Several important human pathogens, including P. aeruginosa (9, 10), A. hydrophila (3, 34), and V. cholerae (22, 28), employ the MTB to transport a variety of survival and virulence proteins to the extracellular milieu. With the possible exception of pullulanase (25) and exotoxin A (10), the current study of ChiA is the most extensive genetic analysis of a secreted protein to identify the amino acid sequences which constitute a bacterial ETS. Our effort to identify the ETS of ChiA is an important step toward the goal of defining the precise molecular interactions that occur between extracellular proteins of V. cholerae and the components of the eps-encoded secretion machinery.
Acknowledgments
We thank Shanmuga Sozhamannan for providing V. cholerae strains N16961 and NS1 and Maria Sandkvist for providing V. cholerae strains TRH7000 and PU3. We also thank Daniel Metzger, Erin Murphy, and Amy Kirby for critical reviews of the manuscript.
This work was supported by research grant R29AI37817 from the National Institutes of Health to T.D.C. and by funds from the School of Medicine and Biomedical Sciences at the State University of New York at Buffalo.
REFERENCES
- 1.Ali, A., J. A. Johnson, A. A. Franco, D. J. Metzger, T. D. Connell, J. G. Morris, Jr., and S. Sozhamannan. 2000. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68:1967-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bally, M., A. Filloux, M. Akrim, G. Ball, A. Lazdunski, and J. Tommassen. 1992. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol. Microbiol. 6:1121-1131. (Erratum, 6:2745.) [DOI] [PubMed] [Google Scholar]
- 3.Bo, J. N., and S. P. Howard. 1991. Mutagenesis and isolation of Aeromonas hydrophila genes which are required for extracellular secretion. J. Bacteriol. 173:1241-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connell, T. D., D. Metzger, C. Sfintescu, and R. T. Evans. 1998. Immunostimulatory activity of LT-IIa, a type II heat-labile enterotoxin of Escherichia coli. Immunol. Lett. 62:117-120. [DOI] [PubMed] [Google Scholar]
- 5.Connell, T. D., D. J. Metzger, J. Lynch, and J. P. Folster. 1998. Endochitinase is transported to the extracellular milieu by the eps-encoded general secretory pathway of Vibrio cholerae. J. Bacteriol. 180:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell, T. D., D. J. Metzger, M. Wang, M. G. Jobling, and R. K. Holmes. 1995. Initial studies of the structural signal for extracellular transport of cholera toxin and other proteins recognized by Vibrio cholerae. Infect. Immun. 63:4091-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corpet, F., J. Gouzy, and D. Kahn. 1999. Recent improvements of the ProDom database of protein domain families. Nucleic Acids Res. 27:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, B. M., E. H. Lawson, M. Sandkvist, A. Ali, S. Sozhamannan, and M. K. Waldor. 2000. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXphi. Science 288:333-335. [DOI] [PubMed] [Google Scholar]
- 9.Filloux, A., M. Bally, G. Ball, M. Akrim, J. Tommassen, and A. Lazdunski. 1990. Protein secretion in gram-negative bacteria: transport across the outer membrane involves common mechanisms in different bacteria. EMBO J. 9:4323-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22:177-198. [DOI] [PubMed] [Google Scholar]
- 11.Hirst, T. R., and J. Holmgren. 1987. Conformation of protein secreted across bacterial outer membranes: a study of enterotoxin translocation from Vibrio cholerae. Proc. Natl. Acad. Sci. USA 84:7418-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ish-Horowicz, D., and J. F. Burke. 1981. Rapid and efficient cosmid cloning. Nucleic Acids Res. 9:2989-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornacker, M. G., and A. P. Pugsley. 1990. The normally periplasmic enzyme beta-lactamase is specifically and efficiently translocated through the Escherichia coli outer membrane when it is fused to the cell-surface enzyme pullulanase. Mol. Microbiol. 4:1101-1109. [DOI] [PubMed] [Google Scholar]
- 14.Lindeberg, M., C. M. Boyd, N. T. Keen, and A. Collmer. 1998. External loops at the C terminus of Erwinia chrysanthemi pectate lyase C are required for species-specific secretion through the out type II pathway. J. Bacteriol. 180:1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu, H. M., and S. Lory. 1996. A specific targeting domain in mature exotoxin A is required for its extracellular secretion from Pseudomonas aeruginosa. EMBO J. 15:429-436. [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis, T. E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 17.Marcus, H., J. M. Ketley, J. B. Kaper, and R. K. Holmes. 1990. Effects of DNase production, plasmid size, and restriction barriers on transformation of Vibrio cholerae by electroporation and osmotic shock. FEMS Microbiol. Lett. 56:149-154. [DOI] [PubMed] [Google Scholar]
- 18.Marsh, J. W., and R. K. Taylor. 1998. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Mol. Microbiol. 29:1481-1492. [DOI] [PubMed] [Google Scholar]
- 19.Missiakas, D., and S. Raina. 1997. Protein folding in the bacterial periplasm. J. Bacteriol. 179:2465-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murata, H., M. Fons, A. Chatterjee, A. Collmer, and A. K. Chatterjee. 1990. Characterization of transposon insertion out mutants of Erwinia carotovora subsp. carotovora defective in enzyme export and of a DNA segment that complements out mutations in E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and Erwinia chrysanthemi. J. Bacteriol. 172:2970-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Callaghan, C. H., A. Morris, S. M. Kirby, and A. H. Shingler. 1972. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overbye, L. J., M. Sandkvist, and M. Bagdasarian. 1993. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene 132:101-106. [DOI] [PubMed] [Google Scholar]
- 23.Palomaki, T., and H. T. Saarilahti. 1995. The extreme C terminus is required for secretion of both the native polygalacturonase (PehA) and PehA-Bla hybrid proteins in Erwinia carotovora subsp. carotovora. Mol. Microbiol. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 24.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugsley, A. P., O. Francetic, K. Hardie, O. M. Possot, N. Sauvonnet, and A. Seydel. 1997. Pullulanase: model protein substrate for the general secretory pathway of gram-negative bacteria. Folia Microbiol. 42:184-192. [DOI] [PubMed] [Google Scholar]
- 26.Sandkvist, M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandkvist, M., L. O. Michel, L. P. Hough, V. M. Morales, M. Bagdasarian, M. Koomey, V. J. DiRita, and M. Bagdasarian. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandkvist, M., V. Morales, and M. Bagdasarian. 1993. A protein required for secretion of cholera toxin through the outer membrane of Vibrio cholerae. Gene 123:81-86. [DOI] [PubMed] [Google Scholar]
- 29.Sauvonnet, N., I. Poquet, and A. P. Pugsley. 1995. Extracellular secretion of pullulanase is unaffected by minor sequence changes but is usually prevented by adding reporter proteins to its N- or C-terminal end. J. Bacteriol. 177:5238-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sauvonnet, N., and A. P. Pugsley. 1996. Identification of two regions of Klebsiella oxytoca pullulanase that together are capable of promoting beta-lactamase secretion by the general secretory pathway. Mol. Microbiol. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sixma, T. K., S. E. Pronk, K. H. Kalk, E. S. Wartna, B. A. van Zanten, B. Witholt, and W. G. Hol. 1991. Crystal structure of a cholera toxin-related heat-labile enterotoxin from Escherichia coli. Nature 351:371-377. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, S. R., and T. J. Trust. 1995. A specific PulD homolog is required for the secretion of paracrystalline surface array subunits in Aeromonas hydrophila. J. Bacteriol. 177:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe, T., Y. Ito, T. Yamada, M. Hashimoto, S. Sekine, and H. Tanaka. 1994. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J. Bacteriol. 176:4465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong, K. R., D. M. McLean, and J. T. Buckley. 1990. Cloned aerolysin of Aeromonas hydrophila is exported by a wild-type marine Vibrio strain but remains periplasmic in pleiotropic export mutants. J. Bacteriol. 172:372-376. [DOI] [PMC free article] [PubMed] [Google Scholar]