Abstract
A total of 37 complete genome sequences of bacteria, archaea, and eukaryotes were compared. The percentage of orthologous genes of each species contained within any of the other 36 genomes was established. In addition, the mean identity of the orthologs was calculated. Several conclusions result: (i) a greater absolute number of orthologs of a given species is found in larger species than in smaller ones; (ii) a greater percentage of the orthologous genes of smaller genomes is contained in other species than is the case for larger genomes, which corresponds to a larger proportion of essential genes; (iii) before species can be specifically related to one another in terms of gene content, it is first necessary to correct for the size of the genome; (iv) eukaryotes have a significantly smaller percentage of bacterial orthologs after correction for genome size, which is consistent with their placement in a separate domain; (v) the archaebacteria are specifically related to one another but are not significantly different in gene content from the bacteria as a whole; (vi) determination of the mean identity of all orthologs (involving hundreds of gene comparisons per genome pair) reduces the impact of errors in misidentification of orthologs and to misalignments, and thus it is far more reliable than single gene comparisons; (vii) however, there is a maximum amount of change in protein sequences of 37% mean identity, which limits the use of percentage sequence identity to the lower taxa, a result which should also be true for single gene comparisons of both proteins and rRNA; (viii) most of the species that appear to be specifically related based upon gene content also appear to be specifically related based upon the mean identity of orthologs; (ix) the genes of a majority of species considered in this study have diverged too much to allow the construction of all-encompassing evolutionary trees. However, we have shown that eight species of gram-negative bacteria, six species of gram-positive bacteria, and eight species of archaebacteria are specifically related in terms of gene content, mean identity of orthologs, or both.
Evolutionary analyses based upon macromolecular sequences were initiated by Zuckerkandl and Pauling (76) with hemoglobin and by Fitch and Margoliash with cytochrome c (15). However, rRNA has since replaced these molecules as a universal indicator of evolutionary relationships among organisms (71). Several thousand RNA sequences have been determined in whole or in part and used to create a “universal tree of life” (48). A major apparent outcome of these analyses has been the division of all organisms but viruses into three domains--bacteria, eukaryotes, and archaea (74)--although not without controversy (23, 40). Whole-genome analysis was ushered in with completion of the sequence of the genome of Haemophilus influenzae (17). Since then, the genome sequences of more than 35 species were reported at the time this study was initiated (34, 45) with many more nearing completion. Whole-genome analysis has either been reported to confirm or to refute the relationships deduced from the analysis of RNA (14, 33, 34, 37, 51, 52, 61). However, it is currently thought that if gene transfer, gene duplication, gene deletion, and functional replacement are as extensive as appears to be the case, then hierarchical classifications based upon single genes (or classes of genes) such as rRNA do not provide a complete and accurate picture of evolution.
Whole-genome analysis is still in its infancy, and various means of analyzing the data are currently being explored (4, 16, 22, 27, 33, 47, 61, 65). Genome sizes range from ca. 0.5 to 10 Mb for bacteria and from 3 Mb to 3 Gb for eukaryotes. The number of genes per genome varies from less than 500 to more than 26,000, and the average gene size is ca. 1 kb. There are very few truly universal genes, and the extent of identity among proteins having similar structures and functions varies from highly conserved to barely recognizable. Among the homologs or similar proteins, there are those that share descent from a common ancestor and presumably have the same function (orthologs) and those that have been duplicated and are more likely to have different functions (paralogs). There is no way to know from sequence comparisons alone which genes are true orthologs and which are paralogs, but it is common practice in whole-genome studies to label as orthologs and assign tentative functions to those homologs with the greatest identity above a certain threshold. Duplicated genes are not uncommon, and the presence of split genes and gene fusions are also not unusual (4, 5, 25). Many genes are arranged in groups or operons and may be cotranscribed, but rearrangement of gene order from one species to the next is more often the rule than the exception (4, 5, 27). The presence of introns makes it difficult to identify eukaryotic genes, and best estimates place the numbers of human genes in the neighborhood of 26,000 to 39,000 (68). Thus, whole-genome analysis is not trivial, and it may take some time before agreement is reached on the best way to analyze the data. Nevertheless, we have now compared the genomes of 37 species in novel ways and have reached conclusions that differ from those in previous studies that considered a smaller number of species.
MATERIALS AND METHODS
Complete genome sequences were downloaded from GenBank (ftp://ftp.ncbi.nlm.nih.gov/GenBank/genomes/). GBK files and were automatically processed. The orthologs were identified by using Goldie 4.0, an extension of the earlier version of Goldie 2.0 (4, 5) (also see http://www.cs.kent.edu/∼arvind/orthos.html). The current version extends the previous versions by identifying the orthologs between multichromosome genomes and the genomes of prokaryotes and provides a new analysis of conserved genes. We briefly describe the methodology here. The number of genes compared in each genome is given in Table 1. A small number of genes had frameshifts in them, and their amino acid version was unavailable. These genes were removed from consideration. Orthologs were identified by the pairwise comparison of two genomes modeled as a weighted bipartite graph matching problem (49). Each genome was treated as a bag of genes in the bipartite graph, genes were treated as nodes of the graph, and identity scores after the Smith-Waterman phase between gene pairs from two genomes were treated as edges of the bipartite graph. The weights of the edges were identified by using a two-step process: NIH-BLAST (2) (see http://www.ncbi.nlm.nih.gov/Tools/index.html) was used for efficiency to prune the dissimilar gene pairs, and the local Smith-Waterman alignment (69) software seqaln_1.16 (see http://hto-13.usc.edu/software/seqaln/) and BLOSUM 62 matrix were used to identify the largest aligned fragment (within two genes being aligned) having highest average identity (i.e., the total number of identical amino acids × 100/the number of amino acids in the aligned fragment). All of the genes of the first genome were matched against the database of genes in the second genome by using a relaxed high-score value (P) of >30 and a chance score (E-value) of <10−3. These scores may seem too low, but Snel et al. (61) used a cutoff of 10−2 which they estimate increased the numbers of orthologs by only 3%. We ran experiments with Escherichia coli and Bacillus subtilis with different E-value cutoffs and found that the difference between the lower cutoff of 10−2 and 10−3 is 5% (1,522 versus 1,438 orthologs) and the difference between a cutoff of 10−3 and 10−5 is only 4% (1,438 versus 1,379 orthologs). These data show that the apparent number of orthologs will change slightly with choice of parameters, but provided that we are consistent in using a single cutoff throughout this study, the comparisons among genomes and with other studies using similar cutoffs should be valid.
TABLE 1.
Genomes analyzed in this study
| Genomea | Abbreviationb | Genome size (Mb) | No. of protein genes | Reference |
|---|---|---|---|---|
| Aeropyrum pernix | AP | 1.67 | 2,694 | 31 |
| Aquifex aeolicus | AQ | 1.55 | 1,522 | 12 |
| Archaeoglobus fulgidus | AF | 2.18 | 2,407 | 32 |
| Bacillus halodurans | BH | 4.20 | 4,066 | 64 |
| Bacillus subtilis | BS | 4.20 | 4,100 | 36 |
| Borrelia burgdorferi | BB | 1.44 | 850 | 19 |
| Buchnera sp. | BU | 0.64 | 564 | 57 |
| Campylobacter jejuni | CJ | 1.64 | 1,634 | 50 |
| Chlamydia pneumoniae CWL029 | CP | 1.07 | 1,052 | 28 |
| Chlamydia trachomatis | CT | 1.04 | 894 | 62 |
| Deinococcus radiodurans | DR | 3.28 | 3,102 | 70 |
| Escherichia coli K-12 | ECK | 4.64 | 4,289 | 6 |
| Escherichia coli O157:H7 EDL933 | ECO | 5.44 | 5,283 | 53 |
| Haemophilus influenzae | HI | 1.83 | 1,717 | 17 |
| Halobacterium sp. | HA | 2.57 | 2,058 | 46 |
| Helicobacter pylori 26695 | HP | 1.67 | 1,566 | 67 |
| Lactococcus lactis | LL | 2.37 | 2,266 | 8 |
| Methanococcus jannaschii | MJ | 1.73 | 1,715 | 9 |
| Methanobacterium thermoautotrophicum | MT | 1.75 | 1,869 | 60 |
| Mycobacterium tuberculosis H37Rv | MTB | 4.41 | 3,918 | 11 |
| Mycoplasma genitalium | MG | 0.58 | 467 | 18 |
| Mycoplasma pneumoniae | MP | 0.82 | 678 | 26 |
| Neisseria meningitidis MC58 | NM | 2.27 | 2,025 | 66 |
| Pasteurella multocida | PM | 2.26 | 2,014 | 39 |
| Pseudomonas aeruginosa | PAE | 6.26 | 5,565 | 63 |
| Pyrococcus abyssi | PA | 1.77 | 1,765 | 38 |
| Pyrococcus horikoshii | PH | 1.74 | 2,064 | 30 |
| Rickettsia prowazekii | RP | 1.11 | 834 | 3 |
| Synechocystis sp. | SS | 3.57 | 3,169 | 29 |
| Thermoplasma acidophilum | TA | 1.56 | 1,478 | 54 |
| Thermotoga maritima | TM | 1.86 | 1,846 | 44 |
| Treponema pallidum | TP | 1.14 | 1,031 | 20 |
| Ureaplasma urealyticum | UU | 0.75 | 611 | 21 |
| Vibrio cholerae | VC | 4.03 | 3,828 | 24 |
| Xylella fastidiosa | XF | 2.68 | 2,766 | 58 |
| Caenorhabditis elegans | CE | 97 | 17,085 | 1 |
| Saccharomyces cerevisiae | SC | 12 | 6,297 | 41 |
Strains are indicated only where more than one was available.
See Fig. 1, 2, and 6.
Since BLAST comparison does not pick up all of the similar genes, we repeated the process by comparing all of the genes in the second genome against the database of the first genome by using the same high-score value and the same chance score. The relaxed cutoff of 10−3 was used since we did not want to bias the results with any preconceived notions of phylogeny and we wanted to capture corresponding gene pairs even when they are evolutionarily distant. The union of all of the gene pairs in two comparisons provided the similar gene pairs for the Smith-Waterman alignment phase which used amino acid matching scores given by the BLOSUM 62 matrix. The penalty score was 15 for the first insertion or deletion and 5 for each successive insertion or deletion. The identity scores from the local Smith-Waterman alignment became the weights of the edges for matching the weighted bipartite graph matching. Preference was given to gene pairs belonging to homologous gene groups (4) by biasing the identity score of such gene pairs by 20%. To separate the best candidate for the corresponding gene pairs, the following criteria were used: the corresponding gene pair (i) was unique, (ii) had an identity score 20% greater than the next similar gene pair, or (iii) had an identity greater than 75%. The last case suggests multiple functionally equivalent genes or gene fusion (fission) and almost replicate genes. In the last case, our cutoff was quite stringent to avoid any paralogs. This choice was based upon the notion that genes with >75% identity are more likely to be functionally equivalent than those that are much less similar. We might have lost some of the true-positive cases of corresponding genes due to this stringent choice of cutoff, but if a gene pair did not meet these three criteria, it was dropped from consideration. The scheme identifies weak similarities in the case a homolog is unique or the similarity scores of two homologs are quite different. However, this technique still misses very weak homology due to the choice of BLAST cutoff needed to prune the search space and due to the inherent inaccuracy caused by the treatment of insertions and deletions in sequence alignment schemes.
We validated our technique with experimentally known orthologs in E. coli and B. subtilis and reported comparisons of E. coli with H. influenzae (see http://www.cs.kent.edu/∼arvind/orthos.html). We refer to these corresponding genes as orthologs throughout this study since they are the best computational approximation to functional equivalence. However, this approach still identifies some closely related paralogs as orthologs, and some highly divergent orthologs may be missed. The multichromosomal genome comparisons such as Vibrio cholerae were handled by using two techniques: the genes in multichromosomal genomes were colored to preserve the identity of chromosomes and merged to form one large ordered set. The genome comparisons were done with respect to this larger set. For very large genomes such as Caenorhabditis elegans, the orthologs were identified in two stages: identifying candidates at the individual chromosome level and then resolving the best score from all chromosomes.
Our analysis showed that the genes in very closely related microbes shared larger alignment fragments, and the fragment size varied from better than 90% of the length of the smaller gene for very closely related genomes to ca. 20% for the most distantly related genomes. This varied for different genes (including ribosomal proteins), making it quite difficult to perform any normalization based upon genome relatedness or to develop a broader classification of the set of genomes. Nevertheless, we felt that this should be examined in greater depth to give the reader a better sense of the problem. For example, more than 80% of one of the fully conserved genes, E. coli rpsK (130-amino-acid ribosomal protein), matched with orthologs in all other genomes, while E. coli pheS (327-amino-acid phenylalanyl-tRNA synthetase alpha chain) matched only a 161-amino-acid fragment of Aeropyrum pernix APE2302 (473-amino-acid protein). For the two Bacillus spp., the two Chlamydia spp., and the two E. coli strains, the median matching length was better than 90%. For Saccharomyces cerevisiae versus Ureaplasma urealyticum, for C. elegans versus Borrelia burgdorferi, or for C. elegans versus Mycoplasma genitalium, the median matching length was in the range of 23% ± 4%. For comparisons among the bacteria or among the archaea, the median matching length was ca. 82% ± 8%. For the most divergent prokaryotes, the median matching length was in the range of 51% ± 8%. The median length between S. cerevisiae and the prokaryotes was 38% ± 8%, with extremes of 21% and 57%. The median length for C. elegans and prokaryotes was ca. 31% ± 7%, with extremes of 19% and 42%. The matching length for S. cerevisiae with C. elegans was a surprisingly low 32%. There is no evidence that the eukaryotes match larger segments with either the archaea or the bacteria. It is also clear that we need to analyze greater numbers of organisms in each of the major categories to refine this avenue of research.
Despite the choice of a relaxed cutoff, the BLAST phase did not pick up every homolog. However, the occurrence of such incidences was very low. This fact was found due to the loss of transitivity identified in ortholog relationships for some genes, that is, if a gene X in a genome A was orthologous to a gene Y in a genome B and gene Y in a genome B was orthologous to gene Z in genome C, then it was not always the case that gene X in genome A was identified as a homolog to gene Z in genome C after BLAST comparison. It was also found that, despite our choosing unique or best homologs and pruning all remaining homologs involving two genes (or gene fragments) in orthologous gene pairs, the group of orthologs clustered after merging the results of all 37 genomes often contained two genes with very similar functionality from the same genomes. This phenomenon of multiple similar genes increased when the definition of orthologs allowed smaller best-matching fragments to be included, suggesting that, for distantly related genomes, pairwise genome comparison may also pick up some paralogs or genes which can substitute for the functionality.
We removed all of the gene pairs that had orthologous gene fragments less than 30% the size of the largest orthologous fragment in the cluster. The cutoff of 30% was guided by the fact that many fully conserved genes are ribosomal proteins with sizes of as low as 93 amino acids (E. coli rpsS). Since the artifact cutoff size was taken as 30 amino acids, smaller gene fragments matching with ribosomal proteins will be treated as artifacts, and the results would be inconsistent if the cutoff ratio was <30%. This cutoff also ensures that we restrict the number of closely related paralogs.
To account for missing orthologs caused by incompleteness of BLAST comparison, we used the transitivity relationship described above. Care was taken so that the same alignment fragments were used for the transitive relationship. However, this sometimes resulted in two “orthologous genes” being identified from the same genome: the first one was identified by direct genome comparison of genomes A and C (in the absence of the missed gene pair from BLAST comparison), and the second one was derived by the transitive relationship. It was very difficult to find the exact identity score for orthologs derived by transitivity, and the presence of any false positives (caused by missing homologs from the BLAST phase) in the transitivity chain may cause a paralog to be picked up. In such cases, the following criterion was used to pick up an ortholog: (i) only one gene, with the largest aligned fragment (which was >20% larger than the nearest candidates) in each genome, was retained if the ratio of the two gene fragments from the same genome was <75% the size of the largest fragment; (ii) the genes using direct pairwise genome comparison were preferred over derived orthologs if the separation between the sizes of the two gene fragments was <20%; and (iii) all genes were retained if their size was >75% of the size of the largest fragment. Derived orthologs (using transitivity) and clusters of orthologs were obtained by using a greedy algorithm with the starting point being a set of clusters of fully conserved genes identified from individual genome comparisons with all other genomes. The clusters with orthologs in the maximum number of genomes were joined first. Two clusters of orthologs (containing common orthologs) were merged by using the following criterion: if the differences in the sizes of the largest gene fragments in both clusters were <20% and both largest genes were orthologs, then both clusters were completely merged; otherwise only genes whose size was within 80% of the sizes of the ortholog were copied from the second cluster and included in the first cluster. The rationale for this criterion is that genomes having orthologs with larger aligned fragments are more probably closely related. However, two genes from two genomes being orthologous to a common smaller fragment of a third genome does not ensure that two genomes having larger fragments are closely related unless it is established by direct pairwise comparison of those two genomes. Genes which are specific only to a set of organisms were found by first identifying all of the clusters of orthologs containing at least one genome in the set and then filtering out all of the clusters that contained the microbes which are not members of the set.
RESULTS
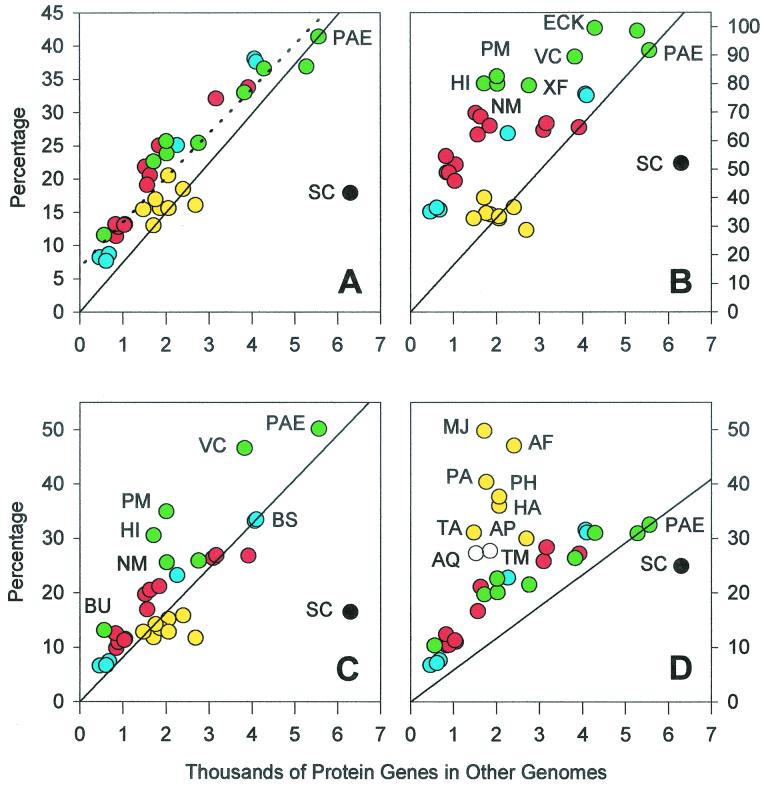
A total of 27 bacterial genomes, 8 archaea, and 2 eukaryotes were chosen for this study (Table 1). At least 20 more genome sequences have been completed (45), although the data had not yet been deposited in GenBank at the time of this study. In any case, 37 genomes are more than sufficient to illustrate our method of analysis. We first determined the numbers of orthologs in pairwise comparisons of the 37 genomes as shown in Fig. 1. Orthologs, for purposes of this study (and consistent with usage in other whole-genome studies), are those homologous genes that show the largest identity of several possibilities above a certain threshold. Because genomes differ so much in size, it is necessary to normalize the data. We have chosen to determine the percentages of genes in each species that are present in each of the other genomes (as orthologs) as shown in Fig. 2. As expected from previous work (4, 61), there is a relationship between the number of orthologs and the genome size as illustrated in Fig. 3A, where the percentage of orthologous Deinococcus radiodurans genes is compared with the total number of genes in the other genomes. Deinococcus has no close relatives in our database; thus, we fit this curve with a straight line, resulting in a slope of 6.7% per 1,000 genes and intercept of 6.8% (excluding only yeast and worm). The intercept could be interpreted as the minimal genome size of ca. 212 genes. The relatively good correlation (0.93) indicates that the microorganisms in our database are able to freely exchange DNA, i.e., the major determinant of gene content appears to be gene transfer. Yeast shares only one-third of the Deinococcus genes expected from the size of its genome. The worm, C. elegans (data not shown), also shares much less than expected of the Deinococcus genome, indicating that they are genetically isolated and that gene transfer is negligible between prokaryotes and eukaryotes. Archaea, on the other hand, cluster with the bacteria. They are slightly below the line (by an average of 3.7%), but this is insignificant in comparison with the average deviation of 2.8%. That is, archaea are part of the same gene pool as the other prokaryotes. For those species of bacteria that are specifically related, they would be expected to deviate from a straight line in a positive direction as shown in Fig. 3B to D. In these cases, the data cannot be fit because we have no way of knowing, a priori, which species are related and which are not. Thus, for purposes of comparison, we drew a line through the origin and the point representing a large presumably unrelated species such as B. subtilis or Pseudomonas aeruginosa. Note that the slope of the arbitrary line (7.4% per 1,000 genes) in Fig. 3A is about the same as that of the fitted line (6.7% per 1,000 genes) and is therefore a reasonable approximation of the size relationships where it is not possible to fit the data. The approximate slope of the two lines in Fig. 3A, 6.7 to 7.4%, represents the approximate percentage of D. radiodurans genes that are likely to be found in any other unrelated prokaryotic genome containing 1,000 genes. The scatter in this plot, 2.8%, represents the amount of variation that is likely for species that are not specifically related (that is, those that do not show a significantly greater than average similarity than for all comparisons).
FIG. 1.
Number of orthologous genes in genome pairs. Abbreviations are as defined in Table 1.
FIG. 2.
Percentage of orthologous genes of each species contained within the other genomes. Each row contains the percentage of orthologous genes on the left contained within the species indicated across the top. Abbreviations are as defined in Table 1.
FIG. 3.
Relationship between gene content and genome size. First, the numbers of orthologous genes in Fig. 1 were divided by the numbers of genes in Table 1, resulting in the percentage of orthologous genes shown in Fig. 2. The percentage of orthologous genes of the first genome of a species pair were then plotted versus the total number of genes in the second genome of the pair for 35 comparisons (because of its exceptionally large size, C. elegans was excluded for clarity). (A) Percentage of D. radiodurans genes in other genomes; (B) percentage of Buchnera sp. genes in other genomes; (C) percentage of E. coli genes in other genomes; (D) percentage of Methanobacterium thermoautotrophicus genes in other genomes. The dotted line is a fit to the data, whereas the solid lines arbitrarily connect the origin to either B. subtilis or P. aeruginosa. The archaebacteria are yellow, the related gram-positive species are blue, the related gram-negative species are green, yeast is black, and the remaining species are red. The two bacteria which appear to be related to the archaea in plot D are shown as open circles.
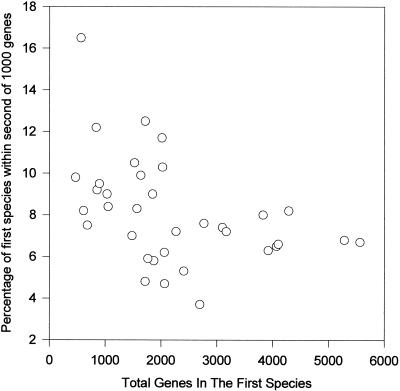
Plots were made for all 37 species to determine whether any specific relationships could be recognized among the bacteria. However, the smaller genomes are expected to be dominated by essential genes and to share a greater percentage of their genomes with the other species, and this is seen by the comparison of Buchnera sp. with E. coli, where 99% of the Buchnera sp. genome is represented by orthologs in the E. coli genome, as shown in Fig. 3B. In this example, it is apparent that Buchnera sp. is specifically related to a number of gram-negative bacteria as indicated by the much greater deviation from the line than observed in the D. radiodurans plot, and Buchnera sp. shares twice as much of its genome with other species (16% per 1,000 total genes). When the slopes of the lines in the gene content plots of Fig. 3 and for all other species were themselves plotted (Fig. 4), it can be seen that below a genome size of ca. 2,000 genes, the percentage of that genome found per 1,000 genes of any other genome more than doubles for the smallest species compared to the largest ones (16% versus 7%). We interpret this to mean that 2,000 genes is the approximate minimal size of a free-living bacterium that is able to make all necessary cofactors.
FIG. 4.
Relationship between the percentage of orthologous genes in other species and genome size. The slopes of the lines in the plots of Fig. 3 and similar bacterial comparisons were plotted versus the genome size of the first species.
E. coli, which has a moderately large genome, shares a smaller percentage of its genes with other species than does Buchnera; in fact, it is more like D. radiodurans in this regard, and the greatest apparent identity, not unexpectedly, is with P. aeruginosa, which is the largest bacterial genome in our study (Fig. 3C). It also appears to share a significantly greater number of genes with H. influenzae and V. cholerae than expected by the size of the genomes alone. On the other hand, E. coli shares only a relatively small percentage of its genes with yeast or worm in spite of the fact that eukaryotic genomes are significantly larger than those of E. coli and P. aeruginosa. This is also true for the comparison of other bacteria with the eukaryotes. As expected, the two eukaryotes are more closely related to one another by this criterion than they are to the bacteria. It is a strong indication that eukaryotes belong in a separate kingdom or domain from the prokaryotes as a whole and are genetically more isolated. This may appear to be a trivial conclusion since few people would disagree, but it is important in the context of the following analysis.
E. coli does not appear to be specifically related to B. subtilis, but ca. 34% of its genome is represented by orthologs in B. subtilis. If a line is drawn from the origin of Fig. 3C through the point representing B. subtilis, it may be taken to represent the approximate empirical relationship between orthologs and genome size. Thus, ca. 8.2% of the E. coli genes are expected to be present as orthologs in a bacterial genome of 1,000 genes. H. influenzae contains 1,717 protein coding genes; thus, E. coli should share ca. 14% of its genome with H. influenzae, based on the sizes of their genomes, if they are not specifically related. In fact, 31% of the E. coli genome is represented by orthologs in H. influenzae, more than twice the number expected by size alone, indicating that they are in fact specifically related. Likewise, V. cholerae should share 32% of the E. coli genome based on size but actually shares 47%, indicating that they, too, are related. Tiny Buchnera sp. should share <5% of the E. coli genome by this measure but contains 13%, again indicating a specific relationship. Neisseria meningitidis appears to be related to E. coli by this criterion but to a lesser extent, as seen by only a slightly greater percentage than represented by the scatter in the line. A self-consistent picture is obtained when other species are plotted in a similar manner as those in Fig. 3. Thus, for those species apparently related to E. coli, a greater-than-expected percentage of, e.g., V. cholerae genes is present in E. coli, H. influenzae, Pasteurella multocida, N. meningitidis, and Buchnera sp. Likewise, the largest percentage of H. influenzae genes is present in P. multocida, E. coli, V. cholera, N. meningitidis, and Buchnera sp. and so on.
It has been argued on the basis of 16S rRNA comparisons that archaebacteria are so different that they belong in a separate domain equivalent in rank to that of the bacteria and of the eukaryotes (74). If archaebacteria do belong in a separate domain, then that difference should be reflected in the gene content as was found above for the eukaryotes in comparison with bacteria. In fact, the archaea group with the bacteria in all cases, that is, within error limits they share as many of their genes with the bacteria as distantly related bacteria of the same size share with one another and on this basis should not be considered to be in a separate domain. Mayr (40) previously argued on different grounds that archaebacteria did not deserve the status of a higher taxon, and our data on gene content are consistent with that conclusion. The archaebacteria fall on the line defined by B. subtilis in Fig. 3C, indicating that, on a normalized basis, they share no more nor less of the E. coli genes than do B. subtilis, P. aeruginosa, Mycobacterium tuberculosis, or Synechocystis sp. Aeropyrum pernix is an exception in that it consistently has somewhat fewer of the bacterial genes than do the other archaebacteria although it is still within error limits of being the same. The plot of the percentage of the Methanobacterium thermoautotrophicum genes present in other species in Fig. 3D shows that the archaebacteria are specifically related to one another as first proposed by Woese (71). Although they do not belong in a separate domain according to our analysis, they do represent a major subdivision of bacteria such as the coliforms, the actinomycetes, and the bacilli. As expected by its lifestyle, Methanococcus jannaschii shares significantly more genes with Methanobacterium thermoautotrophicum than do other species. Archaeoglobus fulgidus also shares a surprising number of genes with the methanogens. Pyrococcus abyssi and Pyrococcus horikoshii are specifically related, as are Halobacterium sp. and Thermoplasma acidophilum, but to a lesser extent. Aeropyrum pernix is only slightly above the scatter in the line defined by P. aeruginosa and the other species of bacteria and in fact Aquifex aeolicus and Thermotoga maritima show greater apparent similarity in their gene content to the methanogens than does Aeropyrum pernix as previously observed by Nelson et al. (44). This is yet another indication that the archaebacteria are not deserving of placement in a higher taxon.
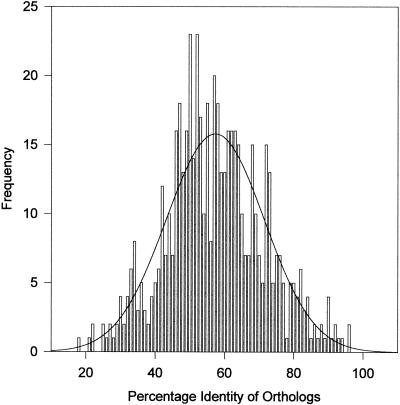
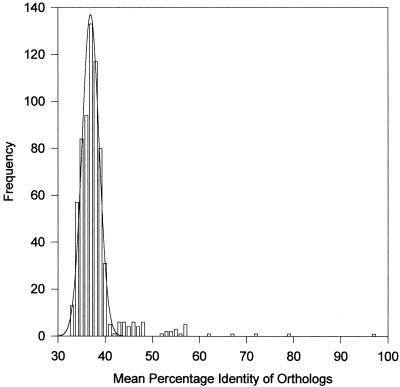
The above analysis is based upon gene content. However, another, independent, criterion of relatedness is the degree of similarity in pairs of orthologs. One should observe approximately the same relationship among species using any gene, provided it is neither a paralog nor resulting from recent gene transfer. When all of the orthologs in a pair of species are aligned and the mean percentage identities are determined, they provide an overall measure of relatedness that is superior to any single gene comparison. By simultaneously comparing all orthologs, any errors resulting from misidentification and misalignment are minimized. As shown in Fig. 5, the orthologous proteins range from highly conserved to barely recognizable. However, the percent identity of orthologs is more or less normally distributed except where the species are very closely or very distantly related, in which cases the distributions are markedly skewed and the median identity becomes a better measure of relatedness than does the mean. Such skew does not qualitatively affect the conclusions and was not taken into account because those comparisons are obvious. The mean of the distribution was calculated for all orthologs of each species pair as shown in Fig. 6. These data are plotted in Fig. 7, where it is apparent that the mean of the mean percent identity for recognizable orthologs in all species is 36.9% (a number very similar to the 36.2% median identity observed by Nolling et al. [47] for Clostridium acetobutylicum in comparison to other species). The standard deviation for this plot is 1.8; thus, species beyond two or three standard deviations (95.4 to 99.7% of the comparisons) or 40.5 to 42.3% mean identity in their orthologs may be considered to be significantly more closely related than the average as highlighted in Fig. 6.
FIG. 5.
Mean identity of orthologs. Distribution of the percentage identities of the 561 orthologous genes shared by E. coli and Buchnera sp. The solid line is a Gaussian fit to the data. The mean is 57.3%, and the standard deviation is 14.2. All such distributions were fitted, and the results are presented in Fig. 6.
FIG. 6.
Mean identity of orthologs in genome pairs. Numbers thought to be significant are in boldface. Abbreviations are as defined in Table 1.
FIG. 7.
Distribution of the mean identity of orthologous genes from Fig. 6. The solid line is a Gaussian fit to the data below 43% identity. The mean of the mean is 36.9%, and the standard deviation is 1.79. All data at or beyond two standard deviations are highlighted in Fig. 6 to indicate which are significant.
Based upon the mean identity of orthologs, it can be seen that most of the same species that have a significantly greater percentage of their genomes represented in other species than can be expected by size of the genome alone also show significantly greater similarity in their orthologs. This is in itself a significant finding, i.e., that two independent methods of analysis give consistent results, something that is not often observed in evolutionary studies. Thus, M. genitalium, which has 97% of its genes in common with Mycoplasma pneumoniae (15 times as much as expected), also shows 67% mean identity of its orthologs as shown in Fig. 6. Likewise, Pyrococcus abyssi, which has 82% of its genes in common with Pyrococcus horikoshii (or more than seven times the expected amount), also shows more than 79% mean identity in its orthologs (ignoring skew). Chlamydia trachomatis, which has 92% of its genes in common with Chlamydia pneumoniae (or nine times what is expected), also shows 62% mean identity in its orthologs. B. subtilis and Bacillus halodurans share 56% of their genes with one another (twice what is expected based on size), and their orthologs show 54% mean identity. E. coli strain K-12 has 91% of its genes in the larger E. coli strain O157 (twice as much as expected), and strain O157 has 74% of its genes in strain K-12, but their orthologs average an impressive 97% mean identity (ignoring skew). Individual species may acquire or delete a larger or smaller number of genes than average; hence, we should not expect a strong correlation between the two measures of relatedness, but the two should be qualitatively consistent, and that is what we have observed.
More distantly related species than those in the same genus can also be recognized by the mean percentage identity in their orthologs, a finding which is consistent with the fraction of the genome present in other species. Thus, E. coli, H. influenzae, P. multocida, Buchnera sp., and V. cholerae are specifically related to one another by both criteria. Buchnera sp., which has 99% of its genome represented in E. coli (1.5 times expected), shows 57% mean identity in its orthologs. H. influenzae, which has 76% of its genes represented in E. coli (1.5 times expected), also shows 57% identity in its orthologs. P. multocida has 75% of its genome in E. coli (1.5 times expected) and shows 57% mean identity in its orthologs. V. cholerae has 52% of its genome in E. coli (1.5 times expected) and shows 55% mean identity in its orthologs. H. influenzae has 81% of its genome in P. multocida (three times expected) and shows 72% mean identity in its orthologs. In fact, P. multocida and H. influenzae are similar enough to be considered members of the same genus by this measure. P. multocida and H. influenzae are slightly closer to E. coli than to V. cholerae, with 70% of their genomes in V. cholerae (1.5 times expected) and 55% mean identity of orthologs. Buchnera sp. is also closer to E. coli than to either H. influenzae or V. cholerae. Although the fraction of the smaller genomes present in E. coli is not very large (ca. 1.5 times), it is more significant for comparisons among the smaller species (up to three- to fourfold). The gram-negative bacteria P. aeruginosa, N. meningitidis, and Xylella fastidiosa are somewhat more divergent from E. coli and relatives, with ca. 48 to 44% mean identity of their orthologs, respectively. These comparisons are still quite significant at more than four standard deviations from the mean, and the gene contents for many of the comparisons for the group as a whole are greater than that expected by size alone. In this instance, the mean identity of orthologs appears to give a clearer picture of relatedness than does gene content.
There are species that show even less similarity to the coliforms than do P. aeruginosa, N. meningitidis, and X. fastidiosa and thus fall into a marginal category. Although individual comparisons are not in themselves very significant since they are within one standard deviation of the normal distribution, the Rickettsia prowazekii orthologs are consistently more like the coliforms and relatives than they are to other species, i.e., with 39 to 40% mean identity and as many as twice the number of expected orthologs in some cases. It could be argued that D. radiodurans is slightly closer to this group of species and even shows 41% mean identity in orthologs to P. aeruginosa, although it is not obvious from gene content. Campylobacter jejuni and Helicobacter pylori are clearly related to one another at the level of 47% mean identity in their orthologs and show more than three times greater similarity in gene content than expected. They consistently show slightly greater similarity to the coliforms and relatives than to other species (with twice the expected gene content but with only an insignificant 38 to 39% mean identity). Aquifex aeolicus and T. maritima are related to one another at the level of 41% mean identity and 2.5 to 3 times the expected gene content. They also seem to be related to C. jejuni and H. pylori, with 2 to 2.5 times the expected gene content but with only 38 to 39% mean identity of orthologs. This similarity appears to extend to P. multocida, H. influenzae, and N. meningitidis. However, these results need to be corroborated by additional studies.
Treponema pallidum and Borrelia burgdorferi show marginal similarity to one another at 40% mean identity in orthologs, but with more than five times the expected similarity in gene content. They also show greater-than-expected similarity in gene content to a number of both gram-negative and gram-positive species, but they cannot be assigned to a specific group at present. Although there is no specific relationship to the coliforms or to the spirochaetes, it is also possible to show a weak relationship between U. urealyticum and the mycoplasmas (M. genitalium and M. pneumoniae) at 40 to 41% mean identity in orthologs and a much more significant nine times the expected gene content. However, Thermoplasma acidophilum is not part of this small group. Lactococcus lactis shares twice as many genes as expected with B. subtilis and shows 44% mean identity in its orthologs. The Mycoplasma spp. and U. urealyticum share more genes with B. subtilis and L. lactis than expected by factors of 2 to 3, but the mean identity of orthologs is not significant (37 to 39%). In this instance, gene content appears to be more informative than does the mean identity of orthologs. The two measures of relatedness need not necessarily give the same result, although it is more believable when they track together. In fact, there may be some limitations on the utility of mean identity of orthologs as discussed below.
Of the eight archaebacteria analyzed, six can be clearly related to one another, and the remaining two are marginally related. The two Pyrococcus spp. (Pyrococcus abyssi and Pyrococcus horikoshii) are the most closely related species at more than 79% mean identity of orthologs and with more than seven times as many shared genes as expected. The two methanogens show the second most significant relationship to one another at 46% mean identity of orthologs and more than five times as many shared genes as expected. They are slightly more distant from the Pyrococcus species at 44% mean identity and with three to five times as many shared genes as expected. Archaeoglobus fulgidus is more distant at 43 to 44% mean identity to the first four and with three to four times as many shared genes as expected. Aeropyrum pernix is even less related to these five species at 39 to 41% mean identity, with slightly greater similarity to Pyrococcus and three to four times as many shared genes as expected. Halobacterium sp. and Thermoplasma acidophilum are the most distant to the other archaebacteria in that they show a marginal 38 to 40% mean identity in orthologs and share about three times as many genes as expected based upon size alone.
The evolutionary position of Aquifex aeolicus and T. maritima is very interesting for several reasons. They are reported to be among the most ancient of bacteria and share many biochemical similarities with the archaebacteria. For example, they are extreme thermophiles and contain ether-linked lipids. As indicated above, their orthologs average 41% identity to one another, and they share nearly three times as many genes as expected. They also appear to share some similarity to the coliforms and related gram-negative bacteria. All of the archaebacteria show two- to fourfold more shared genes with Aquifex aeolicus and T. maritima than expected, although Pyrococcus abyssii is the only species in which mean identity of orthologs is slightly closer than average (to T. maritima, 40%). On the other hand, from the standpoint of Aquifex aeolicus and T. maritima, neither show significantly greater similarity to the archaebacteria than to the other bacteria. Thus, these two species appear to share characteristics of both gram-negative bacteria and of archaebacteria, but further study is required to determine their precise relationships.
An interesting observation from whole-genome analysis is that not all species can be specifically related to the others in the sense that they are no closer to one another than average by either criterion such as Mycobacterium tuberculosis or Synechocystis sp. Three relatively large groups of related species do stand out: the coliforms and certain other gram-negative species, B. subtilis and certain other gram-positive species, and the archaebacteria. The “tree of life” based upon 16S rRNA purports to relate all species in a hierarchical fashion. However, we know that there is significant gene transfer and duplication among species of bacteria that might compromise single gene comparisons. The simultaneous analysis of all orthologs should minimize the negative effects of gene transfer, duplication, and misalignment on evolutionary studies, but even that does not allow all species to be specifically related to one another since we have found that the mean percent identity of orthologs approaches a limit of ca. 37% for the most distantly related species and is not reliable for taxa much above the family level.
It is thought that slowly evolving genes (those showing the largest overall percent identity) can reveal relationships for the most distantly related species. However, we believe that the slowly evolving genes only make it easier to align sequences and that all orthologous genes will give virtually the same result barring unforeseen gene transfer or paralogy and provided that they can be aligned unequivocally. There is an implied assumption that there is a positive correlation between slowly evolving genes (those with high sequence identity) and conserved genes (that are found in the majority of species), but neither this assumption nor its inverse is necessarily correct. It is also commonly assumed but not necessarily true that the most highly conserved genes are unlikely to be duplicated or transferred and therefore result in more reliable evolutionary trees. The individual comparison of more than a dozen proteins that are found in all species in our study (data not shown) revealed that species which are clearly related as deduced from whole-genome analyses are also obvious from single-gene comparisons. However, the distantly related and marginal species from whole-genome comparisons cannot be precisely and consistently positioned in single-gene trees. The single-gene analysis was particularly uncertain concerning the position of the two methanogens with respect to Archaeoglobus, but mean identity of orthologs and gene content clearly showed that the methanogens are more closely related to one another than to Archaeoglobus, as they should be considering their lifestyles. Halobacterium is generally placed among the methanogens in rRNA trees, and its position is uncertain from the other single-gene comparisons, but it is clearly one of the most divergent of the archaebacteria from whole-genome analysis. Aeropyrum pernix is thought to be the most divergent of the archaebacteria included in this study but is closer to the methanogens than is Halobacterium sp. based upon whole-genome analysis. Thus, we believe that whole-genome analysis can resolve at least some of the uncertainties from single-gene analyses and should be the method of choice where whole-genome sequences are available.
DISCUSSION
Whole-genome analysis is still new enough that there is as yet no consensus on what is the best way to compare genomes. One way is to add up the number of orthologous proteins, which we have operationally defined as homologs having the greatest similarity and presumably the same function. This is in itself not a trivial task because gene duplication, gene transfer, gene deletion, gene fusion, and gene splitting are all common. Due to fusion and splitting, as well as frameshifts, we cannot compare whole genes but must search protein domains, which generally become smaller as species diverge. Homologous domains having the greatest percent identity are then defined as orthologs. True functional identity can only be established by enzymatic activity and gene expression, something that is not likely to be measured for a whole genome. The concept of orthology in whole-genome analysis is thus an inexact quantity and must be viewed as hypothetical in the majority of cases. Once having added up the number of orthologs, we cannot compare species directly because they have variably sized genomes. The orthologs should be expressed as some percentage of the genes being compared. We could use the total number of unique genes in the two genomes (43) or the average of the two genomes, but that would underestimate the similarity between species in which the genes of one of the two are essentially contained within the other, such as M. genitalium within M. pneumoniae or Buchnera sp. within E. coli. The larger species might have acquired a block of genes through gene transfer, or the smaller genome may have lost a block of nonessential genes in a single event that may not be particularly significant. One way around this difficulty is to divide the orthologs by the number of genes in the smaller of the two genomes (4, 61), but that provides little information about the larger genome of the pair. Thus, we followed the example of Tekaia et al. (65) and separately divided the orthologs by the number of genes in each species of the pair. Twice as many numbers are generated that way, but we learn what fraction of a genome is contained as orthologs within any other species. We consider this to be the most meaningful measure of relatedness.
It was previously shown that the percentage of orthologs in one species is related to the size of the other genome (4, 61), although it was not quantified. We can empirically determine that relationship for each species by comparison with a large, presumably distantly related species. We chose B. subtilis for comparison of most of the gram-negative bacteria and P. aeruginosa for most of the gram-positive species. Synechocystis sp., Mycobacterium tuberculosis, or D. radiodurans would also work since they do not appear to have any near relatives in our database. Thus, ca. 8% or 340 of the E. coli genes should be found in unrelated species for every 1,000 genes they contain. We have found that this holds true for most of the larger bacterial species. However, as the genome size of the first species drops below ca. 2,000 genes, then the percentage of genes found in the other species increases to twice that frequency for the smallest genomes sequenced to date. That is probably because there is a greater percentage of essential genes in the smaller genomes. Thus, ca. 16% or 90 of the Buchnera sp. genes should be found among every 1,000 genes of other species based upon the size of the genomes alone. Either way it is viewed, from the Buchnera sp. or the E. coli standpoint, these two species share more genes than expected by factors of 1.4- to 2.9-fold (i.e., an actual 13.1% of the E. coli genome versus an expected 4.5% or an actual 99.5% of the Buchnera sp. genome versus an expected 70%), which suggests that they are specifically related. We can obtain some measure of the significance of this comparison by estimating the deviation from the fitted line in the D. radiodurans plot (2.8%, which is about the same as apparent for the E. coli plot and about half of that in the Buchnera sp. plot). This indicates that the greater-than-expected similarity between E. coli and Buchnera sp. is also significantly greater than the average deviation.
Although Snel et al. (61) recognized the effects of genome size on the numbers of shared genes, they nevertheless constructed a gene content tree, apparently without correction for size, that they found to be similar to rRNA trees. We did not construct a tree because the corrections for genome size are not sufficiently precise and because not all species can be specifically related by this measure. That is, some species do not deviate significantly more than the scatter in the plot. However, we were able to determine that eight species of coliforms and gram-negative bacteria are specifically related to one another, that six species of bacilliform and gram-positive bacteria are related, and that the eight species of archaebacteria are also specifically related to one another. Even so, that is hardly sufficient information to build a tree or trees. We have also established that the archaea are in fact just bacteria in terms of their gene content. Eukaryotes are clearly different from both archaebacteria and other bacteria in terms of the numbers of shared orthologs. That is, the eukaryotes have far fewer genes in common with the bacteria or prokaryotes as a whole than expected based upon the size relationship established above. If eukaryotes freely shared genetic information with bacteria to the same extent that bacteria do among themselves, then ca. 50% of the E. coli genome should be present in S. cerevisiae (2,145 genes as opposed to the actual 706 orthologs or one-third of the expected amount). The observation by Olsen et al. (48) that the distinction between eukaryotes and prokaryotes has become blurred as a result of rRNA comparisons is clearly not supported by whole-genome analysis. There is in fact a very distinct separation in terms of gene content.
Although the method we used to normalize the gene content data was also used by Tekaia et al. (65), they came to very different conclusions than we do. This could be because they did not take into account the effect of size of the genomes on the apparent similarity. In addition to normalization of the data to the percentage of the genome present in other species, they performed correspondence analysis and constructed several trees. Unfortunately, their genomic trees changed topology depending upon whether or not Mycoplasma was included. The number of organisms in the data set should have no effect on topology at all, but this is another common problem with evolutionary studies. In addition to the lack of consistency in their trees, the initial apparent relationships they found were not plausible. We know from other studies that E. coli and H. influenzae are specifically related, but they appear on different branches of the first two genomic trees of Tekaia et al. (65). When only the unique genes of each genome were compared (duplications ignored), E. coli and H. influenzae were properly clustered. However, the lack of consistency in the three reported trees is disturbing. It was noted that there was a strong resemblance between the genomic trees and the 16S rRNA trees, but one has to ask which RNA trees because it is also true that the various published rRNA trees lack consistency and change topology as new species are added to existing trees or as the order of addition is changed.
Another approach to whole-genome analysis is the comparison of clusters of orthologous groups of proteins (COGs) employed by Natale et al. (43). Although their focus was on archaebacteria, these authors considered many of the same species as in our study and constructed trees based upon the co-occurrence of COGs or families of proteins rather than on individual genes. These trees showed several improbably close relationships such as between E. coli and B. subtilis or between H. influenzae and H. pylori which these authors recognized as anomalous but explained that it was due to a mixed reflection of phylogenetic relationships and similarities in gene repertoires related to the lifestyles of the organisms. However, those are largely the same thing; one determines phylogenetic relationships based upon similarities in gene content which are related to lifestyles. If all bacteria had the same lifestyle, they would largely have the same gene content and vice versa. Natale et al. (43) normalized their data by dividing the identities by the number of unique COGs but failed to account for the effect of genome size, which is basically the same problem as with the Snel et al. (61) and Tekaia et al. (65) analyses. Natale et al. also failed to explain how the bacteria acquired similar lifestyles, by common descent, by gene transfer, or by a combination of the two. Thus, we believe that gene content is a mixed reflection of phylogenetic relationships and gene transfer.
Yet another approach to whole-genome comparison is through determination of the “genomic signature” (22). Evolutionarily isolated species, e.g., the archaebacteria, are expected to contain a number of unique genes. However, genomic signature analysis presupposes that one already knows how the organisms are related before the analysis is performed. It is therefore not entirely objective, which is the same problem as with 16S rRNA signature analysis (73). It is possible to find this type of “signature” for any grouping of organisms one wishes to emphasize. It can be useful for designing oligonucleotide probes for ecological studies, but it has little or no evolutionary value. Nevertheless, Graham et al. (22) found 351 clusters of 1,149 genes that were present in at least two of six species of archaebacteria but not in other kinds of bacteria or eukaryotes. These authors apparently found fewer clusters in all nine species considered in their study. Our analysis shows that there are 490 orthologs involving 1,694 genes which are specific to two or more of eight archaebacterial genomes. At least 22 orthologs are specifically conserved in all eight species of archaebacteria but not in other species. This is to be contrasted with the 45 orthologs that are conserved in all 37 species considered in our study, including the archaebacteria and eukaryotes. Characteristics previously considered to be unique to archaebacteria are the enzymes and cofactors necessary for methanogenesis, which Graham et al. (22) included in their study but which are also found in the aerobic methylotrophic bacteria that carry out the reverse reaction, the oxidation of methane to CO2 (10). Another such characteristic is the presence of ether-linked lipids which we now know are also found in Aquifex aeolicus and T. maritima and are fairly common in nature (56). For example, they are even present in the mesophilic sulfate-reducing bacteria, Desulfosarcina and Desulforhabdus spp. (55). Their presence in archaebacteria is probably related to the need to maintain the integrity of membranes at hyperthermophilic growth temperatures. We have in fact found distant relationships between the Aquifex aeolicus and T. maritima genomes and with the archaebacteria (also observed by Nelson et al. [44]) that may in part account for the shared presence of ether-linked lipids. There is also evidence that some of the other supposedly unique coenzymes of archaebacteria such as 2-mercaptoethanesulfonate are present in other kinds of bacteria (35). The cell walls of archaebacteria are supposed to be unique but, in fact, the single-constituent glycoprotein cell wall is common in the more complex cell walls of other bacteria (59). For example, gram-positive and gram-negative bacteria contain peptidoglycan as well as glycoprotein, and gram-negative bacteria contain lipopolysaccharide in addition to peptidoglycan and glycoprotein. Thus, the supposedly unique biological characteristics of archaebacteria are in fact not unique when examined in more detail.
There have been a number of studies with single genes and proteins other than 16S rRNA (7). It is likely that they give conflicting results partly because of the problem of paralogy and partly due to the inherent imprecision in current alignment techniques. Alignment is one of the most important but neglected variables in sequence comparisons. In fact, we believe that alignment or more precisely misalignment is one of the main causes for the lack of consistency in previously published trees. That is, variations caused by approximations in the choice of penalty for gaps and mismatches in alignment matrices is likely to be one reason that the topology of trees change when additional species are added or when the order of addition is altered. Furthermore, we cannot completely eliminate paralogous genes, even for 16S rRNA, for which it is assumed to be rarely, if ever, transferred between species (72). At least one instance of gene transfer of 16S rRNA has in fact been observed (75). Furthermore, there are two disparate rRNA operons in Haloarcula marismortui (13) and more are likely to become apparent as the number of whole-genome sequences increases.
Yet another problem is that all proteins and rRNA should have a limit to change that is determined by the structure-function relationship (42). As two species diverge, their proteins should asymptotically approach such a limit to change. At the limit, divergence is equally balanced by convergent mutations (the sum of back and parallel mutations), resulting in a steady state that is characteristic for each protein having a well-defined function. Changes in the steady state can result from duplications and alterations in function which we recognize as one form of paralogy. Practically speaking, trees cannot be constructed for data near the limit of change for a given gene. The limit of change for genes and proteins has to be empirically determined but, to date, has only been established for some cytochromes (42). It is expected that as the numbers of different genes compared increases, then limits to change will become generally recognized and errors due to paralogy and misalignment should be minimized. We cannot say how many genes are necessary, but the number of orthologs found in whole-genome comparisons should be more than sufficient. When we plotted the frequency of percent identity of orthologs in pairs of species, we found a more or less normal distribution for most comparisons. Only the most similar and most divergent species were markedly skewed. The mean of the distribution for all orthologs in a species pair provides a measure of how closely related the species are. When the frequencies of the means for all species pairs were plotted, they, too, could be fit by a normal distribution. We postulate that the distribution of the mean more or less defines the average limit to change for all orthologous proteins as defined in this study. This is ca. 37% identity, which is significantly greater than the 5% expected from amino acid composition alone but very similar to the 36% median identity determined by Nolling et al. (47) for a smaller number of species than was considered here. Only species comparisons significantly outside the normal distribution permit reasonable inferences about specific relationships, which we have taken to be two or more standard deviations above the mean or 40% identity. We found relatively few such exceptional comparisons. However, they are consistent with the analysis we performed on gene content. Our study is also in agreement with that of Nolling et al. (47), which was published after we completed our analysis and had no influence on the outcome. Our results strongly suggest that all-encompassing evolutionary trees cannot or should not be constructed for bacterial comparisons that approach such a limit to change. Gene content does not have the same limitations on its use and that may be why some specific relationships were observed from gene content but not indicated by mean identity of orthologs. Nevertheless, using our approach to whole-genome analysis, considering both gene content and mean identity in orthologous genes, we have found specific relationships among the majority of the 37 species considered in this study.
Acknowledgments
This work was supported in part by grant GM-21277 from the National Institutes of Health.
REFERENCES
- 1.Ainscough, R., et al. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012-2018. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. M. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 4.Bansal, A. K. 1999. An automated comparative analysis of 17 complete microbial genomes. Bioinformatics 15:900-908. [DOI] [PubMed] [Google Scholar]
- 5.Bansal, A. K., P. Bork, and P. J. Stuckey. 1998. Automated pair-wise comparisons of microbial genomes. Math. Model. Sci. Comput. 9:1-23. [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 7.Bocchetta, M., S. Gribaldo, A. Sananelantoni, and P. Cammarano. 2000. Phylogenetic depth of the bacterial genera Aquifex and Thermotoga inferred from analysis of ribosomal protein, elongation factor, and RNA polymerase subunit sequences. J. Mol. Evol. 50:366-380. [DOI] [PubMed] [Google Scholar]
- 8.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenback, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, et al. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 10.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olsen, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 13.Dennis, P. P., S. Ziesche, and S. Mylvaganam. 1998. Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 180:4804-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doolittle, W. F. 2000. Uprooting the tree of life. Sci. Am. 2:90-95. [DOI] [PubMed] [Google Scholar]
- 15.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 16.Fitz-Gibbon, S. T., and C. H. House. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27:4218-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 18.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 19.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 20.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochaete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 21.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, E. Y. Chen, and G. H. Cassell. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 22.Graham, D. E., R. Overbeek, G. J. Olsen, and C. R. Woese. 2000. An archaeal genomic signature. Proc. Natl. Acad. Sci. USA 97:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta, R. S. 1998. What are archaebacteria: life's third domain or monoderm prokaryotes related to gram-positive bacteria? A new proposal for the classification of prokaryotic organisms. Mol. Microbiol. 29:695-707. [DOI] [PubMed] [Google Scholar]
- 24.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henikoff, S., E. A. Greene, S. Pietrokovski, P. Bork, T. K. Attwood, and L. Hood. 1997. Gene families: the taxonomy of protein paralogs and chimeras. Science 278:609-614. [DOI] [PubMed] [Google Scholar]
- 26.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huynen, M. A., and P. Bork. 1998. Measuring genome evolution. Proc. Natl. Acad. Sci. USA 95:5849-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and Chlamydia trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 29.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, et al. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 30.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 31.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, et al. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83-101. [DOI] [PubMed] [Google Scholar]
- 32.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, et al. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 33.Koonin, E. V., A. R. Mushegian, M. Y. Galperin, and D. R. Walker. 1997. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol. Microbiol. 25:619-637. [DOI] [PubMed] [Google Scholar]
- 34.Koonin, E. V., L. Aravind, and A. S. Kondrashov. 2000. The impact of comparative genomics on our understanding of evolution. Cell 101:573-576. [DOI] [PubMed] [Google Scholar]
- 35.Krum, J. G., and S. A. Ensign. 2001. Evidence that a linear megaplasmid encodes enzymes of aliphatic alkene and epoxide metabolism and coenzyme M (2-mercaptoethanesulfonate) biosynthesis in Xanthobacter strain Py2. J. Bacteriol. 183:2172-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed]
- 37.Kurland, C. G. 2000. Something for everyone. Horizontal gene transfer in evolution. EMBO Rep. 1:92-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecompte, O., R. Ripp, V. Puzos-Barbe, S. Duprat, R. Heilig, J. Dietrich, J. C. Thierry, and O. Poch. 2001. Genome evolution at the genus level: comparison to three complete genomes of hyperthermophilic archaea. Genome Res. 11:981-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and B. Kapur. 2001. Complete genomic sequence of Pasteurella multocida Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayr, E. 1998. Two empires or three? Proc. Natl. Acad. Sci. USA 95:9720-9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mewes, H. W., K. Albermann, M. Baehr, D. Frishman, A. Gleissner, J. Hani, K. Heumann, K. Kleine, A. Maierl, S. G. Oliver, F. Pfeiffer, and A. Zollner. 1997. Overview of the yeast genome. Nature 387:7-65. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, T. E., M. A. Cusanovich, and M. D. Kamen. 1986. Evidence against use of bacterial amino acid sequence data for construction of all-inclusive phylogenetic trees. Proc. Natl. Acad. Sci. USA 83:217-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Natale, D. A., U. T. Shankavaram, M. Y. Galperin, Y. I. Wolf, L. Aravind, and E. V. Koonin. 2000. Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs). Genome Biol. 1:9.1-9.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, et al. 1999. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed]
- 45.Nelson, K. E., I. T. Paulsen, J. F. Heidelberg, and C. M. Fraser. 2000. Status of genome projects for nonpathogenic bacteria and archaea. Nat. Biotechnol. 18:1049-1054. [DOI] [PubMed] [Google Scholar]
- 46.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, et al. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olsen, G. J., C. R. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 176:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papadimitrou, C. H., and K. Steiglitz. 1982. Combinatorial optimization: algorithm and complexity. Prentice Hall, New York, N.Y.
- 50.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 51.Pennisi, E. 1998. Genome data shake tree of life. Science 280:672-674. [DOI] [PubMed] [Google Scholar]
- 52.Pennisi, E. 1999. Is it time to uproot the tree of life? Science 284:1305-1307. [DOI] [PubMed] [Google Scholar]
- 53.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-532. [DOI] [PubMed] [Google Scholar]
- 54.Ruepp, A., W. Graml, M. L. Santos-Martinez, K. K. Koretke, C. Volker, H. W. Mewes, D. Frishman, S. Stocker, A. N. Lupas, and W. Baumeister. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508-513. [DOI] [PubMed] [Google Scholar]
- 55.Rütters, H., H. Sass, H. Cypionka, and J. Rullkötter. 2001. Monoalkylether phospholipids in the sulfate-reducing bacteria Desulfosarcina variabilis and Desulforhabdus amnigenus. Arch. Microbiol. 176:435-442. [DOI] [PubMed] [Google Scholar]
- 56.Schouten, S., E. C. Hopmans, R. D. Pancost, and J. S. Sinninghe-Damste. 2000. Widespread occurrence of structurally diverse tetraether membrane lipids: evidence for the ubiquitous presence of low-temperature relatives of hyperthermophiles. Proc. Natl. Acad. Sci. USA 97:14421-14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 58.Simpson, A. J. G., et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-159. [DOI] [PubMed] [Google Scholar]
- 59.Sleytr, U. B. 1997. Basic and applied s-layer research: an overview. FEMS Microbiol. Rev. 20:5-12. [Google Scholar]
- 60.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snel, B., P. Bork, and M. A. Huynen. 1999. Genome phylogeny based on gene content. Nat. Genet. 21:108-110. [DOI] [PubMed] [Google Scholar]
- 62.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 63.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 64.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tekaia, F., A. Lazcano, and B. Dujon. 1999. The genomic tree as revealed from whole proteome comparisons. Genome Res. 9:550-557. [PMC free article] [PubMed] [Google Scholar]
- 66.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, et al. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1820. [DOI] [PubMed] [Google Scholar]
- 67.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed]
- 68.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 69.Waterman, M. S. 1984. General methods for sequence comparison. Bull. Math. Biol. 46:473-500. [Google Scholar]
- 70.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woese, C. R. 1998. The universal ancestor. Proc. Natl. Acad. Sci. USA 95:6854-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Woese, C. R., J. Maniloff, and L. B. Zablen. 1980. Phylogenetic analysis of the mycoplasmas. Proc. Natl. Acad. Sci. USA 77:494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yap, W. H., Z. Zhang, and Y. I. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zuckerkandl, E., and L. Pauling. 1965. Molecules as documents of evolutionary history. J. Theor. Biol. 8:357-366. [DOI] [PubMed] [Google Scholar]