Abstract
The minimal replication region of the mycobacterial plasmid pAL5000 encompasses the replication origin (ori) and two tandemly organized replication genes, repA and repB, the functions of which are not clearly known. It was observed that when the repA and repB genes were expressed in Escherichia coli, a strong ori binding activity was generated in the host cells. Inactivation of repB led to a complete loss of activity, whereas inactivation of repA had a partial effect, indicating that while repB plays an important role in the process, its activity is stimulated through coexpression of repA. However, this stimulatory effect could be demonstrated only when expression of repA and that of repB were coupled. At a relatively high concentration (1,000 nM), the purified RepB protein was found to form an ori complex with low specificity, which was sensitive to high salt concentrations and challenge by a nonspecific competitor. In contrast, the complex formed by an extract of repA-repB-expressing cells was highly specific and was resistant to both types of challenges. At a 10-fold-lower concentration, RepB did not exhibit ori binding activity, but it could nevertheless form a salt-resistant ori complex in vitro, provided that host factors were present. Antibody supershift experiments indicated that RepB is a key component of the specific complex formed by extracts prepared from E. coli cells expressing the repA and repB genes and also from mycobacterial cells harboring pAL5000-derived vectors. The results indicate that in vivo RepB interacts with host factors and forms an ori complex, but this activity is maximal only when there is coupled expression of repA.
The reemergence of mycobacterial diseases, particularly tuberculosis, is a matter of great public health concern (2, 12). In recent years, significant efforts have been made to understand the molecular biology of Mycobacterium tuberculosis, with the objective of developing newer methods for prevention and cure of the disease (9, 10). The availability of Escherichia coli-mycobacterium shuttle vectors based on the Mycobacterium fortuitum plasmid pAL5000 (16) has been very useful in this context (6, 7, 18). Although these vectors have been used often, the mechanisms of their replication and stability are not known in detail, which limits their usage.
Plasmid pAL5000 is a 4.8-kb plasmid (16), and it is believed that a 1.8-kb fragment spanning the two open reading frames, ORF1 and ORF2, which overlap by one nucleotide (Fig. 1), is necessary and sufficient for replication (14, 18). This region encompasses a 600-bp cis-acting region which has been shown to function not only as a replication origin but also as a divergent promoter element, possibly controlling expression of flanking genes, which include ORF1 and ORF2 (19, 21). ORF1 and ORF2 can potentially produce two polypeptides (RepA and RepB, respectively), but there is no direct evidence indicating that these polypeptides are expressed in mycobacterial cells harboring pAL5000 vectors. Indirect attempts to assign specific functions to these polypeptides have led to identification of homologs in the database. Although several homologies have been discovered (14, 15), the functions of these polypeptides remain undefined due to a lack of experimental support. Nevertheless, it appears that pAL5000 and possibly several other bacterial plasmids of mycobacterial origin (1, 15) have common replication mechanisms involving evolutionarily conserved replication proteins (14).
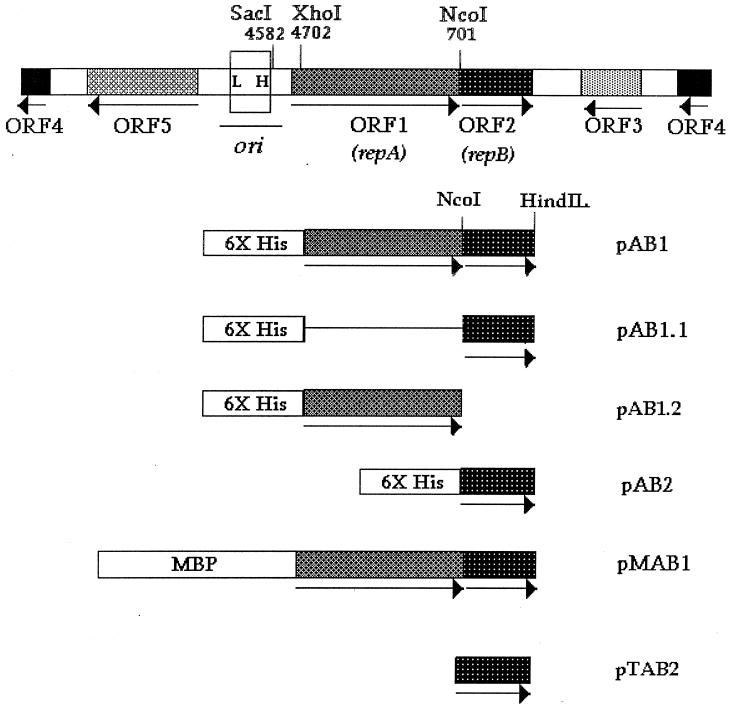
FIG. 1.
Linear map of pAL5000 (4,837 bp), showing the locations of the open reading frames (ORF1 to ORF5), the origin of replication (ori), and the relevant restriction sites. The locations of the low-affinity (L) and high-affinity (H) binding sites of RepB described previously (20) are indicated. The region enclosed in a box (200 bp) was used as a probe in EMSA. The in-frame fusions were fusions either with the six-histidine tag (6X His) or with MBP. In pTAB2 the expressed protein lacks any extra amino acid residues at the N terminus.
In a previous study Stolt and Stoker expressed orf2 (repB) in E. coli and demonstrated binding of the RepB polypeptide to the plasmid ori (20). Similarly, the same authors also expressed the orf1 (repA) region, but no ori binding activity could be associated with the RepA polypeptide. Although the ORF1 and ORF2 polypeptides have been designated replication proteins A and B (RepA and RepB, respectively) (19, 20), their role in the replication process is not clearly understood.
In this study, we examined the role played by RepA and RepB in the formation of an ori complex. The results presented here suggest that while RepB plays a central role in the formation of an ori complex, its function is dependent on host factors and on coupled expression of repA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli XL1 Blue was used for routine manipulation of plasmid DNA (17). The same strain was used for expression of proteins fused to maltose-binding protein (MBP). For expression of fusion proteins tagged with six histidine residues, E. coli strain SG13009(pREP4) obtained from Qiagen Inc. (Valencia, Calif.) and E. coli XL1 Blue were used. To express proteins from the vector pT7-7 (22), E. coli BL21(DE3) was used (17). E. coli cells were grown with vigorous aeration at 37°C in Luria broth (17) supplemented with kanamycin (25 μg/ml) and/or ampicillin (100 μg/ml) as appropriate. Mycobacterium smegmatis LR222 harboring plasmid pMC2 (4) was grown in 2XYT medium supplemented with 0.5% glycerol and 0.2% Tween 80 in the presence of kanamycin (50 μg/ml) as described previously (6, 7).
Construction of expression plasmids.
The various fusion constructs used in this study are shown schematically in Fig. 1, and their important features are summarized in Table 1. The RepA expression plasmid pAB1 was constructed by cloning a 1.4-kb XhoI-HindIII fragment from pMC2, containing the repA-repB region, into the SalI-HindIII sites of pQE31 (Qiagen) (Fig. 1). In this construct the repA gene was fused in frame with the six-His tag. In another construct (pAB1.1), the same fragment was inserted into the SalI-HindIII site of pQE32. This construct was similar to pAB1 except that the repA fusion was not in frame. Construct pAB1.2 was also derived from pAB1. In this case the repB sequence was deleted by removing the NcoI-HindIII repB fragment from pMC2 and replacing it with a double-stranded oligonucleotide in order to regenerate the C terminus of RepA. The RepA expression plasmid pMAB1 (RepA expressed as an MBP fusion protein) was constructed first, by cloning a 1.5-kb SacI-PstI fragment from pMC2 into the SacI-PstI sites of pUC19, giving rise to pMC2a. This vector was partially digested with EcoRI and PstI, and a 1.5-kb fragment encompassing the repA-repB region was purified and subcloned into the EcoRI-PstI sites of pMAL-c2 (New England Biolabs, Beverly, Mass.) in order to obtain an in-frame fusion. The RepB expression plasmids pAB2 and pTAB2 were constructed by inserting a PCR fragment containing only the repB region into the BamHI-PstI sites of pQE30 (Qiagen) and the NdeI-PstI sites of pT7.7 (22), respectively. The primers used were S1 (5′-TTCGGATCCCATATGAGCGACGGCTACAGC-3′) (the BamHI and NdeI sites are underlined) derived from the junction of the repA-repB regions and the universal reverse primer for plasmid pUC19 (5′-AGCGGATAACAATTTCACACAGG-3′). Plasmid pMC2 was used as the template in PCR. The PCR product was digested with BamHI or NdeI and PstI for cloning in the appropriate vector. The vector pAB2a was constructed from pAB2 by excising an XhoI-XbaI fragment containing the repB expression sequence and ligating it with a SalI-XbaI fragment from pSD4 (7) containing the p15A origin of replication.
TABLE 1.
Vectors used in this study
| Plasmid(s) | Description |
|---|---|
| pMC2 | Contains the 2.56-kb EcoRV-HpaI fragment (nucleotides 3895 to 1257) derived from the pAL5000 replication region |
| pQE30, -31, -32 | Expression vectors for E. coli; polypeptides expressed from these vectors contain an affinity tag of six histidine residues fused to the N terminus |
| pMALc2 | Expression vector for E. coli; polypeptides expressed from this vector contain MBP as the affinity tag |
| pT7.7 | T7 promoter-based expression systema |
| pAB1 | repA and repB genes expressed from pQE31; repA is fused in frame with the polyhistidine tag |
| pAB1.1 | Similar to pAB1, but repA is fused out of frame with the polyhistidine tag; does not express repA |
| pAB1.2 | repB region deleted from pAB1 |
| pAB2 | repB gene expressed from pQE30 |
| pAB2a | Same as pAB2, but carrying a p15A origin of replication instead of the usual ColE1 origin; vector pTAB2a is compatible with pAB1.2 in E. coli |
| pTAB2 | RepB expressed as untagged protein from vector pT7.7 |
| pMAB1 | repA and repB genes expressed from pMA1-c2; repA is fused in frame with MBP |
See reference 22.
Preparation of bacterial cell extract.
Bacterial cell extracts were prepared from E. coli XLI Blue and SG13009(pREP4) cells transformed with various constructs and from M. smegmatis LR222 harboring pMC2. E. coli cells were grown in Luria broth, and protein expression was induced by adding isopropyl-β-d-thiogalactoside (IPTG) at a concentration of 1 mM and incubating the preparation for 4 h at 37°C. Mycobacterial cells were grown to the late log phase. In both cases, cells from a 200-ml culture were harvested by centrifugation at 8,000 × g for 15 min at 4°C, washed with 0.9% NaCl, and stored frozen at −80°C overnight. Frozen cells were thawed in lysis buffer (20 mM Tris-HCl [pH 8], 100 mM NaCl, 1 mM EDTA, 5% glycerol) in ice, sonicated, and clarified by centrifugation at 12,000 × g for 30 min at 4°C. The clarified supernatants obtained in this way were referred to as extracts.
Purification of recombinant RepA and RepB proteins.
Recombinant RepA and RepB polypeptides were overproduced in E. coli. The cells were grown at 37°C in 200 ml of Luria broth containing the appropriate antibiotic to an optical density at 600 nm of 0.5. Expression was induced by adding 1 mM IPTG, and the organisms were allowed to grow for 4 h. RepA and RepB were purified either on an Ni-nitrilotriacetic acid resin column (Qiagen) or on an amylose resin column (New England Biolabs), depending on whether the protein was tagged with six His residues or MBP. Untagged RepB was purified by cation-exchange chromatography with a fast protein liquid chromatograph using a Mono S column (Amersham Pharmacia Biotech). About 5 mg of extract was loaded onto the column in buffer A (50 mM phosphate buffer [pH 7.0], 0.01 mM dithiothreitol, 0.1 mM EDTA) containing 50 mM KCl. Fractions (0.5 ml) were eluted with a 60-ml 200 to 400 mM KCl gradient in buffer A at a flow rate of 1 ml/min and were assayed for binding activity. The RepB protein eluted at about 240 to 300 mM KCl. The purity of the protein was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis followed by Coomassie blue staining.
Immunological techniques.
For immunization of rabbits the affinity-purified six-His-tagged RepB polypeptide was subjected to SDS-PAGE. After mild staining of the gel, the appropriate band was excised, crushed, and injected into rabbits. Antiserum was raised by using a standard protocol (8). About 10 μg of the extracted protein was loaded into each lane of a minigel (Bio-Rad Mini-PROTEAN II). Western blotting was performed by using this antiserum at a dilution of 1:5,000. In the case of anti-RepB antiserum, the lower limit of detection of RepB antigen was about 100 ng per lane. To detect expression of His fusions, a commercially available mouse antibody (QIAexpress detection reagent; Qiagen) targeted against the Arg-Gly-Ser-His epitope present at the N terminus of the six-His-tagged protein was used.
Electrophoresis techniques.
For the electrophoretic mobility shift assay (EMSA), a 200-bp DNA fragment derived from the pAL5000 origin of replication (nucleotides 4459 to 4663) encompassing the high- and low-affinity binding sites for RepB described by Stolt and Stoker (20) was PCR amplified by using primers S6 (5′-GGATCCTGGTTGGTACAGGTGGTTGGG-3′) and S7 (5′-GCTGCTCAAATTCGTCGGCG-3′). Ten picomoles of S7 was 5′ end labeled with [γ-32P]ATP (BRIT, Bombay, India) and T4 polynucleotide kinase (New England Biolabs) and directly used in PCR. The PCR product was purified using a Qiagen column. Unless indicated otherwise, the binding reaction mixtures (final volume, 30 μl) contained 1 μg of protein extract, 3 μl of 10× binding buffer (100 mM Tris [pH 8], 600 mM NaCl, 30 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 20% glycerol), 1 μg of salmon sperm DNA, and 10,000 cpm of labeled DNA. The reaction mixtures were preincubated for 10 min and then for an additional 20 min on ice after the probe was added. The DNA-protein complexes were separated on a 4% native polyacrylamide gel by electrophoresis in 0.5× Tris-borate buffer (50 mM Tris-borate, 1 mM EDTA) at 200 V for 3 to 5 h at 4°C. Following electrophoresis, the gel was vacuum dried, and the bands were visualized by autoradiography. For the competition EMSA, the unlabeled 200-bp ori fragment was used. In some cases nonspecific DNA, either salmon sperm DNA or poly(dI-dC) (Amersham Pharmacia Biotech), was used. Antibody supershift assays (13) were performed by adding 1 μl of anti-RepB serum to the binding reaction mixture. For each experiment the corresponding preimmune serum was used as a control. Two independently derived anti-RepB antisera and their preimmune counterparts were used.
RESULTS
DNA binding activities in E. coli cells expressing the repA-repB region.
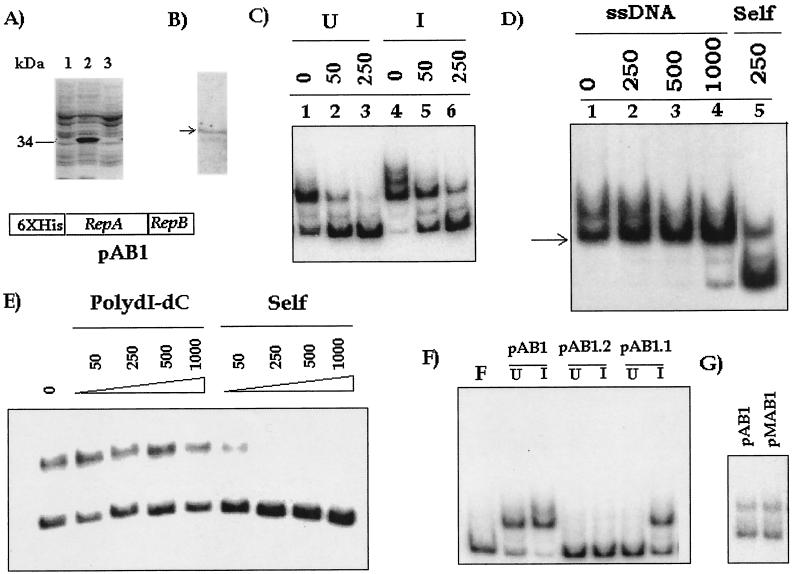
The recombinant plasmid pAB1 (Fig. 1) was transformed into E. coli, and expression was monitored by SDS-PAGE. Since repA was fused in the zero frame to the six-His tag, we expected that following induction with IPTG high levels of a 34-kDa polypeptide would be produced. SDS-PAGE analysis (Fig. 2A, lane 2) confirmed that such a protein was present, but the protein was not efficiently solubilized (Fig. 2A, lane 3). Trace amounts were detected in the soluble form, as indicated by Western blotting with anti-RGS-His monoclonal antibody (Fig. 2B). The repB region was also expressed in these cells at a low level (Fig. 3A). The soluble fraction was tested for the presence of any ori binding activity by using the 200-bp origin sequence as the probe. DNA binding activity was observed in the extract even without induction with IPTG (Fig. 2C, lane 1). Induction with IPTG, however, increased the extent of retardation (Fig. 2C, lane 4). The complexes were outcompeted by a self-competitor, indicating that they were specific (Fig. 2C, lanes 2, 3, 5, and 6). The specificity of the retarded complex was also demonstrated by a competition EMSA performed with nonspecific competitors, salmon sperm DNA and poly(dI-dC). When salmon sperm DNA was used, we observed that the lower retarded complex was stable to challenge with high doses of this nonspecific competitor. The complex was not eliminated when 1 μg of the nonspecific competitor was used, whereas 250 ng of the self-competitor almost completely eliminated complex formation (Fig. 2D). This complex was also resistant to challenge with the other nonspecific competitor, poly(dI-dC) (Fig. 2E). These results indicated that the major complex formed by the extract was specific.
FIG. 2.
Origin binding activity in extracts of pAB1-expressing cells. (A) SDS-PAGE analysis of SDS lysates from uninduced cells (lane 1) and IPTG-induced cells (lane2). Lane 3, SDS-PAGE analysis of extract. The position of the band produced by the overexpressed 34-kDa fusion protein is indicated. (B) Western blot of lane 3 in panel A, obtained by using anti-RGS-His antibody. The position of the cross-reactive 34-kDa band is indicated by an arrow. (C) EMSA with 1 μg of pAB1 extract (uninduced [U] and induced [I]) and competition with no competitor (lane 1) or with 50 or 250 ng of self-competitor (lanes 2, 3, 5, and 6). The probe used was the end-labeled 200-bp amplicon derived from the origin of replication of plasmid pAL5000. (D) Competition EMSA with 1 μg of pAB1 extract in the absence (lane 1) or in the presence of either salmon sperm DNA (ssDNA) (250 to 1,000 ng) (lanes 2 to 4) or ori DNA (250 ng) (lane 5). The position of the most stable complex is indicated by an arrow. (E) Competition EMSA with no competitor or with 50 to 1,000 ng of poly(dI-dC) or 50 to 1,000 ng of self-competitor. A smaller amount of extract (0.2 μg) was used in this experiment to ensure that only the major complex was formed. (F) EMSA with uninduced (U) and IPTG-induced (I) extracts of pAB1 and mutants pAB1.2 and pAB1.1. Lane F, free probe. (G) Comparison of the sizes of complexes formed by pAB1 and pMAB1 extracts. Complex formation was kept at a low level so that an accurate comparison could be made.
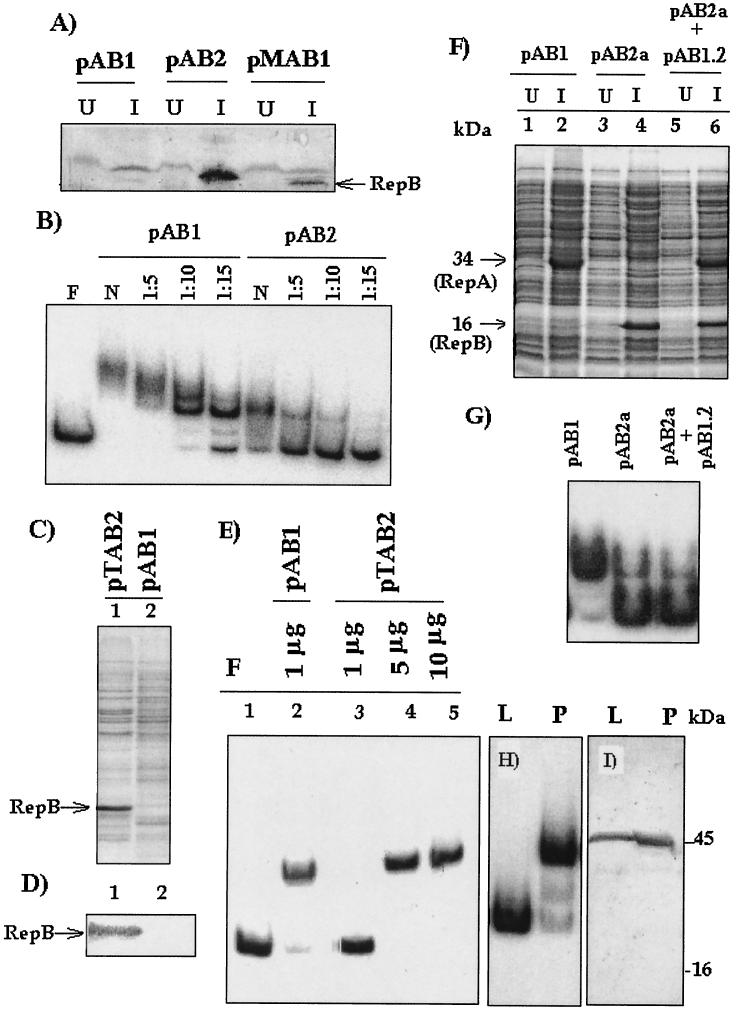
FIG. 3.
Origin binding activities in extracts of pAB1-expressing cells, pAB2-expressing cells, pTAB2-expressing cells, and M. smegmatis pMC2-harboring cells. (A) Detection and estimation of the RepB polypeptide in pAB1, pAB2, and pMAB1 extracts. Identical amounts of extracts (10 μg) were subjected to Western blotting with anti-RepB antiserum. U, extract from uninduced cells; I, extract from IPTG-induced cells. The position of the RepB-specific band is indicated by an arrow. The relative intensities of the anti-RepB cross-reactive bands were taken into account when differences in binding efficiency between extracts of pAB1- and pAB2-expressing cells were calculated. (B) Relative ori binding efficienciesof extracts from pAB1- and pAB2-expressing cells when the labeled 200-bp origin-derived fragment was used as the probe. One microgram of extract was used for EMSA either neat (N) or after dilution. Lane F contained free probe. (C and D) SDS-PAGE analysis of pTAB2 and pAB1 extracts (10 μg each) (C), followed by Western blotting with anti-RepB antiserum (D). (E) ori binding activity in pAB1 and pTAB2 extracts using the amounts indicated. F, free probe. (F and G) In vivo complementation test for RepA and RepB. (F) SDS-PAGE analysis of SDS lysates of pAB1-, pAB2a-, and pAB2a/pAB1.2-containing cells before and after induction. The positions of the RepA- and RepB-specific bands are indicated. (G) ori binding with the induced extracts. (H) ori binding activity in M. smegmatis LR222 cells that were either vector free (lane L) or harbored pMC2 (lane P). In each case 1 μg of extract was used. (I) Western blotting with anti-RepB antiserum. The lanes each contained 10 μg of extract from M. smegmatis cells that were either vector free (lane L) or harbored pMC2 (lane P).
In contrast to pAB1, the repB deletion derivative pAB1.2 did not give rise to any activity (Fig. 2F), indicating that expression of repB was absolutely essential for binding to occur. In the case of IPTG-induced pAB1.1 (mutant with inactivated repA) extract, the activity was diminished, and no activity was observed in the uninduced extract (Fig. 2F). An alternative fusion construct (pMAB1) was made with MBP. The pMAB1 extract exhibited activity similar to that of the pAB1 extract (data not shown). When the sizes of the complexes formed by the pMAB1 and pAB1 extracts at subsaturation levels were compared (Fig. 2G), no difference was observed, although the MBP fusion with RepA (80 kDa) was substantially larger than the His fusion (34 kDa). This indicates that most likely RepA is not part of the final complex.
DNA binding activities in repB-expressing cells.
To investigate the contribution made by RepB to generation of ori binding activity in pAB1-expressing cells, this protein was expressed as a six-His fusion protein in the vector pQE30. Expression from pAB2 could be induced with IPTG, but the expressed product was poorly solubilized, as described above. Antiserum was raised against six-His-tagged RepB isolated from the insoluble fraction, and this antiserum was used to estimate the amount of RepB in pAB1-expressing cells, as well as in pAB2-expressing cells. We found that RepB was also present in pAB1- and pMAB1-expressing cells (Fig. 3A), but the level of this protein in the cells was about 5 to 10 times less than the level in pAB2-expressing cells. However, no RepB could be detected in the uninduced cells. When the levels of ori binding activity were compared, it was found that pAB1-expressing cells had about 30-fold more binding activity than pAB2-expressing cells (Fig. 3B). If the values were normalized with respect to the level of RepB, the activity in repA-repB (pAB1) extracts was about 150 times greater than the activity in RepB-expressing cells. Similar observations were obtained with untagged RepB-expressing cells (pTAB2) in which RepB was effectively solubilized (Fig. 3C, lane 1). In this case, even though RepB could not be detected either by staining or by Western blotting (Fig. 3C and D, lanes 2), the activity in the pAB1 extract could be detected with 1 μg of protein, compared to 5 μg of protein in the case of pTAB2 (Fig. 3E, lanes 2 and 4). This shows that to form an equivalent complex, repA-repB-expressing cells require much less RepB than cells expressing RepB alone require.
To investigate this phenomenon further, we attempted to find out whether uncoupled expression of repA and expression of repB from two compatible plasmids could complement each other in trans. Plasmids pAB1.2 (the construct containing only repA) and pAB2a (the compatible construct containing only repB) were cotransformed into E. coli XL1 Blue cells. Induction resulted in uncoupled synthesis of both RepA and RepB in the cotransformed cells (Fig. 3F, lane 6). When pAB1, pAB2a, and pAB2a/pAB1.2 extracts were tested, the activity in the pAB2a/pAB1.2 extracts was same as the activity in the pAB2a extracts, and no apparent stimulation in trans was observed (Fig. 3G). This result indicates that high-level activity could be produced only if RepA synthesis and RepB synthesis were coupled, as in the case of pAB1.
A binding activity similar to that of pAB1-expressing cells was observed with extracts of M. smegmatis LR222 harboring the pAL5000-derived vector pMC2 (Fig. 3H, lane P). No activity was observed in the plasmid-free cells (lane L) when an identical amount of extract was used. No RepB-specific band in the expected 16-kDa region was detected by Western blot analysis (Fig. 3I, lane P), indicating that the level of expression must have been below the level of detection. A cross-reactive band at 45 kDa was also present in plasmid-free cells and therefore was nonspecific.
DNA binding activity of purified RepB.
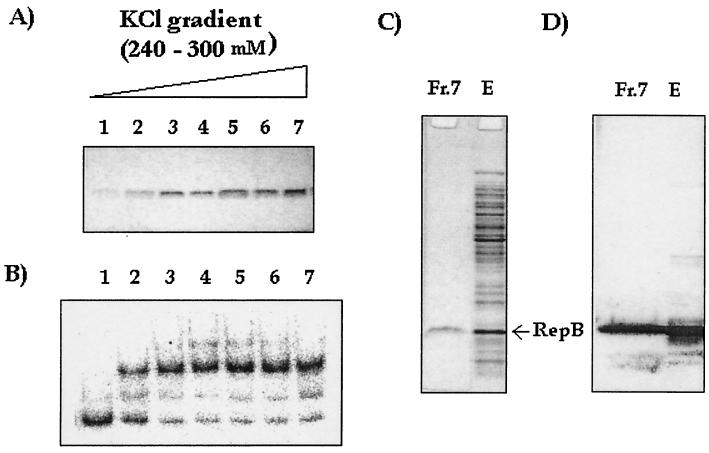
Extracts of pTAB2-expressing cells were fractionated on a Mono S column. The fractions containing the purified protein eluted at 240 to 300 mM KCl (Fig. 4A). Aliquots (20 μl) of these fractions were assayed directly, but no activity was obtained (data not shown); however, when the fractions were dialyzed against the low-salt buffer (50 mM KCl in buffer A) and 20-μl aliquots were tested, activity was detected in fractions that contained approximately 1 μg or more of the protein per 20 μl of sample (Fig. 4B, lanes 2 to 7). Fraction 7, containing RepB protein at the maximum concentration, was further characterized by SDS-PAGE and Western blotting. A single band that had the right mobility and cross-reacted with an anti-RepB serum was observed (Fig. 4C and D).
FIG. 4.
Purification of RepB expressed from pTAB2 with a Mono S column. The active fractions were eluted by using a 240 to 300 mM KCl gradient in buffer A. (A) SDS-PAGE analysis of eluted fractions. (B) Binding activity after dialysis against low-salt buffer (50 mM KCl in buffer A). (C) SDS-PAGE analysis of fraction 7 (lane Fr.7) and the unfractionated extract (lane E). Lane Fr.7 contained 1 μg of fraction 7, and lane E contained 10 μg of the extract. (D) Corresponding Western blot with anti-RepB antiserum.
Differences in the complexes formed by purified RepB and extracts of pAB1-expressing cells.
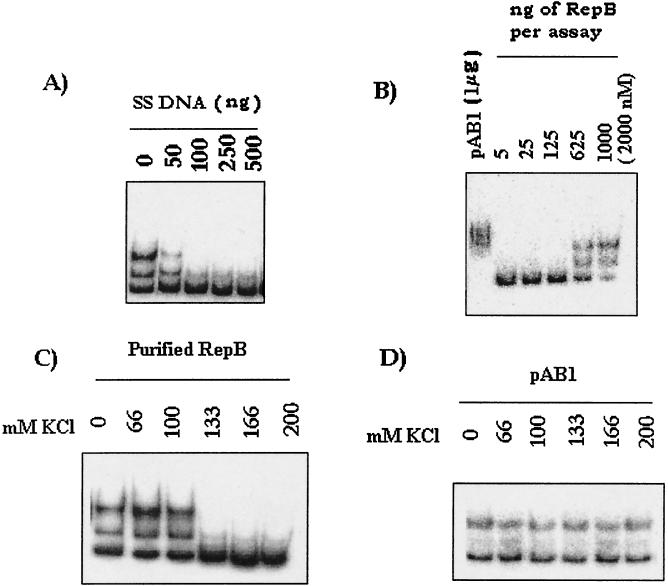
The binding activity observed when purified RepB was used was distinct from that observed with cells expressing repA and repB (RepB) in three respects. First, the complex formed by purified RepB was easily outcompeted by increasing amounts of a nonspecific competitor (salmon sperm DNA) (Fig. 5A), whereas the complex formed by the repA-repB (pAB1) extract was stable in the presence of as much as 1 μg of the same DNA (Fig. 2D). Second, titration of the amount of RepB required for binding revealed that nearly 1 μg of the protein, at a concentration of 2,000 nM, was needed for about 50% binding to occur, whereas 1 μg of a total pAB1 extract with no visible band corresponding to RepB (Fig. 3C) resulted in complete retardation (Fig. 5B). Third, as mentioned above, ion-exchange chromatography revealed that the presence of high salt concentrations in the eluates hampered ori binding of RepB. Therefore, the effect of increasing the concentration of salt on the complex formed directly by RepB was tested. We found that 133 mM KCl completely eliminated formation of the RepB complex (Fig. 5C), whereas the major complex formed by the repA-repB (pAB1) extract was stable in the presence of 200 mM salt (Fig. 5D). These results indicate that the RepB-dependent complex formed by repA-repB-expressing cells was different from the complex formed by the purified protein in vitro.
FIG. 5.
ori binding properties of RepB. (A) Competition EMSA with the 200-bp ori fragment as the probe and different amounts of nonspecific competitor salmon sperm DNA (SS DNA). (B) Determination of the minimal amount of RepB required for binding. Different amounts of RepB (5 to 1,000 ng) were added. The molar concentration of RepB in the assay mixture containing 1,000 ng of RepB is indicated in parentheses. The binding obtained with 1 μg of pAB1 extract is shown for comparison. (C and D) Effects of different concentrations of KCl on complex formation by purified RepB and pAB1 extract. In the case of pAB1 extract the extent of complex formation was deliberately maintained at a low level in order to test rigorously the salt resistance of the complex.
Effect of host factors on ori complex formation by RepB.
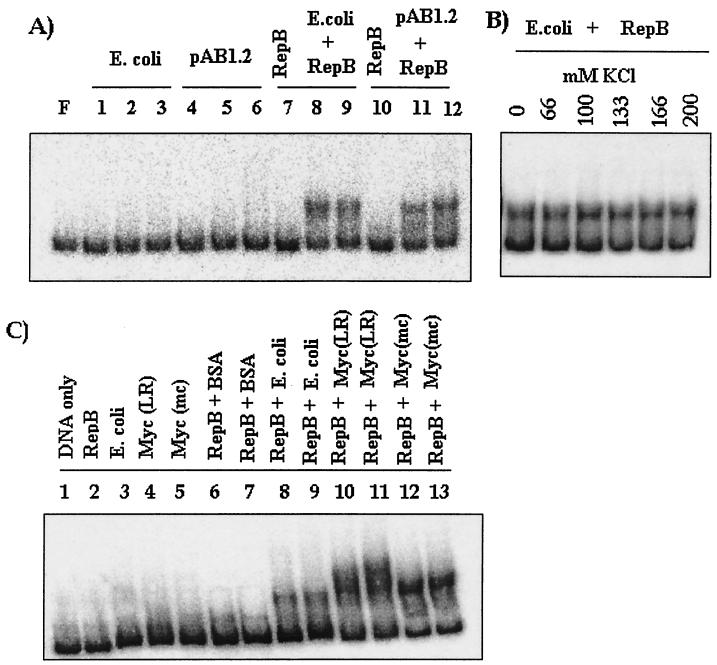
Since the results described above indicate that in vivo the mechanism of action of RepB must be distinct from the direct binding activity observed in vitro, the possibility that host factors play a role in the formation of the complex by RepB was investigated. Purified RepB protein was mixed with various amounts of extracts of either the host (E. coli XL1 Blue) or E. coli cells expressing repA (pAB1.2). Individually, these extracts were inactive (Fig. 6A, lanes 1 to 6). When 125 ng of RepB was used, no binding was observed (Fig. 6A, lanes 7 and 10), but when RepB was used in combination with the extracts (lanes 8, 9, 11, and 12), activity was detected. These results indicate that in the presence of host factors RepB forms the ori complex at concentrations lower than the concentration required for direct binding. However, the presence of repA-expressing cells (pAB1.2) did not result in further enhancement of binding (Fig. 6A, lanes 11 and 12) over the binding obtained with the E. coli extract. This is consistent with the observation made in the in vivo complementation experiment (Fig. 3G), in which repA had an effect only when its expression was coupled to expression of repB, as in the case of pAB1. To examine whether the complex formed in vitro by RepB in the presence of host factors resembles the salt-resistant type of complex characteristic of pAB1-expressing cells, a salt challenge experiment was performed. We found that the complex was salt resistant (Fig. 6B), as in the case of pAB1-expressing cells. Similar complexes were also observed when mycobacterial extracts were used (Fig. 6C, lanes 10 to 13), indicating that such interactions also occur with mycobacterial proteins. These interactions are specific since when a nonspecific protein, such as bovine serum albumin, was used along with RepB, it did not give any binding activity (Fig. 6C, lanes 6 and 7).
FIG. 6.
(A) ori binding activity of RepB with extracts obtained from either E. coli cells or E. coli cells expressing pAB1.2 (RepA). Lanes 1 to 6, ori binding with 10, 20, and 30 μg of extract from either E. coli cells (lanes 1, 2, and 3) or E. coli/pAB1.2 cells (lanes 4, 5, and 6); lanes 7 to 12, ori binding activity of purified RepB (125 ng) either alone (lanes 7 and 10) or in combination with extracts (20 and 30 μg) (lanes 8, 9, 11, and 12). (B) Effect of salt concentration on the complex formed by RepB (125 ng) and E. coli extract (20 μg). (C) ori binding activity in combination with E. coli and mycobacterial extracts. The following protein samples were used for ori binding: lane 2, RepB (125 ng); lane 3, E. coli (20 μg); lane 4, M. smegmatis LR222 [Myc(LR)] (20 μg); lane 5, M. smegmatis mc26 [Myc(mc)] (20 μg); lane 6, RepB (125 ng) plus nonspecific protein bovine serum albumin (BSA) (20 μg); lane 7, RepB (125 ng) plus nonspecific protein bovine serum albumin (30 μg); lane 8, RepB (125 ng) plus E. coli extract (20 μg); lane 9, RepB (125 ng) plus E. coli extract (30 μg); lane 10, RepB (125 ng) plus M. smegmatis LR222 (20 μg); lane 11, RepB (125 ng) plus M. smegmatis LR222 (30 μg); lane 12, RepB plus M. smegmatis mc26 (20 μg); and lane 13, RepB plus M. smegmatis mc26 (30 μg).
RepB participates directly in formation of the ori complex.
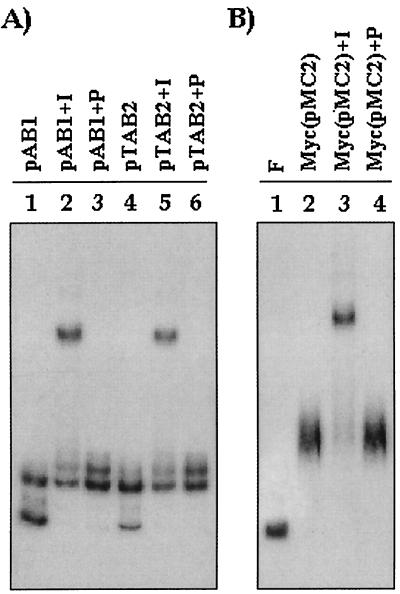
To specifically investigate whether RepB was involved directly in formation of the complex, an antibody supershift assay was designed by using an antiserum specific to RepB. Positive supershifts were observed both with pTAB2 extracts and with pAB1 extracts, indicating that in both cases RepB was directly involved in complex formation (Fig. 7A, lanes 2 and 5). Preimmune sera did not give rise to supershifts (Fig. 7A, lanes 3 and 6), indicating that the experiment was specific. The same experiment was repeated with mycobacterial extracts, and the results showed that although no RepB was detected in Western blots (Fig. 3I), a specific supershift occurred with anti-RepB immune serum (Fig. 7B, lane 3) but not with preimmune serum (lane 4). The results indicate that in M. smegmatis, which can be a productive host for pAL5000, RepB is directly involved in complex formation.
FIG. 7.
Antibody supershift assays performed with either pAB1 or pTAB2 extracts (A) and with extracts of mycobacterial cells harboring pMC2 [Myc(pMC2)] (B). I, 1 μl of anti-RepB immune serum was added to the assay mixture; P, 1 μl of preimmune serum was added to the assay mixture. F, free probe.
DISCUSSION
We found that expression from the repA-repB region of pAL5000 results in a strong origin binding activity. The activity is maximal when the complete repA-repB region is expressed, but expression of repB alone results in a basal level of activity. Undoubtedly, RepB plays a central role, as expression of this protein was found to be necessary for generation of binding activity in repA-repB-expressing cells. Moreover, antibody supershift experiments using anti-RepB antibody indicated that RepB was present in the bound complex. In a previous investigation (20) it was found that the optimum amount of purified RepB required in a standard assay mixture for direct binding was about 1 μg (concentration, 1,000 to 2,000 nM). This binding activity was reproduced in our study, but the novel feature in this study was that RepB-dependent binding activity was observed at much lower concentrations (100- to 1,000-fold lower) when unfractionated extracts of repA-repB- and repB-expressing cells were used. The difference was not only quantitative but also qualitative, as we observed that the direct RepB-ori complexes were sensitive to high salt concentrations and competition with a nonspecfic competitor, whereas the complexes formed by the extracts were resistant. Therefore, the specificity of the direct binding by RepB was low, whereas the RepB-mediated binding activity observed when the extracts were used was highly specific.
The observations made in this study make a strong case for host factor involvement. In vitro mixing experiments clearly indicated that host factors play a major role in determining the efficiency of RepB binding to the origin of replication. The formation of a salt-labile DNA-protein complex by purified RepB alone can be explained on the basis of the pI of RepB, the predicted value of which is 9. RepB is a basic protein which can bind to negatively charged molecules, as shown by its binding to a cation-exchange column, like the Mono S column. The binding of RepB to ori, therefore, is mostly, if not entirely, due to ionic interactions. This is evident from the deleterious effect that increasing the salt concentration has on formation of the RepB-ori complex. We concluded that since the complex formed by the extracts is salt resistant, this complex, although directly involving RepB, is distinct from the complex formed by purified RepB alone. Apparently, the salt-resistant complex is more specific and is stabilized by structure-based interactions (3) rather than ionic interactions.
Involvement of host factors in plasmid replication is a common feature in various E. coli plasmids, although such involvement in mycobacterial plasmid replication has not been described yet. Thus, in the case of plasmid P1 the affinity of RepA is increased through association with DnaK and DnaJ (5, 23). It is possible that in pAL5000 chaperons present in the extract associate with RepB and enable it to interact more efficiently with the ori. Yet another possibility is the involvement of a histone-like protein, HU (11), which may be needed to maintain the conformation of the DNA to facilitate RepB binding. Whatever the nature of the host factor is, the factor is apparently a global factor and is present in mycobacterial systems as well as in E. coli systems.
In addition to host factors, expression of repA also played a role in modulating the activity of RepB. The complexes formed by extracts of repA-repB- and repB-expressing cells were similar, and we found that the complex formed by a pTAB2 extract (untagged RepB-expressing cells) was as salt resistant as the complex formed by a pAB1 extract (data not shown). In both cases RepB was directly involved in complex formation, as shown by the antibody supershift assays. Only the amount of RepB required differed. The activity of a repB extract (pAB2 or pTAB2) could be made comparable to that of a repA-repB extract (pAB1) if the level of RepB could be increased, as observed with the pTAB2 extract. A loss of RepA can therefore be compensated for by an increase in the level of RepB. Similarly, a decrease in the level of RepB can be compensated for by coupled expression of repA. However, RepA itself is not an origin binder, as has been reported previously (20), and similar results were obtained in our laboratory when an MBP-RepA fusion was tested. It also does not appear that RepA associates with other proteins and then forms the complex; no differences between the sizes of the complexes formed by MBP-RepA (pMAB1) and His-RepA (pAB1) extracts were observed, although MBP-RepA is nearly twice the size of His-RepA. However, the influence of repA expression is observed only when there is strict coupling of expression. One advantage of coupled expression could be maintenance of a precise ratio of the level RepA to the level of RepB, which could be crucial for optimal activity. This could be one of the reasons behind our inability to reproduce the stimulatory effect either in vivo or in vitro when RepA synthesis and RepB synthesis were uncoupled. Although our analysis did indicate that coupled expression of the repA gene had a positive effect, it has not been possible to precisely delineate the function of the repA gene product.
The observations made with E. coli cell extracts are consistent with the data obtained with corresponding extracts from mycobacteria. First, the complexes are similar. Second, supershift experiments with anti-RepB antibody indicated that RepB participates in complex formation, although the RepB antigen could not be detected in Western blots. Prior to this study there was no evidence which suggested that any of the putative mycobacterial plasmid-encoded replication proteins identified either in pAL5000 or in related plasmids (1, 14, 15) are actually expressed in mycobacteria and are involved in complex formation. This is the first time that direct involvement of a mycobacterial plasmid replication protein in ori complex formation has been demonstrated in mycobacteria using RepB as a model.
Acknowledgments
This work was funded by a grant from CSIR, Government of India. The Department of Biotechnology, Government of India, is acknowledged for a postdoctoral fellowship awarded to S.C. A.B. and M.C. are grateful to CSIR for fellowships.
We are grateful to P. Parrack, P. Roy, S. Roy, and A. Sil for carefully reading the manuscript and giving their comments. We are deeply indebted to our technicians, P. Halder and D. Majumder, for their excellent technical assistance.
REFERENCES
- 1.Beggs, M. L., J. T. Crawford, and K. D. Eisenach. 1995. Isolation and sequencing of the replication region of Mycobacterium avium plasmid pLR7. J. Bacteriol. 177:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloom, B. R., and C. J. L. Murray. 1992. Tuberculosis: commentary on a reemergent killer. Science 257:1055-1064. [DOI] [PubMed] [Google Scholar]
- 3.Calladine, C. R., and H. R. Drew. 1997. Specific DNA-protein interactions, p. 177-203. In Understanding DNA, the molecule and how it works, 2nd ed. Academic Press, New York, N.Y.
- 4.Chawla, M., and S. K. Das Gupta. 1999. Transposition induced instability in E. coli-mycobacteria shuttle vectors. Plasmid 41:135-140. [DOI] [PubMed] [Google Scholar]
- 5.DasGupta, S., G. Mukhopadhyay, P. P. Papp, M. S. Lewis, and D. K. Chattoraj. 1993. Activation of DNA binding by the momomeric form of P1 replication initiator RepA by heat shock proteins DnaJ and DnaK. J. Mol. Biol. 232:23-34. [DOI] [PubMed] [Google Scholar]
- 6.Das Gupta, S. K., M. D. Bashyam, and A. K. Tyagi. 1993. Cloning and assessment of mycobacterial promoter by using a plasmid shuttle vector. J. Bacteriol. 175:5186-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das Gupta, S. K., D. Kaushal, and A. K. Tyagi. 1998. Expression systems for study of mycobacterial gene regulation and development of recombinant BCG vaccine. Biochem. Biophys. Res. Commun. 246:797-804. [DOI] [PubMed] [Google Scholar]
- 8.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 9.Hatfull, G. F. 1993. Genetic transformation of mycobacteria. Trends Microbiol. 1:310-314. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs, W. R., Jr. 2000. Mycobacterium tuberculosis: a once genetically intractable organism, p. 1-16. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 11.Kar, S., and S. Adhya. 2001. Recruitment of HU by piggyback: a special role of GalR in repressosome assembly. Genes Dev. 15:2273-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kochi, A. 1991. The global tuberculosis situation and the new control strategy of the World Health Organization. Tubercle 72:1-6. [DOI] [PubMed] [Google Scholar]
- 13.Kristie, T. M., and B. Roizman. 1986. a4, the major regulatory protein of herpes simplex virus type 1, is stably and specifically associated with promoter-regulatory domains of a gene and/or selected viral genes. Proc. Natl. Acad. Sci. USA 83:3218-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pashley, C., and N. G. Stoker. 2000. Plasmids in mycobacteria, p. 55-67. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 15.Qin, M., H. Taniguchi, and Y. Mizuguchi. 1994. Analysis of the replication region of a mycobacterial plasmid, pMSC262. J. Bacteriol. 176:419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauzier, J., J. Moniz-Pereira, and B. Gicquel-Sanzey. 1988. Complete nucleotide sequence of pAL5000, a plasmid from Mycobacterium fortuitum. Gene 71:315-321. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 19.Stolt, P., and N. G. Stoker. 1996. Functional definition of region necessary for replication and incompatibility in the Mycobacterium fortuitum plasmid pAL5000. Microbiology 142:2795-2802. [DOI] [PubMed] [Google Scholar]
- 20.Stolt, P., and N. G. Stoker. 1996. Protein DNA interactions in the ori region of the Mycobacterium fortuitum plasmid pAL5000. J. Bacteriol. 178:6693-6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stolt, P., and N. G. Stoker. 1997. Mutational analysis of the regulatory region of the Mycobacterium plasmid pAL5000. Nucleic Acids Res. 25:3840-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickner, S., J. Hoskins, and K. McKenney. 1991. Function of DnaJ and DnaK as chaperons in origin-specific binding by RepA. Nature 350:165-167. [DOI] [PubMed] [Google Scholar]