Abstract
Alanine scanning of the Escherichia coli RNA polymerase α subunit C-terminal domain (αCTD) was used to identify amino acid side chains important for class I cyclic AMP receptor protein (CRP)-dependent transcription. Key residues were investigated further in vivo and in vitro. Substitutions in three regions of αCTD affected class I CRP-dependent transcription from the CC(−61.5) promoter and/or the lacP1 promoter. These regions are (i) the 287 determinant, previously shown to contact CRP during class II CRP-dependent transcription; (ii) the 265 determinant, previously shown to be important for αCTD-DNA interactions, including those required for class II CRP-dependent transcription; and (iii) the 261 determinant. We conclude that CRP contacts the same target in αCTD, the 287 determinant, at class I and class II CRP-dependent promoters. We also conclude that the relative contributions of individual residues within the 265 determinant depend on promoter sequence, and we discuss explanations for effects of substitutions in the 261 determinant.
The α subunits of Escherichia coli RNA polymerase holoenzyme (RNAP) play a key role in transcription initiation and activation (reviewed in references 5 and 11). Each α subunit consists of two independently folded domains connected by a flexible linker (4, 12, 13). The N-terminal domain (αNTD) is critical for the assembly of the core RNAP complex (5). The C-terminal domain (αCTD) binds to A/T-rich sequence elements (UP elements) at many promoters (11, 22) and is also a target for transcription activators, with many activators interacting directly with αCTD and recruiting it, and consequently the rest of RNAP, to target promoter DNA (5). The structure of αCTD has been determined by nuclear magnetic resonance spectroscopy (8, 12). The aim of this study was to identify the contact target in αCTD for the transcription activator cyclic AMP (cAMP) receptor protein (CRP; also referred to as catabolite activator protein).
Transcription activation by CRP provides an important model system for understanding mechanisms of bacterial transcriptional regulation (reviewed in reference 6). CRP is a homodimer that binds to DNA in the presence of cAMP. Simple CRP-dependent promoters can be grouped into two classes, depending on the location of the DNA binding site for CRP. At class I CRP-dependent promoters, CRP binds upstream of RNAP, at sites centered near position −61, −71, −82, or −92 upstream from the transcription start site. The best-characterized class I CRP-dependent promoters are lacP1 and a semisynthetic derivative of the melR promoter, CC(−61.5) (9), each of which contains a CRP-binding site centered at position −61.5. At class II CRP-dependent promoters, the CRP-binding site overlaps the binding site for RNAP. The best-characterized class II CRP-dependent promoters are galP1 and a semisynthetic derivative of the melR promoter, CC(−41.5) (9), each of which contains a CRP-binding site centered at position −41.5.
At both class I and class II CRP-dependent promoters, CRP interacts with αCTD, facilitating the binding of αCTD to the DNA segment adjacent to the CRP binding site. CRP-αCTD interaction is mediated by a surface-exposed loop comprising residues 156 to 164 of CRP (designated activating region 1 [AR1]). At class I promoters, AR1 is functionally presented by the downstream subunit of the CRP dimer and interacts with αCTD located downstream of CRP, whereas at class II promoters, AR1 is functionally presented by the upstream subunit of the CRP dimer and interacts with αCTD located upstream of CRP. Substitutions within AR1 disrupt CRP-αCTD interactions at both class I and class II promoters, but quantitative effects of alanine substitutions for individual residues differ from promoter to promoter (30).
In previous work, we used random mutagenesis and alanine scanning to identify amino acid side chains of αCTD required for CRP-dependent transcription activation at a class II promoter, CC(−41.5) (23). We identified two determinants that play a key role. The first determinant, termed the 287 determinant, included residues T285, E286, V287, E288, and R317. Substitutions in the 287 determinant did not affect sequence-specific αCTD-DNA interactions but did reduce cooperativity of DNA binding by α subunits and CRP (23). The 287 determinant of αCTD therefore was proposed to be the surface directly contacted by AR1 of CRP during transcription activation at class II promoters. The second determinant, termed the 265 determinant, included R265 and other amino acids previously shown to be required for binding of αCTD to UP elements (8, 11, 20). The 265 determinant was proposed to mediate αCTD-DNA interactions at class II promoters.
In this work, we systematically tested the effects of alanine substitutions throughout the αCTD on CRP-dependent transcription at the class I CRP-dependent promoter CC(−61.5). In addition, we evaluated the roles of key residues of αCTD in transcription at CC(−61.5) and at a second class I CRP-dependent promoter, lacP1, in vivo using strains lacking wild-type α subunits and in vitro using RNAP derivatives reconstituted with α subunits carrying alanine substitutions. We discuss our results in the light of previous reports.
MATERIALS AND METHODS
Strains, plasmids, and promoters.
Bacterial strains and plasmids used in this study are listed in Table 1. Promoter fragments were cloned as EcoRI-HindIII fragments. Strains RLG4650 and RLG4651 were constructed by the method of Simons et al. (24), as described previously (23). Strains WAM140 and WAM144 were constructed by exploiting the observation that the rpoA341 mutation in strain WAM105 results in a Cym− Mel− phenotype (26). WAM140 and WAM144 were obtained as Cym+ Mel+ revertants resulting from recombination with plasmid-borne 3′ segments of rpoA encoding the EA261 and VA287 substitutions, respectively. After curing the rpoA plasmid, the chromosomal rpoA gene was checked by DNA sequencing.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference(s) |
|---|---|---|
| Strains | ||
| M182Δcrp | Δcrp derivative of M182 (E. coli Δlac K-12 strain) | 7 |
| RLG4650 | M182 carrying λ prophage with CC(−61.5)::lacZ fusion | This work |
| RLG4651 | M182 carrying λ prophage with lacP1::lacZ fusion | This work |
| WAM105 | rpoA341 derivative of MJF1 | 26 |
| WAM106 | rpoA+ derivative of MJF1 | 26 |
| WAM140 | Derivative of WAM106 with rpoA EA261 mutant | This work |
| WAM144 | Derivative of WAM106 with rpoA VA287 mutant | This work |
| Plasmids | ||
| pSR | pBR322 derivative containing transcription terminator | 16 |
| pSR/CC(−61.5) | pSR derivative carrying CC(−61.5) promoter fragment (−101 to +35) | This work |
| pSR/lacP1 | pSR derivative carrying lacP1 promoter fragment (−140 to +63) | This work |
| pSR/lacUV5 | pSR derivative carrying lacUV5 promoter fragment (−59 to +37) | 23 |
| pSR/lacUV5(−140/63) | pSR derivative carrying lacUV5 promoter fragment (−140 to +63) | This work |
| pREIIα and derivatives | Plasmid carrying rpoA encoding RNAP α subunit and derivatives carrying alanine substitutions at positions 273-329, except 302 | 4, 8, 14, 25, 28 |
| pHTf1α and derivatives | Plasmid carrying rpoA encoding RNAP α subunit and derivatives carrying alanine substitutions at positions 255-271 and 302 | 8, 25 |
| pDCRP | pBR322 derivative carrying crp | 3 |
| pDCRP HL159 | Derivative of pDCRP encoding CRP HL159 | 3 |
| pDU9 | Derivative of pDCRP with crp gene deleted | 3 |
| pRW50 | Broad-host-range lac expression vector | 17 |
| pRW50/CC(−61.5) | pRW50 derivative carrying CC(−61.5) promoter fragment (−101 to +266) | 27 |
| pRW50/lacP1 | pRW50 derivative carrying lacP1 promoter fragment (−140 to +63) | This work |
| pRW50/lacUV5 | pRW50 derivative carrying lacUV5 promoter fragment (−59 to +37) | This work |
Strain RLG4650 and plasmid pSR/CC(−61.5) contain positions −101 to +35 of CC(−61.5), a promoter that carries a consensus DNA site for CRP centered 61.5 bp upstream of the pmelR core promoter elements (9). Expression from CC(−61.5) is completely dependent on the interaction between αCTD and AR1 of CRP (3). Strain RLG4651 and plasmids pSR/lacP1 and pRW50/lacP1 contain positions −140 to +63 of the wild-type lacP1 promoter (15, 16). Plasmid pSR/lacUV5(−140/63), which was used to generate the DNA fragment used in the experiments described in Fig. 4, contains positions −140 to +63 of the lacUV5 promoter and includes a functional CRP-binding site (15, 16). The lacUV5 promoter is identical to the lacP1 promoter except for two substitutions in the −10 hexamer that improve interactions with the σ subunit of RNAP holoenzyme and make the promoter CRP independent without altering the positioning of αCTD or CRP within the initiation complex (15). Plasmids pSR/lacUV5 and pRW50/lacUV5, used in the experiments described in Fig. 2, contain positions −59 to +37 of the lacUV5 promoter and do not contain a functional CRP-binding site.
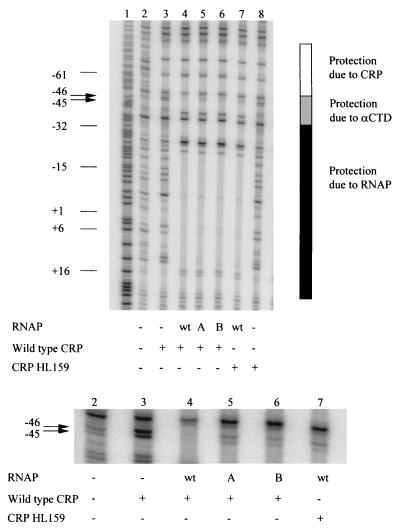
FIG. 4.
Substitutions in the 287 determinant alter the DNase I footprint of a class I transcription initiation complex. DNase I footprints of complexes containing the lacUV5 promoter (end labeled on the template strand), CRP or CRP derivative (100 nM), and reconstituted wild-type (wt) or mutant RNAPs. A, TA285:VA287 α RNAP; B, VA287:RA317 α RNAP. Lane 1 of the autoradiogram shows a Maxam-Gilbert G + A sequencing reaction. The shaded bars indicate protection by α, CRP, and the rest of RNAP. Arrows indicate DNase I-hypersensitive sites at positions −45 and −46, immediately downstream of the CRP binding site. The lower part of the figure shows a close-up of this part of the gel.
FIG. 2.
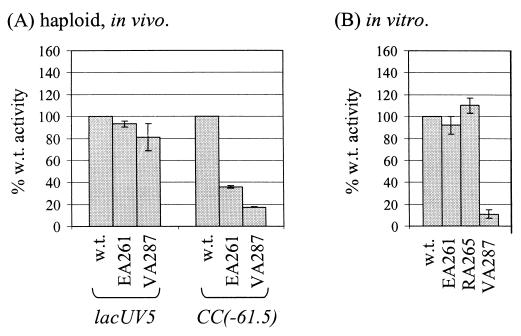
Residues of αCTD important for activation by CRP at CC(−61.5): in vivo transcription assays of haploid strains and in vitro transcription assays. (A) Strains haploid for rpoA (WAM106, wild-type chromosomal rpoA; WAM140, chromosomal rpoA-EA261; and WAM144, chromosomal rpoA-VA287) were transformed with the CC(−61.5)::lacZ fusion plasmid pRW50/CC(−61.5) or the lacUV5::lacZ fusion plasmid pRW50/lacUV5. β-Galactosidase activities are expressed as percentages of the activity obtained with transformed WAM106 [100%= 340 Miller units for CC(−61.5) and 1156 Miller units for lacUV5]. (B) Multiple-round in vitro transcription experiments were performed using supercoiled pSR/CC(−61.5) template, wild-type CRP, and RNAP reconstituted with hexahistidine-tagged α derivatives containing alanine substitutions at the indicated positions. Purified RNAPs were normalized as described in Materials and Methods. Values (with standard deviation) are expressed as percentages of the yield of transcript with wild-type RNAP and wild-type CRP.
Measurement of β-galactosidase activity.
Cultures were inoculated to an A600 of approximately 0.007 and grown to mid-log phase (A600 of approximately 0.35 to 0.40) at 37°C with vigorous aeration in L broth (20 g of tryptone, 10 g of yeast extract, and 10 g of NaCl per liter) containing antibiotics where appropriate. β-Galactosidase activities were determined by the method of Miller (19). Results presented are averages of at least three independent assays and are shown with standard deviations.
Protein purification.
N-terminally hexahistidine-tagged α subunit derivatives were prepared and reconstituted into RNA polymerase as described previously (23). Wild-type CRP and CRP HL159 were purified by the method of Ghosaini et al. (10) from M182 Δcrp cells transformed with plasmid pDCRP and pDCRP HL159, respectively.
In vitro transcription assays.
Reactions (25-μl final volume) were performed with 0 to 12.5 nM RNAP, 20 nM CRP, 0.2 mM cAMP, 0.2 nM supercoiled plasmid templates, 200 μM ATP, 200 μM CTP, 200 μM GTP, 10 μM UTP, and 5 μCi of [α-32P]UTP in 100 mM KCl-40 mM Tris-acetate (pH 7.9)-10 mM MgCl2-1 mM dithiothreitol-100 μg of bovine serum albumin per ml as described previously (23). Templates were prepared using a QIAgen miniprep kit and contained promoters cloned as EcoRI-HindIII fragments into plasmid pSR. Reactions were started by the addition of RNAP, and after 15 min at 22°C, products were analyzed by denaturing gel electrophoresis and quantified by phosphorimaging (Molecular Dynamics) with ImageQuant software. RNAP preparations were used at 2.4 nM (wild-type RNAP), 2.2 nM (αEA261 RNAP), 12.5 nM (αRA265 RNAP), or 2.6 nM (αVA287 RNAP), concentrations that resulted in the same amounts of transcription from the lacUV5 promoter of pSR/lacUV5 in the absence of CRP.
DNase I footprinting.
DNase I footprinting studies of purified CRP, CRP HL159, and reconstituted RNAP binding to a PstI-HindIII fragment of pSR/lacUV5(−140/63) were performed as described previously (2). The template strand was end labeled at the HindIII end using [γ-32P]ATP. Products of Maxam-Gilbert G + A sequencing reactions were used as markers.
RESULTS
Residues of αCTD important for activation by CRP at CC(−61.5). (i) In vivo transcription assays. (a) Merodiploid strains.
As an initial screen to identify residues of αCTD important for activation of a class I CRP-dependent promoter, we analyzed the effects of 69 alanine substitutions, spanning residues 255 to 329 of α, in vivo in merodiploid assays (8, 23, 25). A reporter strain carrying a single-copy chromosomal CC(−61.5)::lacZ fusion was transformed individually with plasmids encoding α derivatives, and levels of β-galactosidase expression were determined (Fig. 1A).
FIG. 1.
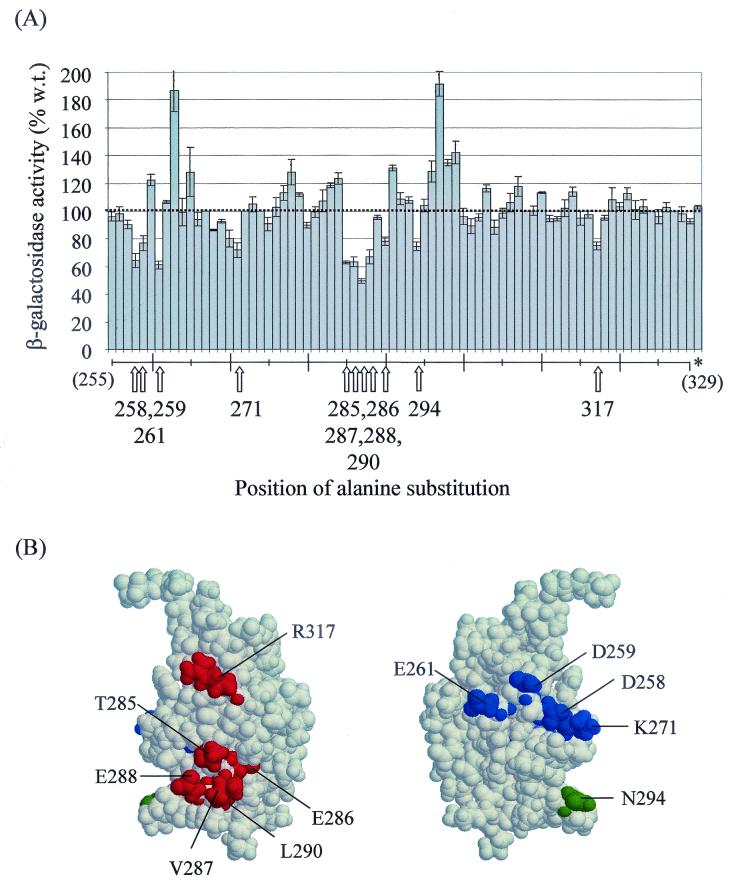
Residues of αCTD important for activation by CRP at CC(−61.5): in vivo transcription assays of merodiploid strains. (A) RLG4650 cells carrying a chromosomal CC(−61.5)::lacZ fusion were transformed with derivatives of plasmids pHTf1α (substitutions at 255 to 271 and 302) or pREIIα (substitutions at remaining positions). Each plasmid encoded an α derivative with a single alanine substitution between residues 255 and 329 as indicated in the figure. β-Galactosidase activities are expressed as percentages of the activity obtained with cells transformed with plasmids encoding wild-type α (100% = 143 Miller units). Positions at which alanine substitution decreased activity by >20% are indicated by arrows. Residues 267, 272, 274, 308, 324, and 327 are alanines in the wild-type protein. The β-galactosidase activity obtained with cells transformed with a vector-only control plasmid (pDU9) is indicated by an asterisk. (B) Structure of αCTD (12), showing side chains identified as critical for CRP-dependent transcription at CC(−61.5). Two views of the structure, related by a 180° rotation on the vertical axis, are shown. Side chains of residues belonging to the 261, 287, and 265 determinants are shown in blue, red, and green, respectively.
Overproduction of α derivatives with alanine substitutions at positions 258, 259, 261, 271, 285 to 288, 290, 294, and 317 decreased CRP-dependent transcription from the CC(−61.5) promoter by 20 to 50%, with substitution of V287 and E261 causing the largest decreases (Fig. 1A). These positions can be divided into three groups (6). Residues T285, E286, V287, E288, L290, and R317 cluster on one face of αCTD and correspond to the 287 determinant previously shown to be required for class II CRP-dependent transcription (Fig. 1B) (23). Residues D258, D259, E261, and K271 cluster on the opposite face of αCTD and correspond to the 261 determinant previously shown to be required for class I CRP-dependent transcription at the lacP1 promoter (6, 25) (see below). The remaining residue, N294, is part of the 265 determinant previously shown to be required for both class II CRP-dependent transcription (6, 23) and class I CRP-dependent transcription at the lacP1 promoter (6, 20, 25). (Although N294 is not critical for UP element-dependent transcription at rrnB P1 [8], it contacts the DNA backbone in an α-DNA complex [29].) The results indicate that the side chains of residues 258, 259, 261, 271, 285 to 288, 290, 294, and 317 of α may make interactions that are important for class I CRP-dependent transcription at CC(−61.5).
Overproduction of certain alanine-substituted α derivatives, most notably TA263 and KA297, increased CRP-dependent transcription from the CC(−61.5) promoter. T263 and K297 are surface exposed in (or adjacent to) the 265 determinant important for αCTD-DNA interactions (8, 20). Preliminary results suggest that these residues are involved in αCTD-αCTD interactions that compete with αCTD-DNA interactions in the activation complex and that the alanine substitutions increase CRP-dependent transcription by eliminating this competition (H. Chen and R. H. Ebright, unpublished results). It is also possible that these residues in the wild-type protein interfere directly with αCTD-DNA interactions at CC(−61.5).
(b) Haploid strains.
In the merodiploid assays described in the previous section, the effects of substitutions in the plasmid-encoded α subunits were moderated by the presence of wild-type α subunits encoded by the chromosomal rpoA gene (8, 23, 25). To confirm the effects of the substitutions that caused the largest defects in the merodiploid assays, VA287 and EA261, we also performed in vivo assays in strains haploid for mutant rpoA (Fig. 2A). In these experiments, we measured transcription from a plasmid-borne CC(−61.5)::lacZ fusion in strains in which VA287 or EA261 mutant rpoA alleles were substituted on the chromosome for the wild-type rpoA allele.
The results indicate that alanine substitutions at V287 and E261 reduced CRP-dependent transcription at CC(−61.5) by 83 and 64%, respectively (Fig. 2A). As expected, the defects in promoter activity were greater than those observed in the presence of plasmid-borne mutant rpoA alleles expressed in trans to wild-type rpoA. Control experiments indicated that the VA287 and EA261 substitutions had little or no effect on CRP-independent transcription from the lacUV5 promoter (Fig. 2A).
(ii) In vitro transcription assays.
While the effects of alanine substitutions in αCTD in vivo suggested that the 261 and 287 determinants are important for CRP-dependent transcription from CC(−61.5), these results did not prove that the effects were direct. Therefore, we measured the effects of the EA261 and VA287 substitutions on CRP-dependent transcription at CC(−61.5) in vitro using reconstituted RNAP derivatives (Fig. 2B). We also examined the effects of the RA265 substitution, the substitution in αCTD that causes the largest defect in DNA binding to UP elements (8, 20).
The VA287 substitution resulted in an 89% reduction in CRP-dependent transcription at CC(−61.5) in vitro. In contrast, neither the EA261 substitution nor the RA265 substitution caused significant changes in CRP-dependent transcription at CC(−61.5) in vitro under the assay conditions used. None of the RNAP derivatives tested produced a detectable level of transcript from the CC(−61.5) promoter in the absence of CRP. However, as the assays used RNAP concentrations that gave identical yields of transcript from the CRP-independent lacUV5 promoter, the observed defects are most likely attributable to decreases in activation by CRP. These results confirm the direct role of the 287 determinant in CRP-dependent transcription from CC(−61.5).
Residues of αCTD important for activation by CRP at lacP1.
Previous investigations of activation of promoters where CRP binds at position −61.5 focused on the E. coli lacP1 promoter (20, 25, 31). These studies employed both random and site-directed mutagenesis approaches and concluded that key determinants of αCTD for class I CRP-dependent activation of lacP1 are the 261 determinant (25) and the 265 determinant (20, 25). The previous studies did not identify the 287 determinant (20, 25). In order to compare directly the role of the key residues in each determinant in class I CRP-dependent transcription activation at lacP1 and CC(−61.5), we measured transcription from lacP1 in vivo and in vitro using the same experimental conditions as for our analysis of CC(−61.5).
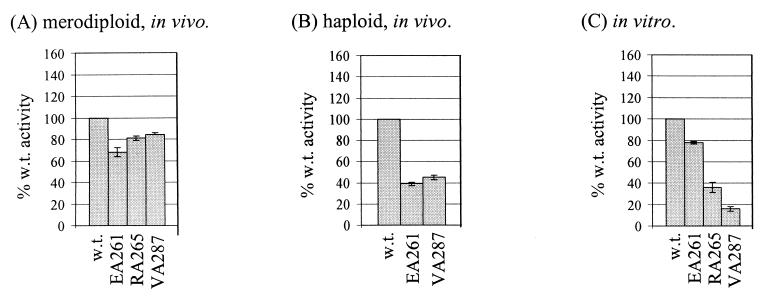
The effects of overproducing α derivatives with single alanine substitutions were determined using a reporter strain carrying a single-copy chromosomal lacP1::lacZ fusion (merodiploid assays; Fig. 3A). The reporter strain was transformed individually with plasmids encoding α derivatives with single alanine substitutions at positions 287, 265, and 261, and levels of β-galactosidase expression were measured. All three substitutions caused small but reproducible decreases in class I CRP-dependent transcription at lacP1.
FIG. 3.
Residues of αCTD important for activation by CRP at lacP1. (A) RLG4651 (chromosomal lacP1::lacZ fusion) was transformed with plasmids encoding mutant α derivatives as indicated. β-Galactosidase activities are expressed as percentages of the activity obtained with cells transformed with plasmids encoding wild-type α (100% = 2,860 Miller units). (B) Strains haploid for rpoA (WAM106, wild-type chromosomal rpoA; WAM140, chromosomal rpoA-EA261; and WAM144, chromosomal rpoA-VA287) were transformed with the lacP1::lacZ fusion plasmid pRW50/lacP1. β-Galactosidase activities are expressed as percentages of the activity obtained with WAM106 transformed with pRW50/lacP1 (100%= 6,770 Miller units). (C) Multiple-round in vitro transcription experiments were performed using supercoiled pSR/lacP1 template, wild-type CRP, and RNAP reconstituted with hexahistidine-tagged α derivatives containing alanine substitutions at the positions indicated. Purified RNAPs were normalized as described in Materials and Methods. Values (with standard deviation) are expressed as percentages of the yield of transcript with wild-type RNAP and wild-type CRP.
Next, transcription from a lacP1::lacZ fusion carried on a plasmid was measured in strains in which VA287 or EA261 mutant rpoA alleles were substituted on the chromosome for the wild-type rpoA allele (haploid assays; Fig. 3B). The results confirm that the VA287 and EA261 substitutions result in defects in class I CRP-dependent transcription at lacP1 (reducing activity by 61 and 55%, respectively). As observed with CC(−61.5), the defects in promoter activity in these strains, haploid for mutant rpoA, were greater than observed in the presence of plasmid-borne mutant rpoA alleles expressed in trans to wild-type rpoA. (Since RA265 α subunits do not support growth in the absence of wild-type α [8], it was not possible to replace the chromosomal rpoA allele with rpoA RA265.)
Finally, we measured CRP-dependent transcription activation from the lacP1 promoter in vitro using RNAP derivatives reconstituted with α derivatives carrying the VA287, RA265, or EA261 substitution (Fig. 3C). The results indicate that all three substitutions directly cause defects in class I CRP-dependent transcription at lacP1, with the VA287 substitution resulting in the most severe defect (reducing activity by 84%). In the absence of CRP, lacP1 promoter activity was very low, preventing precise quantitation. Nevertheless, no effects of the α mutants on basal transcription were detected, and the defects observed in the presence of CRP are likely to result from defects in activation rather than basal transcription, since the assays used RNAP concentrations that gave identical yields of transcript from the CRP-independent lacUV5 promoter.
DNase I footprinting of a class I transcription initiation complex.
We used DNase I footprinting to examine the effects of single and multiple substitutions within the 287 determinant of αCTD on the architecture of a class I CRP-RNAP-promoter complex (Fig. 4). The lacUV5 promoter was used in these experiments in order to increase occupancy of the promoter by RNAP in the absence of CRP-αCTD contacts (15).
In ternary complexes containing both wild-type CRP and wild-type RNAP, protection was observed spanning the CRP and RNAP binding sites (Fig. 4, lane 4). Positions −45 and −46, which are located at the junction between the CRP and αCTD binding sites and are hypersensitive to DNase I cleavage in the complex containing only CRP (Fig. 4, lane 3, arrows), are protected in the ternary complex containing CRP and RNAP (Fig. 4, lane 4, arrows). In contrast, positions −45 and −46 are not protected in ternary complexes containing a CRP mutant with a disruption in AR1 (CRP HL159; Fig. 4, lane 7, arrows). This indicates that protection of positions −45 and −46 is diagnostic of a productive AR1-αCTD interaction. Protection from DNase I cleavage at the junction of the CRP and α binding sites is also diagnostic of productive AR1-αCTD interaction at the class II CRP-dependent promoter galP1 (2).
Protection of positions −45 and −46 is also reduced in complexes containing RNAPs with substitutions in the 287 determinant of αCTD (TA285:VA287 and VA287:RA317; lanes 5 and 6). This result is consistent with direct involvement of the 287 determinant in AR1-αCTD interactions.
DISCUSSION
Our work defines three determinants of αCTD for class I CRP-dependent transcription at CC(−61.5) and lacP1: the 287 determinant, the 265 determinant, and the 261 determinant.
The 287 determinant is required for CRP-dependent transcription in vivo and in vitro at both class I and class II CRP-dependent promoters (23; this work). Substitutions within the 287 determinant do not affect αCTD-DNA interactions (23) but do affect cooperative DNA binding by CRP and α (23) and the architecture of transcription initiation complexes at class I CRP-dependent promoters (Fig. 4). We propose that the 287 determinant of αCTD constitutes the contact surface for AR1 of CRP at both class I and class II CRP-dependent promoters (Fig. 5). We suggest that the 287 determinant can also mediate αCTD-CRP interactions at promoters where the DNA site for CRP is located further upstream than −61.5 (N. J. Savery, unpublished data). The fact that the 287 determinant was not detected in previous searches for residues of αCTD affecting CRP-dependent activation of the lacP1 promoter (20, 25, 31) most likely reflects the fact that single substitutions in the 287 determinant have only modest effects on transcription from lacP1 when expressed from plasmids in trans to wild-type α (Fig. 3A).
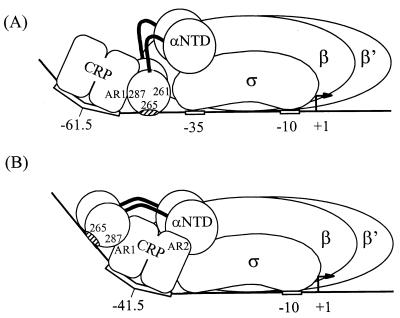
FIG. 5.
Model for class I and class II CRP-dependent promoter complexes. (A) At a class I CRP-dependent promoter with a CRP binding site centered at approximately −61.5 [e.g., CC(−61.5) or lacP1], the αCTD 287 determinant interacts with AR1 of the downstream subunit of the CRP dimer, the 265 determinant interacts with DNA, and the 261 determinant interacts with σ (4). The relative contribution of the αCTD 265 determinant to complex formation (hatched) varies, depending on promoter sequence. (B) At a class II CRP-dependent promoter [e.g., CC(−41.5) or galP1], the αCTD 287 determinant interacts with AR1 of the upstream subunit of the CRP dimer bound at −41.5, and the 265 determinant interacts with DNA. A second activating region of CRP (AR2) interacts with αNTD at class II CRP-dependent promoters (21).
The 265 determinant is also required for CRP-dependent transcription at both class I and class II CRP-dependent promoters 20, 23, 25, 31; this work). This determinant mediates both sequence-specific and nonspecific αCTD-DNA interactions (8). We propose that the 265 determinant mediates interactions between αCTD and the DNA segment adjacent to the binding site for CRP at both class I and class II promoters and that these interactions contribute to the overall stability of the initiation complex. However, our results indicate that the relative contributions of residues within the 265 determinant to CRP-dependent transcription vary from promoter to promoter; for example, R265 contributes significantly to class I CRP-dependent transcription activation at lacP1 but not at CC(−61.5).
The promoter specificity of the contribution of individual residues within the 265 determinant could derive from differences in the α-binding potential of the DNA sequences adjacent to the CRP binding site in different promoters, from differences in the contribution of the kinetic step affected by αCTD-DNA interactions to the overall rate of transcription initiation at different promoters, and/or from subtle variations in the geometry of the specific initiation complex at different promoters. Consistent with the possibility that DNA sequence variation can alter details in DNA recognition by αCTD, alanine substitutions in the DNA-binding determinant of αCTD differentially affect transcription activation of the rrnB P1 versus rrnE P1 promoters by the transcription factor Fis (1).
The 261 determinant is required for class I CRP-dependent transcription but, with the possible exception of residue K271, not for class II CRP-dependent transcription (23). Substitutions within the 261 determinant cause defects in class I CRP-dependent transcription at CC(−61.5) and lacP1 in vivo and, at lacP1, in abortive initiation and multiround transcription experiments in vitro (Fig. 1, 2, and 3) (25). The 261 determinant has been proposed to interact with the σ subunit of RNAP at class I CRP-dependent promoters (where αCTD binds adjacent to the −35 hexamer) (6). In support of this proposal, substitution of E261 does not affect αCTD-DNA interactions or cooperative DNA binding by CRP and α subunits (23), and RNAP containing αEA261 is defective for UP element function at promoters with strong proximal UP element subsites (W. Ross and R. L. Gourse, unpublished data; H. Chen and R. H. Ebright, unpublished data). The observation that KA271 causes modest defects in class II CRP-dependent transcription in vivo and in vitro at CC(−41.5) remains unexplained (23), as does the larger effect of substitutions in the 261 determinant in class I CRP-dependent transcription from CC(−61.5) and lacP1 in vivo than in our in vitro assay (Fig. 2 and 3) (25).
Models for class I and class II CRP-dependent transcription complexes are presented in Fig. 5. The key feature of these models is that the αCTD interaction with CRP involves the same contact surfaces when CRP is located at very different positions, i.e., upstream or downstream of αCTD (6). A similar situation has been reported recently for Fis-dependent activation at the rrn P1 (class I) and proP P2 (class II) promoters, where αCTD interactions with Fis are the same in both cases (1, 18). Furthermore, we speculate that αCTD interacts with both CRP and σ in the class I activation complex and that αCTD-AR1 and αCTD-σ interactions contribute to orienting αCTD on the DNA. Priorities for current work include a detailed analysis of the proposed interaction between the 261 determinant and σ subunit and confirmation of all proposed interactions by determination of high-resolution structures.
Acknowledgments
We thank W. Ross and S. Aiyar for helpful comments on the manuscript.
This work was supported by project grants from the BBSRC to S.B., by project grant 050794 from the Wellcome Trust to S.B. and M.T., by a short-term fellowship from the Human Frontier Science Program to N.J.S., by NIH grants GM37048 to R.L.G. and GM41376 to R.H.E., and by a Howard Hughes Medical Investigatorship to R.H.E.
REFERENCES
- 1.Aiyar, S. E., S. M. McLeod, W. Ross, C. A. Hirvonen, M. S. Thomas, R. C. Johnson, and R. L. Gourse. 2002. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol. 316:501-516. [DOI] [PubMed] [Google Scholar]
- 2.Attey, A., T. Belyaeva, N. Savery, J. Hoggett, N. Fujita, A. Ishihama, and S. Busby. 1994. Interactions between the cyclic AMP receptor protein and the alpha subunit of RNA polymerase at the E. coli gal P1 promoter. Nucleic Acids Res. 22:4375-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, A., K. Gaston, R. Williams, K. Chapman, A. Kolb, H. Buc, S. Minchin, J. Williams, and S. Busby. 1990. Mutations that affect the ability of the Escherichia coli cyclic AMP receptor protein to activate transcription. Nucleic Acids Res. 17:3865-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blatter, E. E., W. Ross, H. Tang, R. L. Gourse, and R. H. Ebright. 1994. Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell 78:889-896. [DOI] [PubMed] [Google Scholar]
- 5.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743-746. [DOI] [PubMed] [Google Scholar]
- 6.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 7.Busby, S., D. Kotlarz, and H. Buc. 1983. Deletion mutagenesis of the Escherichia coli galactose operon promoter region. J. Mol. Biol. 167:259-274. [DOI] [PubMed] [Google Scholar]
- 8.Gaal, T., W. Ross, E. Blatter, H. Tang, X. Jia, V. Krishnan, N. Assa-Munt, R. H. Ebright, and R. L. Gourse. 1996. DNA binding determinants of the alpha subunit of RNA polymerase: novel DNA binding domain architecture. Genes Dev. 10:16-26. [DOI] [PubMed] [Google Scholar]
- 9.Gaston, K., A. Bell, A. Kolb, H. Buc, and S. Busby. 1990. Stringent spacing requirements for transcription activation by CRP. Cell 62:733-740. [DOI] [PubMed] [Google Scholar]
- 10.Ghosaini, L. R., A. M. Brown, and J. M. Sturtevant. 1988. Scanning calorimetric study of the thermal unfolding of catabolite activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry 27:5257-5261. [DOI] [PubMed] [Google Scholar]
- 11.Gourse, R. L., W. Ross, and T. Gaal. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37:687-695. [DOI] [PubMed] [Google Scholar]
- 12.Jeon, Y. H., T. Negishi, M. Shirakawa, T. Yamazaki, N. Fujita, A. Ishihama, and Y. Kyogoku. 1995. Solution structure of the activator contact domain of the RNA polymerase α subunit. Science 270:1495-1497. [DOI] [PubMed] [Google Scholar]
- 13.Jeon, Y. H., T. Yamazaki, T. Otomo, A. Ishihama, and Y. Kyogoku. 1997. Flexible linker in the RNA polymerase α subunit facilitates the independent motion of the C-terminal activator contact domain. J. Mol. Biol. 267:953-962. [DOI] [PubMed] [Google Scholar]
- 14.Kainz, M., and R. L. Gourse. 1998. The C-terminal domain of the alpha subunit of Escherichia coli RNA polymerase is required for efficient rho-dependent transcription termination. J. Mol. Biol. 284:1379-1390. [DOI] [PubMed] [Google Scholar]
- 15.Kolb, A., K. Igarashi, A. Ishihama, M. Lavigne, M. Buckle, and H. Buc. 1993. Escherichia coli RNA polymerase, deleted in the C-terminal part of its α subunit, interacts differently with the cAMP-CRP complex at the lacP1 and at the galP1 promoter. Nucleic Acids Res. 21:319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolb, A., D. Kotlarz, S. Kusano, and A. Ishihama. 1995. Selectivity of the Escherichia coli RNA polymerase Eσ38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 23:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N.-R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 95:271-276. [DOI] [PubMed] [Google Scholar]
- 18.McLeod, S. M., S. E. Aiyar, R. L. Gourse, and R. C. Johnson. 2002. The C-terminal domains of the RNA polymerase α subunits: contact site with Fis and localization during coactivation with CRP at the Escherichia coli proP P2 promoter. J. Mol. Biol. 316:517-531. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Murakami, K., N. Fujita, and A. Ishihama. 1996. Transcription factor recognition surface on the RNA polymerase α subunit is involved in contact with the DNA enhancer element. EMBO J. 16:4358-4367. [PMC free article] [PubMed] [Google Scholar]
- 21.Niu, W., Y. Kim, G. Tau, T. Heyduk, and R. H. Ebright. 1996. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell 87:1123-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross, W., K. K. Gosink, J. Salomon, K. Igarishi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 23.Savery, N. J., G. S. Lloyd, M. Kainz, T. Gaal, W. Ross, R. H. Ebright, R. L. Gourse, and S. J. W. Busby. 1998. Transcription activation at class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 17:3439-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 25.Tang, H., K. Severinov, A. Goldfarb, D. Fenyo, B. Chait, and R. H. Ebright. 1994. Location, structure and function of the target of a transcriptional activator protein. Genes Dev. 8:3058-3067. [DOI] [PubMed] [Google Scholar]
- 26.Thomas, M. S., and R. E. Glass. 1991. Escherichia coli rpoA mutation which impairs transcription of positively regulated systems. Mol. Microbiol. 5:2719-2725. [DOI] [PubMed] [Google Scholar]
- 27.Williams, R. M., V. A. Rhodius, A. I. Bell, A. Kolb, and S. J. W. Busby. 1996. Orientation of functional activating regions in the Escherichia coli CRP protein during transcription activation at class II promoters. Nucleic Acids Res. 24:1112-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood, L. F., N. Y. Tszine, and G. E. Christie. 1997. Activation of P2 late transcription by P2 Ogr protein requires a discrete contact site on the C-terminus of the α subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 274:1-7. [DOI] [PubMed] [Google Scholar]
- 29.Yasuno, K., T. Yamazaki, Y. Tanaka, T. S. Kodama, A. Matsugami, M. Katahira, A. Ishihama, and Y. Kyogoku. 2001. Interaction of the C-terminal domain of the E. coli RNA polymerase α subunit with the UP element: recognizing the backbone structure in the minor groove surface. J. Mol. Biol. 306:213-225. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, Y., T. J. Merkel., and R. H. Ebright. 1994. Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP) II. Role at class I and class II CAP-dependent promoters. J. Mol. Biol. 243:603-610. [DOI] [PubMed] [Google Scholar]
- 31.Zou, C., N. Fujita, K. Igarashi, and A. Ishihama. 1992. Mapping the cAMP receptor protein contact site in the alpha subunit of Escherichia coli RNA polymerase. Mol. Microbiol. 6:2599-2605. [DOI] [PubMed] [Google Scholar]