Abstract
We have previously described a Pseudomonas gene, psrA, which enhances transcription of the rpoS sigma factor gene at stationary phase. We present molecular data which demonstrate that in Pseudomonas putida PsrA binds specifically to the rpoS and psrA promoters in DNA regions having similar palindromic sequences, C/GAAAC N2-4 GTTTG/C, where N is any nucleotide. The position of the initiation of transcription was determined for both promoters, and PsrA binds from positions −59 to −35 in the rpoS promoter and from −18 to +20 in the psrA promoter with respect to the +1 transcription site. Expression studies with a psrA-lacZ transcriptional fusion in wild-type and psrA::Tn5 knockout mutants revealed that psrA was under additional control in response to growth phase. A model for the role of PsrA in the regulation of rpoS and psrA is presented.
Bacteria continuously sense and respond to different environmental stimuli, including several stresses such as starvation, osmosis, oxidation, temperature change, and desiccation. In Pseudomonas spp., as in other gram-negative bacteria, adaptation to these stresses can take place via the changes in gene expression brought about by the stationary-phase sigma factor called RpoS (7, 12, 18). These global transcription responses occur via RpoS, which associates with core RNA polymerase and targets the transcription machinery to specific promoters. Consequently, the levels of RpoS, like those of all other bacterial sigma factors, need to be controlled since perturbations in RpoS concentration can have serious consequences. In Escherichia coli as well as in Pseudomonas, RpoS (also known as σs) levels increase in vivo during stationary phase (3, 6).
In E. coli, RpoS expression is controlled at the levels of transcription, translation, and protein stability (1, 5, 10, 14). The coordination of these different levels of regulation as well as the environmental signals leading to RpoS regulation remain unclear and are the subjects of extensive investigation. For Pseudomonas spp., regulation of rpoS expression has recently been addressed. The first indication that rpoS regulation in Pseudomonas differs from that in E. coli was the observation that the rpoS promoter was not functional in E. coli. Several regulators have been implicated as being responsible for RpoS accumulation, including the gacA/gacS two-component system (20), the cell-density-dependent regulation system known as quorum sensing (11, 21), and, recently, a TetR family regulator (8). The involvement of several regulators highlights the complexity of this process; thus, more studies are required to understand how rpoS expression is regulated in Pseudomonas.
Genetic studies have shown that a TetR family regulator called PsrA (26.3 kDa) is involved in stationary-phase-induced transcriptional regulation of rpoS and in negative autoregulation. This regulator is also present in Pseudomonas aeruginosa, and it has been shown to have a role in stationary-phase rpoS expression (8). This study provides molecular data which demonstrate that PsrA binds to the rpoS and psrA promoters. The PsrA DNA binding regions of both promoters contained palindromic sequences with high levels of identity.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise specified, cultures were grown in Luria-Bertani broth or on Luria-Bertani agar (15). Recombinant DNA techniques involved standard methods (15), and proteins were resolved by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (9). Triparental matings from E. coli to Pseudomonas were performed with an E. coli (pRK2013) helper strain (2). β-Galactosidase activity was measured as previously described (8); all measurements were done in triplicate, and mean values were obtained. Expression and purification of His6-PsrA were carried out as previously described (8). The molecular weight of the purified PsrA protein was determined by high-pressure liquid chromatography with a Progel-TSK G3000SWXL gel filtration column (7.8 mm by 30 cm; Supelco). RNA from a bacterial culture pellet (1 ml) was purified with an RNeasy kit (Qiagen) according to the manufacturer's instructions. Primer extensions were performed as described previously (15) with oligonucleotide rpoS−250 (5′-CCTTGACCTGCTGCCCCTCCC-3′), complementary to nucleotides −223 to −243 from the ATG start codon on the rpoS mRNA, and oligonucleotide psrA+60 (5′-TCTGCAAACCTCTTTCC-3′), complementary to nucleotides +56 to +73 from the ATG codon on the psrA RNA of Pseudomonas putida WCS358. The sequencing ladders presented were generated with the same primers used in the primer extension reaction.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| P. putida WCS358 | Wild type | Geels and Schippers (4) |
| P. putida MT17 | psrA::Tn5 mutant of WCS358, Kmr | Kojic and Venturi (8) |
| E. coli DH5α | Δ(lacZYA-argF) Nxs | Sambrook et al. (15) |
| Plasmids | ||
| pUC18 | Apr, ColE1 replicon | Yanisch-Perron et al. (22) |
| pBluescript KS | Apr, ColE1 replicon | Stratagene |
| pBluescript SK | Apr, ColE1 replicon | Stratagene |
| pREP-4 | lacI Kmr, p15A replicon | Qiagen |
| pRK2013 | Kmr Tra+ Mob+, ColE1 replicon | Figurski and Helinski (2) |
| pMP220 | Promoter probe vector, IncP1 Tcr | Spaink et al. (16) |
| pLAFR3 | Broad-host-range cloning vector IncP1, Tcr | Staskawizc et al. (17) |
| pMK962 | rpoS promoter cloned in pUC18 | Kojic and Venturi (8) |
| pRPO220B | rpoS promoter cloned in pMP220 | Kojic and Venturi (8) |
| pPPSR18 | psrA promoter cloned in pUC18 | Kojic and Venturi (8) |
| pPPSR220 | psrA promoter cloned in pMP220 | Kojic and Venturi (8) |
| pQEPSRA | psrA cloned in pQE30 | Kojic and Venturi (8) |
| pMKP25 | IncQ Cmr, contains psrA gene | Kojic and Venturi (8) |
| pPPSR221 | psrA promoter cloned in pMP220 | This study |
| pBSKPA | rpoS promoter cloned in pBluescript KS | This study |
| pBSKXS | psrA promoter cloned in pBluescript KS | This study |
| pRPO-XX | rpoS promoter cloned in pMP220 | This study |
| pRPO-TT | rpoS promoter cloned in pMP220 | This study |
| pRPO-PT | rpoS promoter cloned in pMP220 | This study |
| pRPO-PA | rpoS promoter cloned in pMP220 | This study |
| pRPO-TE | Part of rpoS promoter cloned in pMP220 | This study |
| pRPO-PE | Part of rpoS promoter cloned in pMP220 | This study |
| pRPO-ET | Part of rpoS promoter cloned in pMP220 | This study |
Apr,Kmr, Tcr, and Cmr, resistant to ampicillin, kanamycin, tetracycline, and chloramphenicol,respectively. Nxs, sensitive to nalidixic acid.
DNA mobility shift assays with purified His6-PsrA were performed as follows. Fragments carrying the psrA (XmnI-ScaI; 198-bp) and rpoS (PstI-AluI; 180-bp) promoters were purified from the plasmid constructs pBSKXS and pBSKPA, respectively, with KpnI-EcoRI and KpnI-HindIII restriction enzymes, respectively. Purified DNA (0.1 pmol) was labeled at its EcoRI and HindIII ends, respectively, with the Klenow fragment of DNA polymerase and [α-32P]dATP. Radiolabeled fragments (1,000 cpm) and various quantities of purified His6-PsrA (from 0 to 500 ng) were incubated for 15 min at room temperature in 20-μl reaction mixtures containing 1× binding buffer (10 mM HEPES [pH 7.5], 10 mM Tris [pH 7.5], 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 10% [vol/vol] glycerol), 20 μg of bovine serum albumin (carrier protein), and 20 μg of salmon sperm (nonspecific competitor) DNA. Supershifting was performed by incubating the reaction mixtures with anti-PsrA antibodies for an additional 15 min at room temperature. Samples were then loaded onto a nondenaturing 6% polyacrylamide 1× TBE (89 mM Tris, 89 mM boric acid, 1 mM EDTA)-7% (wt/vol) glycerol gel, which was prerun for 1 h at 100 V at 4°C, and the samples were run at 150 V.
DNase I protection assays were performed as follows. The 198-bp fragment carrying the psrA promoter was purified after digestion of the plasmid construct pBSKXS with the restriction enzymes EcoRI and KpnI or BamHI and PstI for 3′ end labeling of only one strand. The 180-bp fragment carrying the rpoS promoter was purified after digestion of the plasmid construct pBSKPA with the restriction enzymes HindIII and KpnI or EcoRI and PstI for 3′ end labeling of only one strand. The binding reactions were carried out in a 50-μl mixture with 2.5 μg of PsrA protein and 20,000 cpm of labeled DNA for 15 min at room temperature under the same conditions described above for the band shift mobility assays. To this reaction mixture, a 6-μl solution containing 10 mM MgCl2, 5 mM CaCl2, and 1 U of DNase I (Pharmacia) was added. The reactions were terminated after 1 min by the addition of 140 μl of stop solution (192 mM sodium acetate, 32 mM EDTA, 0.14% [wt/vol] sodium dodecyl sulfate, 64 μl of yeast tRNA). The reaction mixtures were extracted once with 200 μl of phenol-chloroform (1:1), followed by ethanol precipitations and two washing steps. As a sequencing marker for the determination of protected regions on the target DNA, G and A base-specific chemical cleavage was performed on the 3′-end-labeled fragments by the method of Maxam and Gilbert (13). Cleavage of DNA was done with 4% formic acid at 37°C for 30 min and with 1 M piperidine at 90°C for 30 min. The DNA fragments were precipitated and washed twice with n-butanol. The DNA pellets collected by centrifugation were resuspended in formamide loading dye at 1,500 cpm/μl. Denatured samples (2 μl) were loaded on an 8% acrylamide-7 M urea sequencing gel.
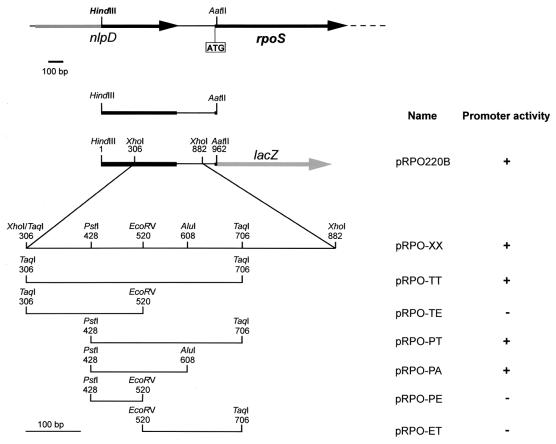
In order to further localize the rpoS promoter, several P rpoS-lacZ transcriptional fusion protein sequences were constructed with the P. putida WCS358 rpoS promoter (Fig. 1). It has been reported previously that the promoter was contained within the 920-bp fragment upstream from the ATG start codon, as demonstrated by use of the pRPO220B transcriptional fusion (Fig. 1) (8). Several subclones in the promoter probe vector pMP220 were further constructed, as shown in Fig. 1. This allowed the localization of the rpoS promoter within a 180-bp PstI-AluI DNA fragment (promoter construct pRPO-PA) (Fig. 1); more precisely, the promoter was positioned between bp 312 and 492 from the translational start codon of the rpoS gene. The transcriptional fusion proteins were either fully active (20,000 to 25,000 Miller units) or silent; in the psrA::Tn5 genomic mutant MT17, all the active fusion proteins had 90% reductions in activity (data not shown). Primer extension analysis revealed a single strong transcription start site, which was localized 373 bp upstream from the rpoS translational start codon (Fig. 2A). Figure 2D shows its position in the promoter sequence and also shows possible −10 and −35 regions.
FIG. 1.
Strategy for construction of rpoS-lacZ fusions. The genetic map shows the location of rpoS in the P. putida WCS358 genome as described previously (8). Plasmid construct pRPO220B (8) is the fusion sequence which was previously constructed and contains the rpoS promoter. Shown are DNA fragments which were cloned in the lacZ promoterless wide-host-range probe vector pMP220. All the fragments were previously cloned in pBluescript KS with the enzymes shown for the fragments and the corresponding sites in pBluescript KS, with the exception of the TaqI and AluI fragment, which was cloned into the ClaI and SmaI sites. From pBluescript KS, all the fragments were removed with BamHI and KpnI and cloned into the BglII-KpnI fragment in pMP220. The name of each resulting transcriptional fusion construct is given along with its promoter activity. +, promoter activity of 20,000 to 25,000 Miller units; −, no activity. No in-between levels of activity were observed.
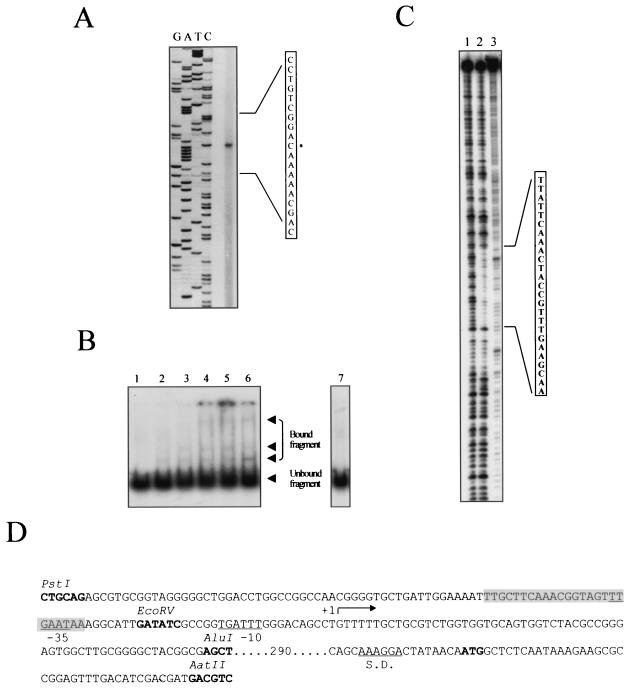
FIG. 2.
rpoS promoter studies. (A) Primer extension of the rpoS gene. The extension product is visible in the unlabeled lane. The DNA region of the extension product is amplified, and the position of the initiation of transcription is marked with an asterisk. (B) Retardation of the movement of a DNA fragment containing the rpoS promoter in gel by purified PsrA protein. The amounts of PsrA protein used were 0, 50, 125, 250, and 500 ng (lanes 1 to 5, respectively), and 250 ng was used with anti-PsrA antibodies (lane 6). A 50-fold excess amount of the same unlabeled DNA fragment was added in lane 7. (C) DNase I footprint of PsrA on the rpoS promoter region. Shown are the patterns of fragments resulting from digestion with DNase I of the 32P-labeled fragment upon binding with no protein (lane 1) and with 375 ng of purified PsrA (lane 2) and resulting from G and A base-specific chemical cleavage sequencing (lane 3). The sequence protected from DNase I is shown. (D) Sequence of the rpoS promoter. The +1, −10, and −35 sites, the ATG translation start codon, and the Shine-Dalgarno sequence (S.D.) are shown. The sequence inside the shaded box represents the region protected from DNase I, as indicated also in panel C.
To test whether the PsrA protein was able to bind to the rpoS promoter, mobility shift assays with the rpoS promoter (a PstI-AluI fragment of 180 bp was used) (see above and Fig. 1 and 2D) and purified PsrA were performed. The activity of the promoter was retarded, and the shift was not observed in the presence of excess unlabeled fragment. A supershift was detected in the presence of anti-PsrA antibodies (Fig. 2B). In order to precisely localize the PsrA DNA-binding region within the rpoS promoter, DNase I footprinting assays were performed on the same DNA fragment used for the gel shift assays. As shown in Fig. 2C, PsrA protected a region of approximately 25 bp covering positions −59 to −35 with respect to the +1 transcription site. This PsrA-protected region includes the palindromic sequence TTCAAACN4GTTTGAA (Fig. 2D), where N is any nucleic acid.
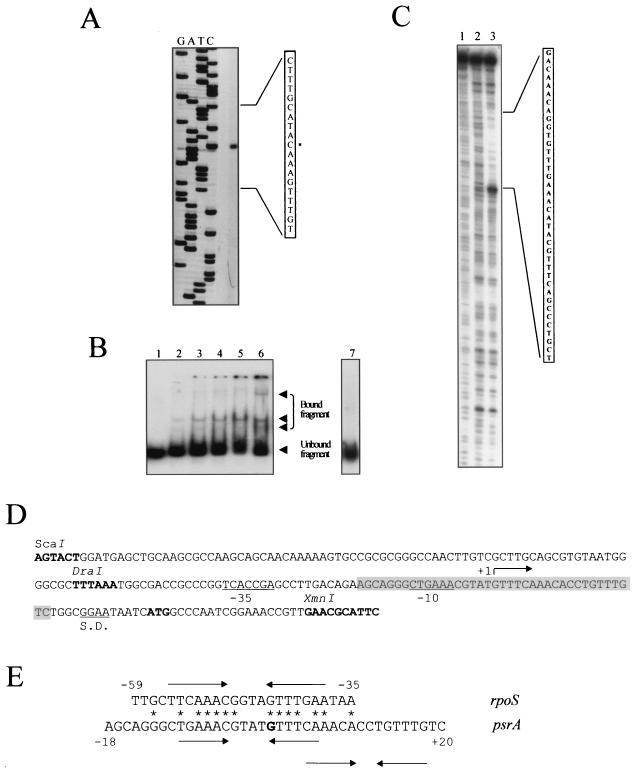
We also performed similar binding studies with the psrA promoter, since genetic studies demonstrated that PsrA negatively influences its own transcription (8). The psrA promoter is located within the 216-bp psrA-lexA intergenic region (8). Primer extension analysis was performed and revealed one strong clear signal, as shown in Fig. 3A, representing a single transcription start site which localized 33 bp upstream from the psrA translational initiation codon (Fig. 3D). The psrA promoter depicted in Fig. 3D as a 198-bp ScaI-XmnI fragment was first cloned in pBluescript (yielding pBSKXS) and then removed as a BamHI-KpnI fragment and cloned into the BglII and KpnI restriction sites of the promoter probe vector pPMP220 to yield pPPSR221. This fragment displayed promoter activity and was regulated by PsrA (see below), as was also observed with the previously reported pPPSR220 (8). The activity of this ScaI-XmnI fragment was retarded by the purified PsrA protein. It was not in the presence of excess unlabeled fragment, and a supershift was detected when anti-PsrA antibodies were added to the mixture (Fig. 3B). DNase I footprinting assays were also performed, and as shown in Fig. 3C, PsrA protected a region of 38 bp covering positions +20 to −18 with respect to the+1 transcription site. This PsrA-protected region included two palindromic sequences with high levels of identity to the one observed in the protected region of the rpoS promoter (Fig. 3C to E).
FIG. 3.
psrA promoter studies. (A) The extension product is visible in the unlabeled lane. The DNA region of the extension product is amplified, and the position of the initiation of transcription is marked with an asterisk. (B) Retardation of the movement of a DNA fragment containing the psrA promoter in gel by purified PsrA protein. The amounts of PsrA protein used were 0, 50, 125, 250, and 500 ng (lanes 1 to 5, respectively), and 250 ng was used with anti-PsrA antibodies (lane 6). A 50-fold excess amount of the same unlabeled DNA fragment was added in lane 7. (C) Patterns of fragments resulting from digestion with DNase I of the 32P-labeled fragment containing the psrA promoter upon binding with no protein (lane 2) and with 375 ng of purified PsrA (lane 3) and resulting from G and A base-specific chemical cleavage sequencing (lane 1). The sequence protected from DNase I is shown. (D) Sequence of the psrA promoter. The +1, −10, and −35 sites, the ATG translation start codon, and the Shine-Dalgarno sequence (S.D.) are shown. The sequence inside the shaded box represents the region protected from DNase I, as indicated also in panel C. (E) DNA comparison of the PsrA-protected region in the rpoS and psrA promoters. An asterisk indicates conservation of identical nucleotides, and arrows denote palindromic sequences. The numbers indicate the positions of the corresponding nucleotides with respect to the +1 transcription site (indicated in bold).
Our results show that PsrA binds specifically to the rpoS (from −59 to −35) and psrA (+20 to −18) promoters, which, together with previously reported genetic data, indicates that PsrA is likely to be an activator of rpoS and a negative autoregulator. Both binding sites contain palindromic sequences with high levels of identity (Fig. 3E), and gel filtration experiments with purified His6-PsrA demonstrated that the protein is a homodimer (data not shown). The PsrA-protected region in the psrA promoter is rather large, with two putative palindromic sequences, which could mean that PsrA cooperatively binds with varying intensities, resulting in different levels of repression.
We searched the P. aeruginosa PAO1 genome for the palindromic sequence G/CAAAC N2-4 GTTTG/C, which has dyad symmetry, in the PsrA-protected regions. The search identified 18 sequences which match the consensus sequence; of these, 11 mapped inside putative open reading frames (ORFs) whereas 7 mapped in putative promoter regions, as they were in intergenic regions upstream of the translation initiation codon (Table 2). Four of these sequences mapped upstream of ORFs which have not yet been studied; however, three have high homologies with known genes and proteins (Table 2). As expected, one sequence is in the rpoS promoter and another is in the psrA promoter of P. aeruginosa; thus, it is very likely that in P. aeruginosa, as in P. putida, PsrA targets the psrA and rpoS promoters. This is not surprising, as it has been shown previously that PsrA positively regulates rpoS expression in P. aeruginosa, and PsrA of P. aeruginosa is 90% identical to PsrA of P. putida (8). A PsrA putative DNA binding region was found in the intergenic region of the ptxR and ptxS regulatory genes involved in the transcriptional regulation of exotoxin A (19). It was reported that PtxS negatively autoregulates its own synthesis by binding to a region in the ptxS promoter; this region happens to have the same palindromic sequence that we report in this paper. It is not known whether PsrA binds to this region or whether PtxS binds to the rpoS and psrA promoters, since PsrA and PtxS belong to different bacterial regulatory families. Future work will test whether the two regulators are part of the same regulatory circuit.
TABLE 2.
PsrA binding sites in P. aeruginosa PAO1
| PsrA binding site | Position in PAO1 genomea | Putative promoterb |
|---|---|---|
| CAAACGCCTGTTTG | 564778-564791 | PA0506 |
| GAAACCGGTTTC | 2487773-2487784 | PA2258-PA2259 (ptxR-ptxS) |
| GAAACCGGTTTC | 2488926-2488937 | PA2260 |
| CAAACACTTGTTTG | 3367686-3367699 | PA3006 (psrA) |
| GAAACCAGCGTTTC | 4029672-4029685 | PA3595 |
| CAAACTTCCGTTTG | 4059323-4059336 | PA3622 (rpoS) |
| GAAACCGGTTTC | 5572071-5572082 | PA4963 |
| C/GAAACN2-4GTTTG/C |
The position in the P. aeruginosa PAO1 genome where the putative PsrA binding site was found. The P. aeruginosa chromosome is 6,264,403 bp in size.
The ORF in P. aeruginosa upstream of which the putative PsrA binding region was found. The name of the gene(s), if known, is given in parentheses.
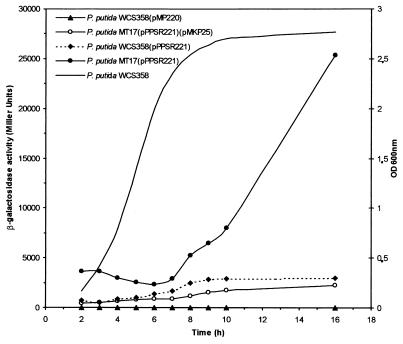
We analyzed the expression of psrA in different growth phases and in different genetic backgrounds by using the transcriptional-fusion construct pPPSR221 (Fig. 4). It was observed that in parent strain P. putida WCS358, in late log phase there was an almost three fold increase in the transcription of psrA; in the psrA::Tn5 mutant MT17, however, this increase in late log or early stationary phase was almost 10-fold. In the complemented MT17 mutant (i.e., harboring a plasmid with the psrA gene), this strong induction was no longer observed (Fig. 4). These results indicate that psrA expression is not constant throughout growth phase but that it is induced when the bacterium enters stationary phase. Furthermore, when PsrA is missing, the induction is much stronger. It is therefore possible that psrA, in addition to being negatively autoregulated, is positively regulated by an activator, which leads to a very strong induction in the absence of PsrA negative regulation. Future work is required to more precisely define psrA regulation.
FIG. 4.
psrA promoter activities. Shown are the activities of wild-type P. putida WCS358 and psrA::Tn5 strain MT17 harboring the psrA promoter-lacZ transcriptional fusion construct pPPSR221. Plasmid pMKP25 contains the intact psrA gene. The line without symbols indicates the P. putida WCS358 growth curve. OD 600 nm, optical density at 600 nm.
Understanding the regulation of rpoS is central to understanding the regulatory network that governs the expression of many stationary-phase-induced genes. It was reported previously that a regulator, PsrA, transcriptionally regulates rpoS in P. putida and P. aeruginosa (8), and in this study we confirm previous genetic evidence with molecular data and present a model (Fig. 5). More studies are required to further understand how the intracellular concentration of RpoS is monitored and why a psrA mutant exhibits an approximately 50% reduction of RpoS levels in stationary phase (8). Future work will also focus on posttranscriptional and posttranslational levels of control.
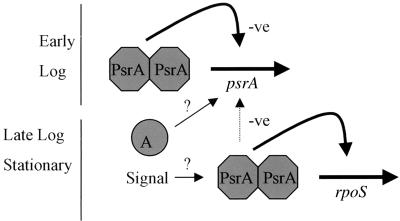
FIG. 5.
Working model for the regulation of rpoS and psrA by PsrA. In early and log phases of growth, PsrA negatively regulates psrA expression. In late log and stationary phases, this repression is partially relieved and rpoS expression is activated by PsrA. It is postulated that an activator protein (A) may be in part responsible for the activation of psrA expression, as deduced from psrA expression studies. It is currently unknown whether PsrA requires a ligand (e.g., a stationary-phase alarmone molecule) in order to be functional in rpoS activation. -ve, negative.
Acknowledgments
M.K. is a scientist on leave from the Institute of Molecular Genetics and Genetic Engineering, Belgrade, Yugoslavia, and is a recipient of an International Centre for Genetic Engineering & Biotechnology (ICGEB) fellowship. C.A. is a recipient of an ICGEB fellowship.
We are grateful to G. Degrassi, I. Bertani, and G. Devescovi for their interest and support. We thank K. Vlahovicek for computer analysis.
REFERENCES
- 1.Cunning, C., L. Brown, and T. Elliott. 1998. Promoter substitution and deletion analysis of upstream region required for rpoS translational regulation. J. Bacteriol. 180:4564-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujita, M., K. Tanaka, H. Takahashi, and A. Amemura. 1994. Transcription of the principal sigma-factor genes, rpoD and rpoS, in Pseudomonas aeruginosa is controlled according to the growth phase. Mol. Microbiol. 13:1071-1077. [DOI] [PubMed] [Google Scholar]
- 4.Geels, F. P., and B. Schippers. 1983. Reduction in yield depression in high frequency potato cropping soil after seed tuber treatments with antagonist fluorescent Pseudomonas spp. Phytopathol. Z. 108:207-221. [Google Scholar]
- 5.Gibson, K. E., and T. J. Silhavy. 1999. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator SprE. J. Bacteriol. 181:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jishage, M., A. Iwata, S. Ueda, and A. Ishihama. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178:5447-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jorgensen, F., M. Bally, V. Chapon-Herve, G. Michel, A. Lazdunski, P. Williams, and G. S. Stewart. 1999. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145:835-844. [DOI] [PubMed] [Google Scholar]
- 8.Kojic, M., and V. Venturi. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 10.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 11.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 12.Loewen, P. C., and R. Hengge-Aronis. 1994. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 13.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 14.Repoila, F., and S. Gottesman. 2001. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 183:4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Spaink, H. P., R. J. H. Okker, C. A. Wijffelmann, E. Pees, and B. J. J. Lugtemberg. 1987. Promoter in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 17.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suh, S.-J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. H. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swanson, B. L., J. A. Colmer, and A. N. Hamood. 1999. The Pseudomonas aeruginosa exotoxin A regulatory gene, ptxS: evidence for negative autoregulation. J. Bacteriol. 181:4890-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whistler, C. A., N. A. Corbell, A. Sarniguet, W. Ream, and J. E. Loper. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor σS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]