Abstract
The population structure of Legionella pneumophila was studied by using partial RNA polymerase gene (rpoB) and DotA gene (dotA) sequences. Trees inferred from rpoB sequences showed that two subspecies of L. pneumophila, Legionella pneumophila subsp. pneumophila and Legionella pneumophila subsp. fraseri, were clearly separated genetically. In both rpoB and dotA trees, 79 Korean isolates used in this study constituted six clonal populations, four of which (designated subgroups P-I to P-IV) were identified in L. pneumophila subsp. pneumophila and two of which (designated subgroups F-I and F-II) were identified in L. pneumophila subsp. fraseri. Although the relationships among subgroups were not identical, such subgrouping was congruent between the rpoB and dotA trees. Type strains of several serogroups did not belong to any subgroup, presumably because isolates similar to these strains were not present among our local sample of the population. There was evidence that horizontal gene transfer or recombination had occurred within L. pneumophila. Contrary to the phylogeny from rpoB and the taxonomic context, subgroups P-III and P-IV of L. pneumophila subsp. pneumophila proved to be closely related to those of L. pneumophila subsp. fraseri or showed a distinct clustering in the dotA tree. It can be inferred that dotA of subgroups P-III and P-IV has been transferred horizontally from other subspecies. The diverse distribution of serogroup 1 strains through the gene trees suggests that surface antigen-coding genes that determine serogroup can be exchanged. Thus, it can be inferred that genetic recombination has been important in the evolution of L. pneumophila.
Legionella pneumophila was first recognized in 1977 following an outbreak of pneumonia at the American Legion Convention in Philadelphia in 1976 (10) and is the causative agent of Legionnaires' disease. More than 40 Legionella species have been characterized, and at least 20 species are known to be pathogenic to humans (21).
DNA-DNA hybridization analyses (2) and an electrophoretic enzyme mobility study (37) have shown that L. pneumophila is not a homogeneous group. L. pneumophila is now classified into three subspecies, Legionella pneumophila subsp. pneumophila, Legionella pneumophila subsp. fraseri, and Legionella pneumophila subsp. pascullei, based on DNA-DNA hybridization tests (2). Serological tests have also shown that L. pneumophila is separated into 15 serogroups (SGs), most of which belong to L. pneumophila subsp. pneumophila. L. pneumophila subsp. fraseri includes strains of SGs 1, 4, 5, and 15 (2). Compared to the former two subspecies, only three strains of SG 5 isolated from water have been classified as L. pneumophila subsp. pascullei (2). L. pneumophila SG 1 is the most frequent human pathogen among the 15 SGs (28).
Although these methods have been useful in providing a basic population structure for L. pneumophila, which is the most important pathogen among Legionella species, fine grouping of the population structure can be different depending on the methods utilized. Moreover, DNA-DNA hybridization and electrophoretic enzyme mobility tests may be ambiguous and may reflect the genetic structure indirectly. Classification by serogrouping is also limiting because SGs 7 to 15 have no available routine serological assay, and its specificity is questionable (49). On the other hand, recent progress with the nucleotide sequencing technique makes gene analysis easier and more convenient. Compared to other methods, nucleotide sequencing has several advantages: (i) it provides direct genetic information and universal criteria, (ii) more variation can be identified than by hybridization and electrophoretic mobility, and (iii) sequences can be compared more easily with those of other sources or laboratories (19, 27). So far, the 16S rRNA gene is the most widely used molecular marker for phylogenetic studies of bacteria (52, 53). However, it was suggested that its lower variation is inadequate for the population study within a species (27), and its utility has been questioned because of its heterogeneity (6). Thus, the use of other protein-coding genes has been suggested as an alternative.
In several other species of pathogenic bacteria, such as Streptococcus pneumoniae (7), Neisseria meningitidis (8), and Helicobacter pylori (43), nucleotide sequence analysis at multiple protein-coding gene loci has helped us to understand the population structure of such pathogens, as well as providing better information on the epidemiology of pathogenic bacteria. In this study, the molecular sequences of a housekeeping gene (rpoB) and a virulence-related gene (dotA) were used to study the genetic population structure of L. pneumophila. rpoB encodes the β-subunit of DNA-dependent RNA polymerase (26, 38). Rifampin resistances of Escherichia coli and Mycobacterium tuberculosis are related to mutations in a particular region of the rpoB (39, 44). Recently, rpoB sequences were used as an alternative tool in the phylogeny and identification of enteric bacteria (22), Mycobacterium (17), spirochetes such as Borrelia (18, 33), and Bartonella (32). It was suggested that the use of rpoB could avoid the limitations of the 16S rRNA gene, which has been used broadly in the phylogeny and identification of bacteria (6), but it has not been used in the study of population structure within particular microbial species until now.
dotA is known to be related to the virulence of L. pneumophila (1, 36, 46, 47, 51) and is regarded as a pathogenicity island, such as cagA in H. pylori, hly in uropathogenic E. coli, or vir complex in Agrobacterium tumefaciens (11, 12, 16). L. pneumophila with a mutation in dotA cannot replicate intracellularly because it is unable to alter the endocytic pathway of macrophages (1, 47). According to Southern hybridization tests, other Legionella species, such as Legionella micdadei, harbor sequences homologous to dotA as well as other genes related to the Icm/Dot transfer system (16), although they have not been identified yet.
MATERIALS AND METHODS
L. pneumophila strains.
Eighteen L. pneumophila reference strains (Table 1) and 79 Korean isolates were used in this study. Of the 79 isolates, three were isolated from lung tissues of pneumonia patients and the others were isolated from air-conditioning cooling tower water between the years 1985 and 2000.
TABLE 1.
L. pneumophila reference strains used in this study
| Subspecies | Serogroup | Strain | Accession no.
|
|
|---|---|---|---|---|
| rpoB | dotA | |||
| L. pneumophila subsp. pneumophila | 1 | ATCC 33152 (Philadelphia-1) | AF367748 | AY036018 |
| L. pneumophila subsp. pneumophila | 1 | ATCC 33153 (Knoxville-1) | AY036036 | AY036019 |
| L. pneumophila subsp. pneumophila | 1 | SF9 | AY036037 | AY036020 |
| L. pneumophila subsp. pneumophila | 1 | ATCC 43109 (OLDA) | AY036038 | AY036021 |
| L. pneumophila subsp. pneumophila | 2 | ATCC 33154 (Togus 1) | AY036039 | AY036022 |
| L. pneumophila subsp. pneumophila | 3 | ATCC 33155 (Bloomington 2) | AY036040 | AY036023 |
| L. pneumophila subsp. fraseri | 4 | ATCC 33156 (Los Angeles 1) | AY036041 | AY036024 |
| L. pneumophila subsp. fraseri | 5 | ATCC 33216 (Dallas 1E) | AY036042 | AY036025 |
| L. pneumophila subsp. pneumophila | 6 | ATCC 33215 (Chicago 2) | AY036043 | AY036026 |
| L. pneumophila subsp. pneumophila | 7 | ATCC 33823 (Chicago 8) | AY036044 | AY036027 |
| L. pneumophila subsp. pneumophila | 8 | ATCC 35096 (Concord 3) | AY036045 | AY036028 |
| L. pneumophila subsp. pneumophila | 9 | ATCC 35289 (International 23) | AY036046 | AY036029 |
| L. pneumophila subsp. pneumophila | 10 | ATCC 43283 (Leiden 1) | AY036047 | AY036030 |
| L. pneumophila subsp. pneumophila | 11 | ATCC 43130 (797-PA-H) | AY036048 | AY036031 |
| L. pneumophila subsp. pneumophila | 12 | ATCC 43290 (570-CO-H) | AY036049 | AY036032 |
| L. pneumophila subsp. pneumophila | 13 | ATCC 43736 (82A3105) | AY036050 | AY036033 |
| L. pneumophila subsp. pneumophila | 14 | ATCC 43703 (1169-MN-H) | AY036051 | AY036034 |
| L. pneumophila subsp. fraseri | 15 | ATCC 35351 (Lansing 3) | AY036052 | AY036035 |
DNA extraction.
DNA was extracted by the bead-beater-phenol extraction method (17). In a 2.0-ml screw-cap microcentrifuge tube, 100 μl of cell suspension was placed with 100 μl of phenol-chloroform-isopropyl alcohol (50:49:1) and 100 μl (packed volume) of glass beads. To disrupt the cells, the tube was oscillated on a mini-bead beater (Biospec Products) for 30 s at 5,000 rpm. The tube was centrifuged at 15,000 rpm for 10 min, and the aqueous phase was transferred into a new tube. The DNA was precipitated with isopropyl alcohol with the same volume and dissolved with 60 μl of deionized distilled water. It was used as a template for PCR.
PCR amplification.
A pair of primers, RL1 (5′-GAT GAT ATC GAT CAY CTD GG-3′) and RL2 (5′-TTC VGG CGT TTC AAT NGG AC-3′), was used to amplify the rpoB DNA (369 bp) containing the highly conserved Rifr region (17, 24, 39). The sequence variations of the corresponding region are very useful for discriminating species of the genera Mycobacterium (17) and Borrelia (18).
Another pair of primers, DL1 (5′-TTG ATT TGG TGA AAC TCA ATG G-3′) and DL2 (5′-CAA TCA AAA TCC TGG TGC TTC-3′), was applied to amplify the dotA DNA (434 bp) (1). Template DNA (ca. 50 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer, Daejeon, Korea), which contained 1 U of Taq DNA polymerase, each deoxynucleoside triphosphate at a concentration of 250 μM, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and gel loading dye (18). The final volume was adjusted to 20 μl with distilled water. The reaction mixture was subjected to 30 cycles for amplification. Each cycle consisted of 30 s at 95°C for denaturation, 30 s at 55°C for annealing, and 1 min at 72°C for extension; a final extension at 72°C was carried out for 5 min (model 9700 Thermocycler; Perkin-Elmer Cetus). The amplified PCR products were detected on 1.5% agarose gels stained by ethidium bromide and were purified for the sequencing process with a QIAEX II gel extraction kit (Qiagen, Hilden, Germany).
Nucleotide sequencing.
The partial rpoB and dotA sequences of the purified PCR products were directly determined with forward and reverse primers by using an Applied Biosystems model 377 automated sequencer and a BigDye terminator cycle sequencing kit (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom). For the sequencing reaction, 30 ng of purified PCR products, 2.5 pmol of primer, and 4 μl of BigDye terminator RR mix (Perkin-Elmer Applied Biosystems; no. 4303153) were mixed and adjusted to a final volume of 10 μl with distilled water. The reaction was run with 5% (vol/vol) dimethyl sulfoxide for 30 cycles of 15 s at 95°C, 5 s at 50°C, and 4 min at 60°C. Both strands were sequenced as a cross-check.
Sequence analysis.
The rpoB and dotA sequences (300 and 360 bp, respectively) were aligned with the multiple-alignment program CLUSTAL X (45). The amino acid sequences were deduced in the MegAlign program (Windows version 3.12e; DNASTAR, Madison, Wis.), and compared with published sequences (1, 24). The phylogenetic trees were inferred from both DNA sequences by the parsimony and neighbor-joining methods in PAUP (Phylogenetic Analysis Using Parsimony, version 4; Sinauer Associates, Sunderland, Mass.). In the rpoB tree, Legionella gormanii was selected as an outgroup, while a midpoint rooting option was applied to root the tree due to the absence of a reliable outgroup in dotA. The branch supporting values were evaluated with 500 bootstrap replications (9, 14). The nucleotide substitution in dotA was analyzed by measuring the ratio of synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dN), which were estimated by using the SNAP program (www.mlst.net) based on the method of Nei and Gojobori (23) and incorporating a statistic developed by Ota and Nei (25).
Nucleotide sequence accession numbers.
The rpoB and dotA DNA sequences determined in this study were submitted to the GenBank database. The accession numbers of 18 reference strains are given in Table 1.
RESULTS AND DISCUSSION
Subspecies of L. pneumophila.
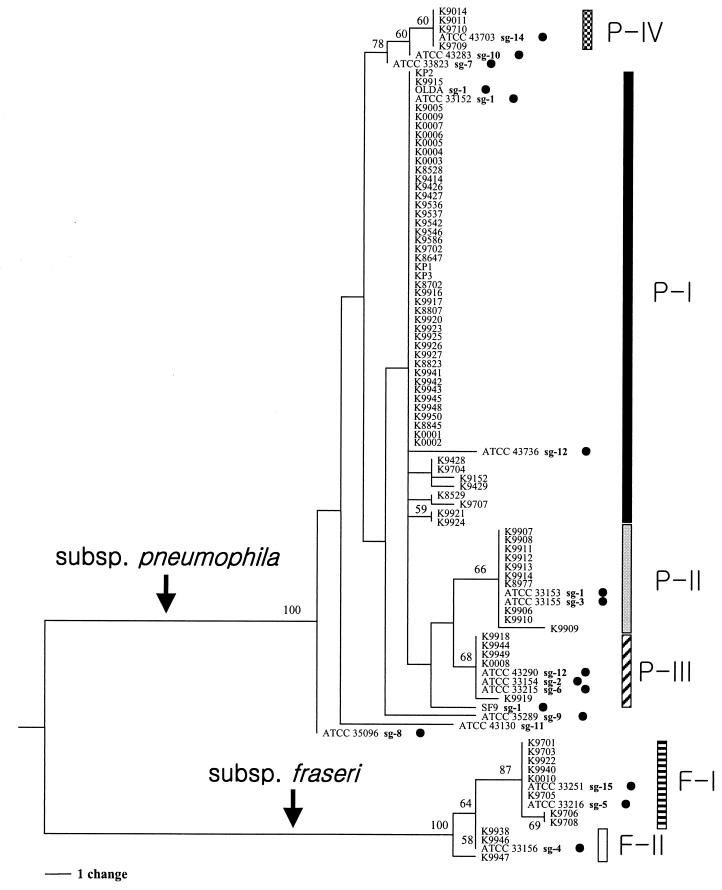
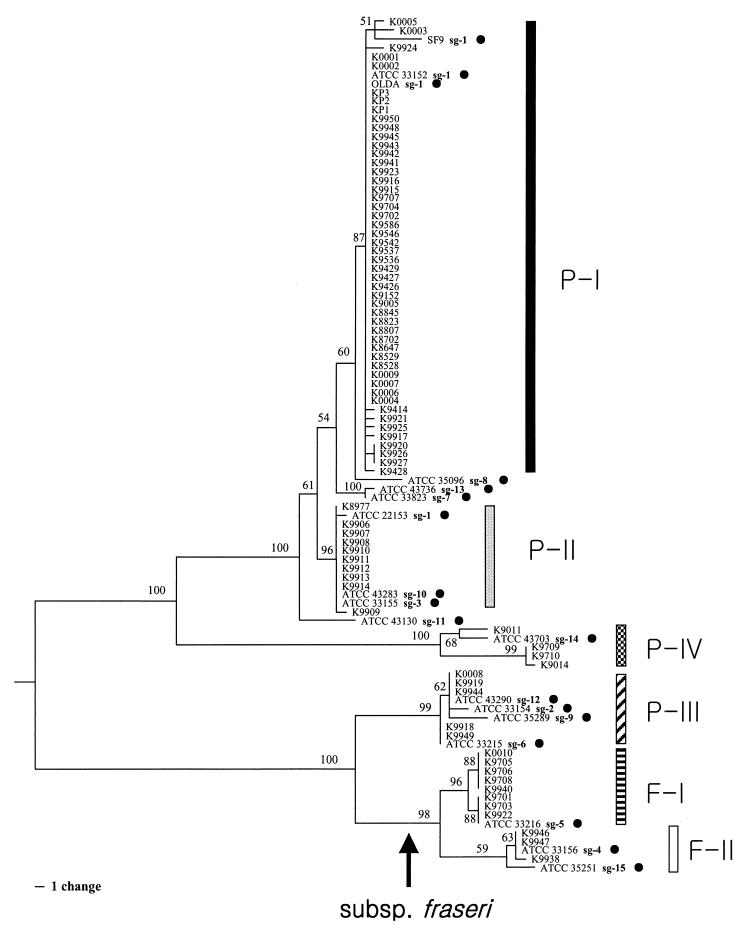
The phylogenetic relationships of 97 L. pneumophila strains including 18 reference strains, which were inferred from the rpoB and dotA sequences by the parsimony method, are shown in Fig. 1 and 2, respectively. Tree topologies inferred by the neighbor-joining method were not significantly different from those inferred by the parsimony method. Both the rpoB and dotA trees showed the existence of six distinct subgroups (designated P-I to P-IV, F-I, and F-II), which were also grouped in the neighbor-joining trees without any exception. Subgroups P-I, P-II, P-III, and P-IV belonged to L. pneumophila subsp. pneumophila, and subgroups F-I and F-II belonged to L. pneumophila subsp. fraseri (2). rpoB gave more reliable relationships within L. pneumophila, because two subspecies were distinctly separated in the rpoB tree but not in the dotA tree. However, L. pneumophila subsp. pneumophila and L. pneumophila subsp. fraseri are separated genetically in both gene trees, despite a discrepancy in the relationships among subgroups. Although subgroup P-III was closely related to subgroups F-I and F-II in the dotA tree (Fig. 2), they did not intermix with L. pneumophila subsp. fraseri.
FIG. 1.
rpoB tree of L. pneumophila inferred by the parsimony method in PAUP. This tree is one of the 82 most parsimonious trees which required 70 steps. The rpoB sequences of L. gormanii were used as an outgroup to root this tree. Subgroups are indicated by specific bars on the right, and branches leading to each subspecies are also included. The branch lengths are proportional to changes in the nucleotides. Branches supported by values higher than 50% in the bootstrap analysis (500 replications) are indicated.
FIG. 2.
dotA tree of L. pneumophila based on nucleotide sequences. This tree is one of the 45 most parsimonious trees, which required 193 steps. It was constructed by the procedure described for Fig. 1. Due to the absence of a reliable outgroup in dotA, the midpoint rooting method was applied to root this tree. Branches supported by values higher than 50% in the bootstrap analysis (500 replications) are indicated.
The DNA-DNA relatedness among the three subspecies (i.e., L. pneumophila subsp. pneumophila, L. pneumophila subsp. fraseri, and L. pneumophila subsp. pascullei) showed 66 to 74% similarities (2), which is very close to the species limit (50). According to 16S rRNA gene analysis, L. pneumophila subsp. pneumophila and L. pneumophila subsp. fraseri showed a 99.2% sequence similarity (15), which is within the species boundaries (41, 50). The difference in biochemical traits among the three subspecies has not been determined (2), but the population genetic analysis using the protein-coding genes rpoB and dotA in this study indicated that they are genetically distinct. The morphological phenotypes or biochemical traits of bacteria do not always coincide with their genotypes. Genetic divergence may not be revealed at the phenotypic level because it occurred too recently and/or it is simply not yet detectable.
Generally, bacterial species have been defined as strains with at least 70% DNA-DNA relatedness and/or sharing more than 97% 16S rRNA gene sequence similarity (41, 50). However, these cutoff values are arbitrary and not guaranteed to identify populations of bacteria that correspond to real ecological units (27). It was reported that the 16S rRNA gene lacks resolving capacity below the species level (35). In addition, individual strains may have two or more 16S rRNA genes with relatively high sequence dissimilarity (35, 48). Thus, clustering based on the sequences of several protein-coding genes has been recommended as a primary criterion for delimiting taxa (27). The results of this study support the statement that it is necessary to use several protein-coding genes in a phylogenetic or population study of bacteria (19, 40).
Comparison of nucleotides and deduced amino acids.
The measure of similarity for rpoB was slightly lower than that for dotA. For the nucleotide sequences of rpoB, 97.0 to 100% similarity was observed among the strains of L. pneumophila subsp. pneumophila and 98.3 to 100% similarity was observed among the L. pneumophila subsp. fraseri strains. The strains that belong to two different subspecies showed 87.3 to 89.7% similarity. However, the deduced amino acids in the rpoB sequence were all the same in spite of such nucleotide differences.
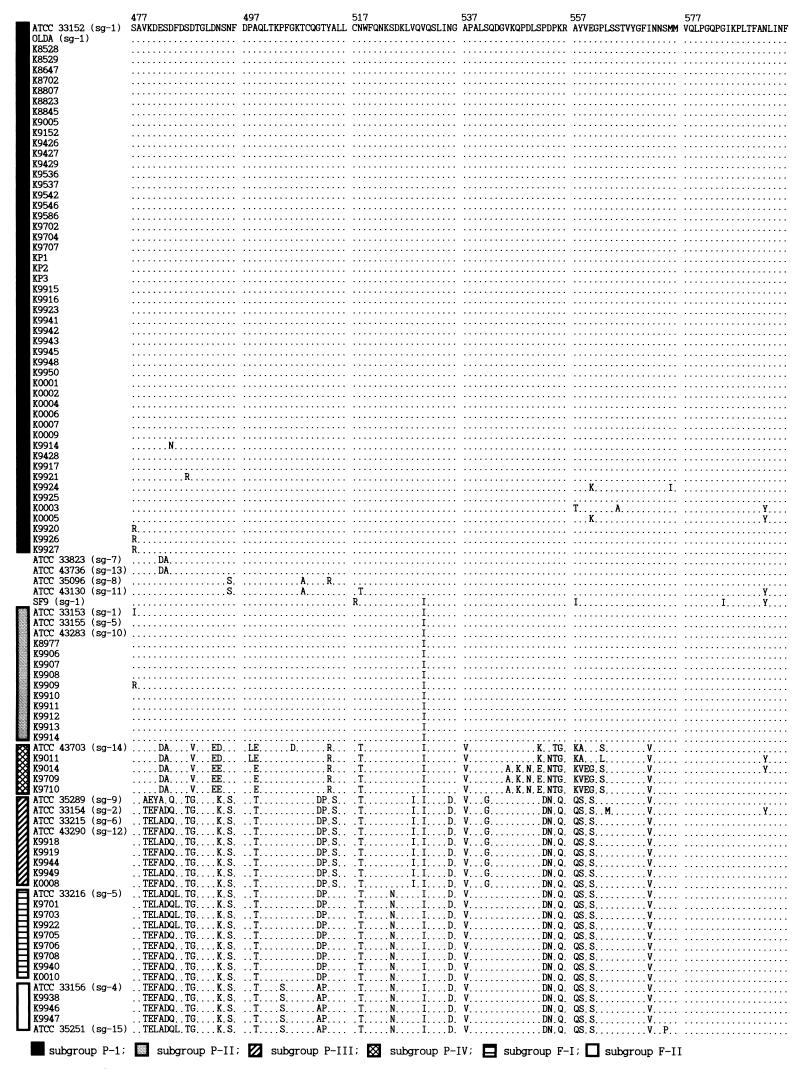
The overall nucleotide sequence similarities of dotA among the 97 strains used in this study were 78.1 to 100%. The nucleotides sequences of dotA within each subgroup were nearly identical, as in rpoB, except for subgroup P-IV (96.1 to 98.3%). However, the deduced amino acid sequences of dotA (S477 to F596 in reference 1) were remarkably different among the subgroups compared to the same amino acid sequences of rpoB. There was a single amino acid variation (V530→I530) between the strain Philadelphia-1 (ATCC 33152) and all of the strains belonging to subgroup P-II except Knoxville-1 (ATCC 33153) and K9909 (Fig. 3). On the other hand, there were 19 to 24 amino acid substitutions between subgroups P-I and P-IV, and there were 27 to 28 substitutions between subgroups P-I and P-III. Subgroups F-I and F-II differed by three or four amino acids. There were 25 to 28 amino acid substitutions between Philadelphia-1 and L. pneumophila subsp. fraseri (subgroups F-I and F-II) (Fig. 3).
FIG. 3.
Deduced amino acid sequences (S477 to F596) in reference 1 of dotA. Bars on the left represent subgroups. Amino acids identical to those in ATCC 33152 are represented by dots. Contrary to dotA, the deduced amino acid sequences of rpoB were identical in all strains of the L. pneumophila used in this study.
This distinction of the amino acid differences between the two protein-coding genes can be explained by their nature. As a housekeeping gene, rpoB encodes the RNA polymerase β-subunit, and there is a strong aversion to alteration of amino acids, so most of the base substitutions are usually synonymous. Contrary to rpoB, dotA is related to the virulence of L. pneumophila (1, 36, 46, 47, 51). Unlike in the RNA polymerase, a diversity of amino acids in DotA occurred via lateral gene transfer and/or point mutation. A high dS/dN ratio (7.29) showed no evidence of positive selection for the amino acid change (25) in dotA.
Distribution of serogroups within the population structure of L. pneumophila.
The positions of the reference strains are presented in Table 2. Of these, the strains of SG 13 (ATCC 43736) and SG 1 (SF9) differed in position between the two trees (Fig. 1 and 2; Table 2). Meanwhile, SG 13 in the rpoB tree and SF9 of SG 1 in the dotA tree showed several nucleotide differences from the others, so they were not likely to belong to subgroup P-I in a strict sense. In L. pneumophila subsp. fraseri, the placement of ATCC 35251 (SG 15 or Lansing 3) was unclear in that it was within subgroup F-I in the rpoB tree and within subgroup F-II in the dotA tree (Table 2).
TABLE 2.
Distribution of serogroups of L. pneumophila
| Gene | SG (strain) belonging to subgroup
|
||||||
|---|---|---|---|---|---|---|---|
| P-I | P-II | P-III | P-IV | F-I | F-II | Othera | |
| rpoB | 1 (Philadelphia-1) | 1 (Knoxville-1) | 2 | 14 | 5 | 4 | 1 (SF9) |
| 1 (OLDA) | 3 | 6 | 15 | 7 | |||
| 13b | 12 | 8 | |||||
| 9 | |||||||
| 10 | |||||||
| 11 | |||||||
| dotA | 1 (Philadelphia-1) | 1 (Knoxville-1) | 2 | 14 | 5 | 4 | 7 |
| 1 (OLDA) | 3 | 6 | 15 | 8 | |||
| 1 (SF9)b | 10 | 9 | 11 | ||||
| 12 | 13 | ||||||
Reference strains not belonging to any subgroup in this study.
Strain not belonging to subgroup P-I in a strict sense.
ATCC 33823 (SG 7), ATCC 35096 (SG 8), ATCC 43283 (SG 10), and ATCC 43130 (SG 11) did not belong to any subgroup in either the rpoB or dotA trees. Strains not belonging to any subgroup may exist because only the Korean strains were used in this study. If more globally collected isolates were included, more than six clonal populations might be present.
Four reference strains of SG 1 were included in this study. They did not cluster into a single subgroup in either of the gene trees. While ATCC 33152 (type strain) and OLDA both belonged to subgroup P-I, Knoxville-1 (ATCC 33153) was part of subgroup P-II. Strain SF9 did not cluster into any subgroup in the rpoB tree (Fig. 1 and 2).
A diverse distribution of SG 1 strains through the L. pneumophila populations was also observed in previous studies using other genes, such as mip (31) and the intergenic 23S-5S ribosomal spacer (34). At times, certain SGs shared identical rpoB and dotA alleles, such as SGs 1 and 3 in subgroup P-II and SGs 2, 6, and 12 in subgroup P-III (Table 2). Such distribution of SGs suggested that serotyping does not accurately correspond to genetic structure in L. pneumophila.
An SG reflects variations in most likely a small number of genetic loci that encode the antigenic protein (37). As each subgroup showed clonality (Fig. 1 and 2), it is not likely that L. pneumophila has evolved in accordance with SG. The most plausible explanation is that the surface antigen-coding genes that determine SG have been transferred horizontally within the population (7, 30). Recombinational exchanges at the serotype-coding capsular genes in S. pneumoniae have been reported (3, 4, 5). Such gene exchanges that alter SG may be strongly selected by the host immune system (7).
Horizontal gene exchange of dotA.
Subgroups P-III and P-IV showed incongruent placements between the rpoB and dotA trees (Fig. 1 and 2). In the rpoB tree, the two subspecies in L. pneumophila were clearly separated into two groups (Fig. 1), precisely reflecting their taxonomic relationships. However, subgroup P-III, which included reference strains of SGs 2, 6, 9, and 12, showed a close relationship with L. pneumophila subsp. fraseri, even though they are L. pneumophila subsp. pneumophila (2, 37). Subgroup P-IV, which included a reference strain of SG 14, also formed a distinct cluster in the dotA tree. The phylogeny inferred from the deduced amino acid sequences of the dotA was very similar to that seen in Fig. 2 (not shown).
As an explanatory hypothesis, it is suggested that the region containing dotA of L. pneumophila subsp. fraseri was horizontally transferred to subgroup P-III of L. pneumophila subsp. pneumophila. L. pneumophila has been reported to be naturally transformable (20, 42), and the competence of the Legionella species makes it possible for them to exchange genes naturally (13, 20, 29). Additionally, the Icm/Dot coding gene cluster has been believed to be a pathogenicity island (11, 16). Pathogenicity islands are often flanked by small directly repeated sequences, are often associated with transfer RNA genes, and/or often carry genes encoding mobility factors, such as integrases, transposases, and insertion sequence elements (11, 12). Therefore, the Icm/Dot coding gene cluster in L. pneumophila can also be horizontally transferred more readily as pathogenicity islands in other pathogens (11).
The origin of dotA of subgroup P-IV, which includes SG 14, was not clear in this study. Its phylogenetic position was distinct in the dotA tree (Fig. 2), and the deduced amino acids were unique at 17 sites from Asp482 to Ser562 or Leu562 (Fig. 3). Most likely, the dotA of subgroup P-IV originated from sources other than L. pneumophila subsp. pneumophila or L. pneumophila subsp. fraseri. Although it was so genetically divergent, it was more closely related to L. pneumophila subsp. pneumophila than to L. pneumophila subsp. fraseri (Fig. 2). Based on the phylogenetic relationships, the origin of dotA of subgroup P-IV might be within the species boundary of L. pneumophila, so it is probable that the dotA of subgroup P-IV originated from another subspecies of L. pneumophila, possibly L. pneumophila subsp. pascullei. Unfortunately, it could be not confirmed because L. pneumophila subsp. pascullei was not included in this study.
In conclusion, the results of this study confirmed that the division of L. pneumophila into two subspecies is supported by the rpoB data, which gave more reliable relationships than those of dotA. It was also evident that recombination has occurred among the populations of L. pneumophila, since there are clear incongruencies in the dotA tree and a poor correlation between serogroup and genotype. These subgroupings based on rpoB and dotA sequences will provide a valuable tool that can supplement identification by culture or serological test in the epidemiological study of Legionnaires' disease.
Acknowledgments
This work was supported by grant 2001 from Seoul National University College of Medicine and the Hospital Research Fund and in part by the BK21 Project for Medicine, Dentistry, and Pharmacy.
REFERENCES
- 1.Berger, K. H., J. J. Merriam, and R. R. Isberg. 1994. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol. Microbiol. 14:809-822. [DOI] [PubMed] [Google Scholar]
- 2.Brenner, D. J., A. G. Steigerwalt, P. Epple, W. F. Bibb, R. M. McKinney, R. W. Starnes, J. M. Colville, R. K. Selander, P. H. Edelstein, and C. Wayne Moss. 1988. Legionella pneumophila serogroup Lansing 3 isolated from a patient with fatal pneumonia, and descriptions of L. pneumophila subsp. pneumophila subsp. nov. L. pneumophila subsp. fraseri subsp. nov., and L. pneumophila subsp. pascullei subsp. nov. J. Clin. Microbiol. 26:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claverys, J.-P., M. Prudhomme, I. Mortier-Barrière, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35:251-259. [DOI] [PubMed] [Google Scholar]
- 4.Coffey, T. J., M. Daniels, M. C. Enright, and B. G. Spratt. 1999. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology 145:2023-2031. [DOI] [PubMed] [Google Scholar]
- 5.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryneiwicz, J.C. Paton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 6.Dahllöf, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 8.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1985. Confidence limits in phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, D. D., D. L. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepard, and P. S. Brachman. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189-1197. [DOI] [PubMed] [Google Scholar]
- 11.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 12.Hacker, J., G. Blum-Oehler, I. Mühldorfer, and H. Tschäpe. 1997. Pathogenicity islands of virulent bacterial: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 13.Håvarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillis, D. M., and J. J. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 15.Hookey, J. V., N. A. Sauders, N. K. Fry, R. J. Birtles, and T. G. Harrison. 1996. Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like amoebal pathogens. Int. J. Syst. Bacteriol. 46:526-531. [Google Scholar]
- 16.Joshi, A. M., and M. S. Swanson. 1999. Comparative analysis of Legionella pneumophila and Legionella micdadei virulence traits. Infect. Immun. 67:4134-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, B.-J., S.-H. Lee, M.-A. Lyu, S.-J. Kim, G.-H. Bai, S.-S. Kim, G.-T. Chae, E.-C. Kim, C.-Y. Cha, and Y.-H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 37:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, S.-H., B.-J. Kim, J.-H. Kim, K.-H. Park, S.-J. Kim, and Y.-H. Kook. 2000.. Differentiation of Borrelia burgdorferi sensu lato on the basis of RNA polymerase gene (rpoB) sequences. J. Clin. Microbiol. 38:2557-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiden, M. C. J., J. A. Bygraves, D. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mintz, C. S. 1999. Gene transfer in Legionella pneumophila. Microb. Infect. 1:1203-1209. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto, H., H. Yamamoto, K. Arima, J. Fujii, K. Maruta, K. Izu, T. Shiomori, and S.-I. Yoshida. 1997. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63:2489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mollet, C., M. Drancourt, and D. Raoult. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005-1011. [DOI] [PubMed] [Google Scholar]
- 23.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, K., P. Hindersson, N. Høiby, and J. M. Bangsborg. 2000. Sequencing of the rpoB gene in Legionella pneumophila and characterization of mutations associated with rifampin resistance in the Legionellaceae. Antimicrob. Agents Chemother. 44:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ota, T., and M. Nei. 1994. Variance and covariances of the numbers of synonymous and nonsynonymous substitution per site. Mol. Biol. Evol. 11:613-619. [DOI] [PubMed] [Google Scholar]
- 26.Ovchinnikov, Y. A., G. S. Monstyrskaya, V. V. Gubanov, S. O. Guryev, O. Y. Chertov, N. N. Modyanov, V. A. Grinkevich, I. A. Makarova, T. V. Marchenko, I. N. Polovnikova, V. M. Lipkin, and E. D. Sverdlov. 1981. The primary structure of Escherichia coli RNA polymerase. Eur. J. Biochem. 116:621-629. [DOI] [PubMed] [Google Scholar]
- 27.Palys, T., K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 28.Pasculle, W. 2000. Update on Legionella. Clin. Microbiol. Newsl. 22:97-101. [Google Scholar]
- 29.Poulsen, K., J. Reinholdt, C. Jespersgaard, K. Boye, T. A. Brown, M. Hauge, and M. Kilian. 1998. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombinational within and between species. Infect. Immun. 66:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratcliff, R. M., J. A. Lanser, P. A. Manning, and M. W. Heuzenroeder. 1998. Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 36:1560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renesto, P., J. Gouvernet, M. Drancourt, V. Roux, and D. Raoult. 2001. Use of rpoB gene analysis for detection and identification of Bartonella species. J. Clin. Microbiol. 39:430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renesto, P., K. Lorvellec-Guillon, M. Drancourt, and D. Raoult. 2000. rpoB gene analysis as a novel strategy for identification of spirochetes from the genera Borrelia, Treponema, and Leptospira. J. Clin. Microbiol. 38:2200-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson, P. N., B. Heidrich, F. Tiecke, F. J. Fehrenbach, and A. Rolfs. 1996. Species-specific detection of Legionella using polymerase chain reaction and reverse dot-blotting. FEMS Microbiol. Lett. 140:111-119. [DOI] [PubMed] [Google Scholar]
- 35.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 36.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 37.Selander, R. K., R. M. McKinney, T. S. Whittam, W. F. Bibb, D. J. Brenner, F. S. Nolte, and P. E. Pattison. 1985. Genetic structure of populations of Legionella pneumophila. J. Bacteriol. 163:1021-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Severinov, K., A. Mustaev, A. Kukarin, O. Muzzin, I. Bass, S. A. Darst, and A. Goldfarb. 1996. Structural modules of the large subunits of RNA polymerase. J. Biol. Chem. 271:27969-27974. [DOI] [PubMed] [Google Scholar]
- 39.Severinov, K., M. Soushko, A. Goldfarb, and V. Nikiforov. 1993. Rifampicin region revisited. J. Biol. Chem. 268:14820-14825. [PubMed] [Google Scholar]
- 40.Spratt, B. G., and M. C. J. Maiden. 1999. Bacterial populations genetics, evolution and epidemiology. Philos. Trans. R. Soc. Lond. 354:701-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 42.Stone, B. J., and Y. A. Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suerbaum, S., J. Maynard Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Telenti, A., N. Honoré, C. Bernasconi, J. March, A. Ortega, B. Heym, H. E. Takiff, and S. T. Cole. 1997. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J. Clin. Microbiol. 35:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogel, J. P., and R. R. Isberg. 1998. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 2:30-34. [DOI] [PubMed] [Google Scholar]
- 47.Vogel, J. P., H. L., Andrews, S. K., Wong, and R. R. Isberg. 1999. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Y., Z. Zhang, and N. Ramanan. 1997. The actinomycete Thermobispora contains two distinct types of transcriptionally active 16S rRNA genes. J. Bacteriol. 179:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterer, G. W., V. S. Baselski, and R. G. Wunderink. 2001. Legionella and community-acquired pneumonia: a review of current diagnostic tests from a clinician's viewpoint. Am. J. Med. 110:41-48. [DOI] [PubMed] [Google Scholar]
- 50.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. International Committee on Systematic Bacteriology: report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 51.Wiater, L. A., K. Dunn, F. R. Maxfield, and H. A. Shuman. 1998. Early events in phagosome establishment are required for intracellular survival of Legionella pneumophila. Infect. Immun. 66:4450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]