Abstract
Overexpression of the EvgA regulator of the two-component signal transduction system was previously found to modulate multidrug resistance of Escherichia coli by increasing efflux of drugs (K. Nishino and A. Yamaguchi, J. Bacteriol. 183:1455-1458, 2001). Here we present data showing that EvgA contributes to multidrug resistance through increased expression of the multidrug transporter yhiUV gene.
For many years, antibiotics have been effective in the treatment of many infectious diseases caused by a range of pathogens. The occurrence of antibiotic resistance, however, has transformed some previously treatable diseases into a new threat to public health. One of the mechanisms underlying antibiotic resistance involves the extrusion of the compounds by an efflux pump or carrier. The most intriguing mechanisms of drug extrusion are those that include a wide variety of structurally unrelated compounds as substrates for multidrug resistance (MDR) transporters (14, 23). MDR transporters are found in a variety of bacterial species (19, 20, 26). Recently we cloned 37 putative drug transporter genes of Escherichia coli and investigated their drug resistance phenotypes (15). During the course of that study, we found that the DNA locus including both the putative drug efflux transporter emrKY genes and the two-component signal transduction system evgSA genes conferred MDR to E. coli. We showed that the MDR phenomenon was not due to the emrKY genes but was due to the response regulator evgA gene (16).
Two-component systems are signal transduction pathways in prokaryotic organisms responding to environmental conditions (4, 18). A typical two-component system consists of two types of signal transducers, a sensory kinase and a response regulator. The sensory kinase monitors some environmental conditions and accordingly modulates the phosphorylation state of the response regulator. The response regulator regulates gene expression and/or cell behavior. The EvgSA two-component system is known to regulate the expression of the putative drug efflux transporter emrKY genes (5).
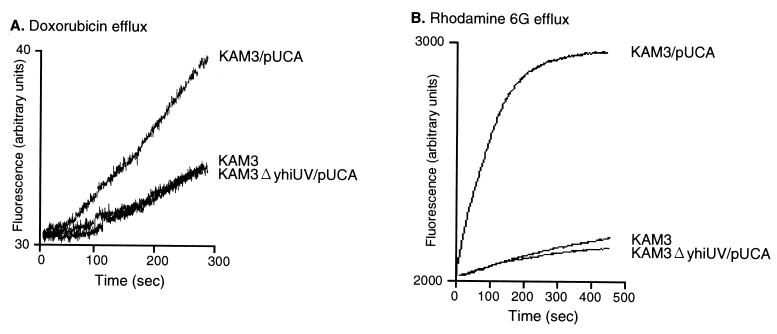
Effect of deletion of yhiUV on MDR induced by overexpression of the EvgA regulator.
In our previous study (16), we found that overexpression of the gene regulator EvgA modulates MDR. E. coli KAM3 cells (13) harboring the pUCA plasmid carrying evgA showed drug resistance against deoxycholate (>32-fold compared to the no-plasmid control level), doxorubicin (64-fold), rhodamine 6G (16-fold), erythromycin (8-fold), crystal violet (8-fold), benzalkonium (8-fold), and sodium dodecyl sulfate (SDS) (4-fold) (Table 1). Since EvgA is known to positively regulate emrKY gene expression (5), we cloned emrKY into the pQE30 expression vector and observed that E. coli cells overexpressing EmrKY acquire resistance only to deoxycholate (eightfold) (Table 1). These results suggest a clear difference between the effects of overexpression of EmrKY and EvgA on MDR (16). One possibility is that EvgA regulates an additional, unknown multidrug efflux system(s), different from EmrKY. Previously, we cloned 37 known and putative drug transporter genes of E. coli and investigated the drug resistance activities (15). In those studies, we found that the drug resistance pattern conferred by overexpression of YhiUV is very similar to that conferred by EvgA (Table 1). In order to investigate whether EvgA regulates the expression of yhiUV, we constructed a yhiUV deletion mutant of E. coli KAM3. Construction of a chromosomal in-frame deletion mutant was performed by the gene replacement method as previously described (8), using plasmid pKO3ΔyhiUV (Table 2). The yhiUV deletion strain itself exhibited no alteration in drug susceptibility compared to the parental strain KAM3 (Table 1), probably because yhiUV are not expressed under normal conditions. This observation is in good agreement with the results of the systematic deletion of putative drug transporter genes in E. coli as reported by Sulavik et al. (27). In contrast to the case for the KAM3 strain, even when the cells were transformed with plasmid pUCA carrying the evgA gene, the mutant that lacks the yhiUV genes exhibited neither increased drug resistance (Table 1) (except for deoxycholate) nor increased drug efflux (Fig. 1). These observations indicate that EvgA-induced MDR is caused by stimulation of yhiUV gene expression. In regard to deoxycholate resistance, EvgA overexpression in the yhiUV deletion mutant caused a moderate increase (fourfold), which is probably due to EvgA-dependent EmrKY expression. Overexpression of either evgA or yhiUV resulted in different levels of resistance to some compounds, probably due to differences in yhiUV expression levels.
TABLE 1.
Resistance of E. coli cells harboring a plasmid carrying evgA, yhiUV, or emrKY
| Compound | MIC (μg/ml)a for strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| KAM3 | KAM3/ pUCA | KAM3ΔyhiUV | KAM3ΔyhiUV/ pUCA | KAM3/ pUCyhiUV | KAM3/ pQE30emrKYb | KAM3/ pUCA-D52A | KAM3/ pUCA-D54A | |
| Doxorubicin | 3.13 | 200 | 3.13 | 3.13 | 25 | 3.13 | 25 | >200 |
| Erythromycin | 3.13 | 25 | 3.13 | 3.13 | 25 | 3.13 | 3.13 | 50 |
| Crystal violet | 1.56 | 12.5 | 1.56 | 1.56 | 3.13 | 1.56 | 3.13 | 12.5 |
| Rhodamine 6G | 6.25 | 100 | 6.25 | 6.25 | 100 | 6.25 | 6.25 | 100 |
| Benzalkonium | 3.13 | 25 | 3.13 | 3.13 | 6.25 | 3.13 | 3.13 | 25 |
| SDS | 100 | 400 | 100 | 100 | 200 | 100 | 100 | >400 |
| Deoxycholate | 1,250 | >40,000 | 1,250 | 5,000 | 5,000 | 10,000 | 1,250 | >40,000 |
TABLE 2.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Genotype or characteristicsa | Reference or source |
|---|---|---|
| E. coli strains | ||
| W3104 | Wild type, used as the donor of chromosomal DNA for PCR amplification | 30 |
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZΔM15], used as the cloning host | 28 |
| DH5α | recA endA1 hsdR17 supE4 gyrA96 relA1 Δ(lacZYA-argF)U169 (φ80dlacZΔM15), used as the cloning host | 24 |
| M15 | recA+ uvr+ F−mtl gal ara lac (pREP4), used for overexpression of hexahistidine-tagged EvgA | Qiagen |
| CJ236 | dut-1 ung thi-1 relA1/pCJ105(Cmr F′), used for mutagenesis by the Kunkel method (7) | 7 |
| ZK796 | Tetr; same as MC4100 but tolC::Tn10 | 32 |
| KAM3 | Derivative of TG1 that lacks a restriction system and acrAB | 13 |
| KAM3ΔyhiUV | Derivative of KAM3 that lacks yhiUV | This study |
| KAM3ΔevgS | Derivative of KAM3 that lacks evgS | This study |
| Plasmids | ||
| pUC119 | Vector; Apr; multiple cloning site in lacZ | 31 |
| pQE30 | His expression vector; Apr; multiple cloning site downstream of T5 promoter | Qiagen |
| pKO3 | repA(Ts) CmrsacB+ | 8 |
| pKO3ΔyhiUV | BamHI-BamHI fragment for yhiUV deletion cloned into pKO3 | This study |
| pKO3ΔevgS | BamHI-BamHI fragment for evgS deletion cloned into pKO3 | This study |
| pUCA | HindIII-SalI fragment containing evgA (gene regulator of two-component system) with 366-bp upstream flanking sequence cloned into pUC119 to be in the same orientation as the lactose promoter; Apr | 16 |
| pUCA-D52A | D52A derivative of pUCA | This study |
| pUCA-D54A | D54A derivative of pUCA | This study |
| pQE30emrKY | SphI-PstI fragment containing emrKY (MFP/ MFS transporter) genes cloned into pQE30; Apr | 16 |
| pQE30evgA | SphI-PstI fragment containing evgA gene cloned into pQE30; Apr | 16 |
| pUCyhiUV | PstI-BamHI fragment containing yhiUV (MFP/ RND transporter) genes with 180-bp upstream flanking sequence cloned into pUC119 to be in the same orientation as the lactose promoter; Apr | 15 |
| Oligonucleotidesb | ||
| evgA-D52A | CCGGGGATGTCGACAGCAATGATGACG, mutagenic primer for D52A of evgA | |
| evgA-D54A | CCGGGGATAGCGACGTCAATGATGACG, mutagenic primer for D54A of evgA | |
| yhiU-No | CGCGGATCCCAGTTCAAAATTATGCAACTGATTCTG, used for crossover PCR for the in-frame deletion of yhiUV | |
| yhiU-Ni | CACGCAATAACCTTCACACTCCAAATTTATAACCATTTTAGTCCCTGAAAATTCTTGAG, used for crossover PCR for the in-frame deletion of yhiUV | |
| yhiV-Co | CGCGGATCCCGTCAAATTCCTCTGCATACTATTGC, used for crossover PCR for the in-frame deletion of yhiUV | |
| yhiV-Ci | GTTATAAATTTCGAGTGTGAAGGTTATTGCGTGTAACGTGTAAATGAGAGTAAGGTTGA, used for crossover PCR for the in-frame deletion of yhiUV | |
| evgS-No | CGCGGATCCGGGTGGAAACACTTAAGCCTGA, used for crossover PCR for the in-frame deletion of evgS | |
| evgS-Ni | CACGCAATAACCTTCACACTCCAAATTTATAACCATGTGGTTAGCCGATTTTGTTAC, used for crossover PCR for the in-frame deletion of evgS | |
| evgS-Co | CGCGGATCCCATGGCACCTTTTGATGTTTTCAATACT, used for crossover PCR for the in-frame deletion of evgS | |
| evgS-Ci | GTTATAAATTTGGAGTGTGAAGGTTATTGCGTGTAAATAGCGGCTCCCACAATGTTC, used for crossover PCR for the in-frame deletion of evgS | |
| yhiUpr-F | CTCTCTACCGCCAGCAATGCCCGC, used for amplification of yhiU probe | |
| yhiUpr-R | CCCGTAATCGGCGAGGTGACATTCGCG, used for amplification of yhiU probe | |
| evgAF | GGGGCATGCAACGCAATAATTATTG, used for amplification of evgA cloned into pQE30 | |
| evgAR | CCCCTGCAGTTAGCCGATTTTGTTACGTTGT, used for amplification of evgA cloned into pQE30 | |
| macAPF | ACATTGAGATTAGGCCAGGGAAAGTTCG, used for amplification of macA promoter region | |
| macAPR | TCCGGGTCATTAACTTCAACGAAATATCAA, used for amplification of macA promoter region | |
| emrKPF | AGAAAATCTGAGCTTCCTTAAG, used for amplification of emrK promoter region | |
| emrKPR | CTGTTCCACTATTATCTCTCATTTC, used for amplification of emrK promoter region | |
| yhiUPF | TCAGGACATAAGCAACTGAAATTG, used for amplification of yhiU promoter region | |
| yhiUPR | TTTAGTCCCTGAAAATTCTTGAG, used for amplification of yhiU promoter region |
Ap, ampicillin; Cm, chloramphenicol; MFP, membrane fusion protein; MFS, major facilitator superfamily; RND, resistance-nodulation-cell division family.
Introduced restriction sites used for cloning are underlined.
FIG. 1.
Active efflux of doxorubicin (A) and rhodamine 6G (B) from E. coli KAM3 and KAM3ΔyhiUV cells overproducing EvgA. Active efflux of doxorubicin and rhodamine 6G from E. coli cells was measured as previously described (16). In order to obtain maximal preloading of the fluorophore, the cells were preincubated with 11.5 μM doxorubicin (A) or 1 μM rhodamine 6G (B) in the presence of the protonophore carbonyl cyanide m-chlorophenylhydrazone (40 μM) at 37°C for 1 h. The cells were then centrifuged and resuspended in the same medium containing 25 mM glucose without fluorescent drugs and carbonyl cyanide m-chlorophenylhydrazone, followed by fluorescence measurement. Doxorubicin transport was measured with excitation at 478 nm and emission at 591 nm. Rhodamine 6G transport was measured with excitation at 529 nm and emission at 553 nm.
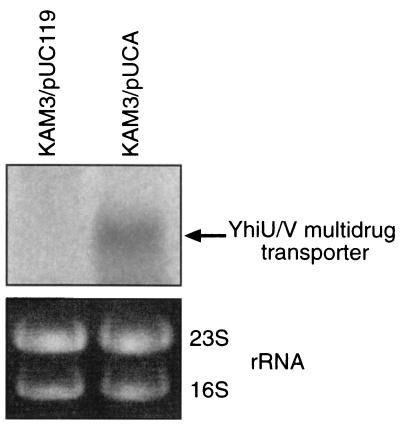
Effect of EvgA on expression of the MDR transporter YhiUV.
The data presented above suggest that the level of the yhiUV transcripts may be increased by EvgA overproduction. To test this hypothesis, total cellular RNA was isolated from pUC119- or pUCA-carrying KAM3 cells. Northern blot analysis using the yhiU probe DNA shows that the yhiUV mRNA is detected in the EvgA-overproducing cells but not in the host KAM3 cells (Fig. 2), indicating that the overproduction of EvgA stimulates yhiUV transcription.
FIG. 2.
Northern blot analysis of mRNA from E. coli cells. KAM3 cells harboring pUC119 or pUCA were grown in Luria-Bertani medium to an optical density at 600 nm of 0.8 at 37°C, and the RNA was isolated by using the SV total RNA isolation system (Promega). The membranes (Hybond-N; Amersham Pharmacia Biotech) were hybridized at high stringency (68°C) with the probe for yhiU labeled with [α-32P]dCTP by random priming. The arrow indicates the main transcript. Each lane contains 15 μg of total RNA.
Determination of drug transporter transcript levels by quantitative real-time PCR.
In our previous study, we found that 20 intrinsic drug transporter genes conferred drug resistance (15). We investigated the changes of the expression levels of these transporter genes (listed in Table 3, except for acrAB) and the evgA-induced yfdX gene (16) when EvgA was overproduced in E. coli KAM3 (lacking acrAB). Quantitative real-time reverse transcription-PCR was used to verify the expression changes. Total RNA was purified from KAM3 cells harboring pUC119 or pUCA, using RNAprotect Bacteria Reagent (Qiagen) and the SV total RNA isolation system (Promega). cDNA samples were synthesized from total RNA by using TaqMan reverse transcription reagents (PE Applied Biosystems) and random hexamers. Specific primer pairs were designed with the ABI PRISM Primer Express software (PE Applied Biosystems), and then real-time PCR was performed with each specific primer pair using SYBR Green PCR Master Mix (PE Applied Biosystems) and run on an ABI PRISM 7000 sequence detection system (PE Applied Biosystems). The results are shown in Table 3. Increases of 470-fold in yhiU expression, 28-fold in emrK expression, and 3,200-fold in yfdX expression were observed upon evgA amplification.
TABLE 3.
Fold induction of transcripts attributed to evgA amplification as determined by amplification of cDNA samples
| Gene | Fold change |
|---|---|
| acrE | 0.9 |
| acrD | 1.0 |
| bcr | 3.0 |
| cusB | 1.6 |
| emrA | 1.6 |
| emrD | 1.3 |
| emrE | 1.2 |
| emrK | 28 |
| fsr | 2.3 |
| mdfA | 1.0 |
| ybjY | 1.1 |
| yceE | 1.6 |
| yceL | 1.0 |
| ydgF | 1.5 |
| ydhE | 1.2 |
| yegM | 1.2 |
| yhiU | 470 |
| yidY | 1.2 |
| yjiO | 0.9 |
| yfdX | 3,200 |
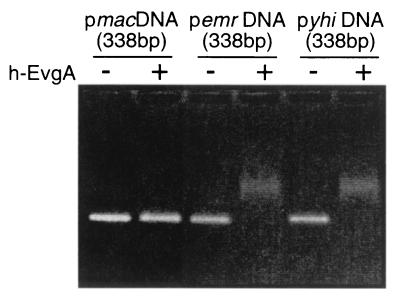
Binding of the EvgA protein to the DNA fragment containing the yhiU promoter region.
If EvgA regulates transcription of yhiUV directly, it may bind to the yhiU promoter region. In order to test this possibility, we constructed plasmid pQE30evgA, which encodes a His6-tagged EvgA (His6-EvgA) under control of the T5 promoter. The His6-EvgA protein was overproduced in E. coli M15(pREP4, pQE30evgA) and purified by Ni-chelating affinity beads (Qiagen). The binding of His6-EvgA to the yhiU and emrK promoter regions was tested by using the electrophoretic mobility shift assay. DNAs of 338 bp containing the yhiU or emrK promoter regions were amplified by PCR and used as target DNAs for the mobility shift assay. The macA (6) promoter region was also amplified as a negative control. Electrophoretic mobility shift assay showed that both the pyhiU and pemrK DNA fragments exhibited changes in mobility in the presence of 5 μg of His6-EvgA (Fig. 3), indicating that EvgA directly binds to both the yhiU and emrK promoter regions.
FIG. 3.
Gel mobility shift profile of EvgA-responsive DNA fragments. DNA fragments of 338 bp including the mac, emr, and yhi promoter regions were prepared by PCR. The binding reaction was done in a total volume of 10 μl of binding buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, and 10% glycerol). Each reaction mixture contained 5 μg of His6-EvgA (+) or bovine serum albumin as a negative control (−) as well as 1 μg of DNA. After they were incubated at 37°C for 30 min, the mixtures were put onto a 4% NuSieve 3:1 (BioWhittaker Molecular Applications) agarose gel, and electrophoresis was done. The gel was stained with ethidium bromide and photographed under UV illumination.
Effect of a mutation at the phosphorylation site of EvgA.
In a two-component regulatory system, a sensor kinase phosphorylates a conserved aspartic acid in a response regulator (25). The phosphorylation is a trigger for a signal transduction event. A phospho-accepting domain of the response regulator is composed of a short consensus motif, ΔΔD (where Δ is a nonpolar amino acid such as I, L, M, or V) (12). The EvgA gene regulator also contains the conserved motif 50IID52, and the aspartate (D) 52 is thought to be a phosphorylation site (29). Another adjacent aspartate, Asp54 in EvgA, which does not seem to be a phosphorylation site, was used as a control in the following experiments. We replaced Asp52 and Asp54 individually with alanine by the Kunkel method (7). The mutant evgA genes were cloned into pUC119 (31), and the resulting plasmids were named pUCA-D52A and pUCA-D54A, respectively. Total cell proteins from E. coli KAM3 cells harboring the constructed plasmids were separated on an SDS-polyacrylamide gel, and EvgA expression was detected by Coomassie brilliant blue staining (data not shown). KAM3 cells transformed by pUCA-D52A were no longer multidrug resistant, except for having resistance to doxorubicin, whereas cells transformed by pUCA-D54A showed resistance similar to that conferred by pUCA (Table 1). pUCA-D52A showed moderate doxorubicin resistance (eightfold), possibly due to phosphorylation of another amino acid in place of Asp52. When the evgA-D52A mutant gene was cloned into pQE30, the cells harboring the mutant plasmid showed gradually increasing moderate MDR depending on the IPTG (isopropyl-β-d-thiogalactopyranoside) concentration (data not shown). Doxorubicin and rhodamine 6G efflux activities of cells carrying pUCA-D52A were significantly lower than those of cells carrying pUCA (data not shown). Thus, phosphorylation of EvgA is required for modulation of MDR.
YhiUV and EmrKY transporters require the outer membrane channel TolC for their function.
The YhiV and EmrY transporter proteins belong to the resistance-nodulation-cell division and major facilitator superfamily transporter families, respectively. YhiU and EmrK both belong to the membrane fusion protein family. AcrA and EmrA, which are typical membrane fusion proteins, show 70 and 65% sequence similarity to YhiU and EmrK, respectively. Membrane fusion protein-dependent drug transporters in general depend on the multifunctional outer membrane channel TolC for their function (3, 6, 9). In order to investigate the role of TolC in the YhiUV and EmrKY systems, the TolC-deficient strain E. coli ZK796 (32) was transformed with pUCyhiUV and pQE30emrKY. The resulting transformed cells showed no increase in resistance (MICs for ZK796 with no plasmid, pUCyhiUV, or pQE30emrKY are 3.13 μg/ml [doxorubicin], 3.13 μg/ml [erythromycin], 1.56 μg/ml [crystal violet], 6.25 μg/ml [rhodamine 6G], 3.13 μg/ml [benzalkonium], 25 μg/ml [SDS], and 156 μg/ml [deoxycholate]), indicating that YhiUV and EmrKY also require TolC for their function.
Conclusions.
Numerous studies on the regulation of multidrug transporter gene expression have been performed with E. coli. For example, expression of the EmrAB multidrug pump (9) is controlled by EmrR, a MarR type of repressor protein (10). The transcription of the acrAB operon in E. coli is regulated by the repressor AcrR (11). Gel mobility shift assays and lacZ transcriptional fusion proteins suggested that the general stress-enhanced transcription of acrAB is mediated primarily by global regulatory pathways such as the mar regulon and that a major function of AcrR is that of a specific secondary modulator (11).
Our results suggest that phosphorylation of EvgA is required for modulation of MDR. Although it was reported that EvgA is phosphorylated by the EvgS sensor protein as determined by phosphotransfer analysis using radiolabeled ATP (21), EvgA also can be phosphorylated by acetylphosphate without EvgS (22). Recently, we have constructed an evgS-deficient strain of E. coli KAM3 and observed that overproduction of EvgA in this strain also causes MDR (data not shown). Thus, in cells overexpressing EvgA, it may be phosphorylated by an EvgS-independent process.
During recent years, it has been found that two-component systems regulate a number of bacterial drug resistance pathways. VncSR in Streptococcus pneumoniae (17) and VanSR in enterococci regulate vancomycin resistance (1). ArlSR in Staphylococcus aureus regulates the MDR pump NorA (2), and we identified EvgSA as the regulator of the MDR pump YhiUV in this study. To the best of our knowledge, this is the first case of a two-component system that directly regulates the expression of MDR transporters in gram-negative bacteria. These findings indicate that two-component system-controlled MDR might become a new threat for chemotherapy against bacterial pathogens in the near future.
Acknowledgments
We thank George M. Church for plasmid pKO3, Hiroshi Nikaido for strain ZK796, and Tomofusa Tsuchiya for strain KAM3. We thank Eitan Bibi for critically reading the manuscript.
K. Nishino is supported by a research fellowship from the Japan Society for the Promotion of Science for Young Scientists. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Arthur, M., C. Molinas, and P. Courvalin. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fournier, B., R. Aras, and D. C. Hooper. 2000. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 5.Kato, A., H. Ohnishi, K. Yamamoto, E. Furuta, H. Tanabe, and R. Utsumi. 2000. Transcription of emrKY is regulated by the EvgA-EvgS two-component system in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 64:1203-1209. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi, N., K. Nishino, and A. Yamaguchi. 2001. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lomovskaya, O., and K. Lewis. 1992. Emr, an Escherichia coli locus for multidrug resistance Proc. Natl. Acad. Sci. USA 89:8938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomovskaya, O., K. Lewis, and A. Matin. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 12.Mizuno, T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli DNA Res. 4:161-168. [DOI] [PubMed] [Google Scholar]
- 13.Morita, Y., K. Kodama, S. Shiota, T. Mine, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishino, K., and A. Yamaguchi. 2001. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak, R., B. Henriques, E. Charpentier, S. Normark, and E. Tuomanen. 1999. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature 399:590-593. [DOI] [PubMed] [Google Scholar]
- 18.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 20.Paulsen, I. T., M. K. Sliwinski, and M. H. Saier, Jr. 1998. Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J. Mol. Biol. 277:573-592. [DOI] [PubMed] [Google Scholar]
- 21.Perraud, A. L., B. Kimmel, V. Weiss, and R. Gross. 1998. Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal HPt domains of the sensor proteins. Mol. Microbiol. 27:875-887. [DOI] [PubMed] [Google Scholar]
- 22.Perraud, A. L., K. Rippe, M. Bantscheff, M. Glocker, M. Lucassen, K. Jung, W. Sebald, V. Weiss, and R. Gross. 2000. Dimerization of signalling modules of the EvgAS and BvgAS phosphorelay systems. Biochim. Biophys. Acta 1478:341-354. [DOI] [PubMed] [Google Scholar]
- 23.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Stock, J. B., A. M. Stock, and J. M. Mottonen. 1990. Signal transduction in bacteria. Nature 344:395-400. [DOI] [PubMed] [Google Scholar]
- 26.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 27.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor, J. W., J. Ott, and F. Eckstein. 1985. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 13:8765-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utsumi, R., S. Katayama, M. Taniguchi, T. Horie, M. Ikeda, S. Igaki, H. Nakagawa, A. Miwa, H. Tanabe, and M. Noda. 1994. Newly identified genes involved in the signal transduction of Escherichia coli K-12. Gene 140:73-77. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto, T., M. Tanaka, C. Nohara, Y. Fukunaga, and S. Yamagishi. 1981. Transposition of the oxacillin-hydrolyzing penicillinase gene. J. Bacteriol. 145:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 32.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 182:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]