Abstract
The TrcRS two-component system of Mycobacterium tuberculosis is comprised of the TrcS histidine kinase and the TrcR response regulator, which is homologous to the OmpR class of DNA binding response regulators. Reverse transcription-PCRs with total RNA showed that the trcR and trcS two-component system genes are transcribed in broth-grown M. tuberculosis. Analysis of the trcR and trcS genes using various SCOTS (selective capture of transcribed sequences) probes also confirmed that these genes are expressed in broth-grown cultures and after 18 h of M. tuberculosis growth in cultured human primary macrophages. To determine if the TrcR response regulator is autoregulated, a trcR-lacZ fusion plasmid and a TrcR expression plasmid were cotransformed into Escherichia coli. Upon induction of the TrcR protein, there was a >500-fold increase in β-galactosidase activity from the trcR-lacZ fusion, indicating that TrcR is involved in transcriptional autoactivation. Gel mobility shift assays with the trcR promoter and TrcR established that the response regulator was autoregulating via direct binding. By use of a delimiting series of overlapping trcR PCR fragments in gel mobility shift assays with TrcR, an AT-rich region of the trcR promoter was shown to be essential for TrcR binding. Additionally, this AT-rich sequence was protected by TrcR in DNase I protection assays. To further analyze the role of the AT-rich region in TrcR autoregulation, the trcR promoter was mutated and analyzed in lacZ transcriptional fusions in the presence of TrcR. Alteration of the AT-rich sequence in the trcR promoter resulted in the loss of trcR transcriptional activation in the presence of TrcR. This report indicates that the M. tuberculosis TrcR response regulator activates its own expression by interacting with the AT-rich sequence of the trcR promoter.
Mycobacterium tuberculosis persists as the leading cause of death from an infectious agent, accounting for nearly 2.4 million deaths annually (20). With the proliferation of multidrug-resistant strains and the association with AIDS, this global impact of tuberculosis will inevitably continue. As an intracellular pathogen, M. tuberculosis must be able to survive and adapt to various environmental conditions encountered during host aerosolization, macrophage phagocytosis, latency, and reactivation. In response to changing environmental conditions, bacteria commonly utilize two-component sensor histidine kinase/response regulator networks to modulate appropriate gene expression (for a review, see reference 30). Since it is now widely accepted that pathogenic bacteria use two-component regulatory systems to regulate expression of virulence factors (5, 16, 17), it is believed that M. tuberculosis will also utilize this mechanism to help fulfill its pathogenic lifestyle.
The complete sequence of the M. tuberculosis genome reveals the presence of 11 paired two-component regulatory systems, two unlinked, orphan histidine kinase genes, and six unlinked, orphan response regulator genes (3). To date, five systems, MtrA/MtrB (33, 35), TrcR/TrcS (9), SenX3/RegX3 (11), DevR/DevS (Rv3133c/Rv3132c) (4, 28), and PhoP/PhoR (23), have been partially characterized. The mtrA response regulator was deemed essential due to the inability to generate an M. tuberculosis mtrA mutant without an extrachromosomal copy of the mtrA gene (35). Recently, the Rv3133c/Rv3132c two-component system genes were shown to be induced under hypoxic conditions, and the Rv3133c response regulator was shown to be involved in the regulation of α-crystallin (acr) expression (28). In another recent report, an M. tuberculosis phoP mutant was impaired in its ability to multiply in mouse bone marrow-derived macrophages and was attenuated in mice (23).
Phosphorylation signal transduction between a sensor/histidine kinase and its cognate DNA binding response regulator triggers the regulation of genes involved in a particular adaptive response (30). It has previously been shown that the M. tuberculosis TrcR/TrcS two-component system communicates via cognate phosphorylation and thus constitutes a functional signal transduction circuit (9). However, the genes regulated by the TrcRS proteins have not yet been identified.
TrcR belongs to the response regulator protein subfamily that also includes OmpR, PhoB, PhoP, and VirG (22, 34). Response regulators from other bacteria classified within this subgroup regulate σ70 promoters and contain a C-terminal helix-turn-helix motif responsible for DNA binding (22). While many two-component systems have been implicated in bacterial pathogenesis and in the regulation of multiple virulence factors, these same systems are also often involved in autoregulation. For example, while the PhoPQ system of Salmonella enterica serovar Typhimurium is important in intramacrophage survival (6) and is predicted to control the expression of up to 40 proteins (7, 19), the phoPQ operon has also been identified as a PhoP-activated locus (29). The BvgA response regulator from Bordetella pertussis controls the expression of many virulence-associated genes, such as pertussis toxin and filamentous hemagglutinin (26), and also regulates the synthesis of the bvgAS operon (27).
Here, we present data regarding the expression of the trcR and trcS genes in broth-grown cultures and in macrophages as well as the expression of the TrcR protein in M. tuberculosis cultures. Transcriptional analysis of the trcR promoter in the presence of TrcR indicates that the TrcR response regulator regulates its own expression. Biochemical characterization of TrcR DNA binding activity reveals that the response regulator binds to the trcR promoter by interacting with an AT-rich region of DNA, and this interaction is essential for TrcR autoregulation.
MATERIALS AND METHODS
Media and bacterial growth conditions.
Escherichia coli cultures were grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C. M. tuberculosis cultures were grown under standard conditions in Middlebrook 7H9 liquid medium (Difco Laboratories, Detroit, Mich.) with or without 0.2% glycerol and supplemented with Middlebrook ADC enrichment (Becton Dickinson, Franklin Lakes, N.J.) and 0.05% Tween 80 at 37°C. The following concentrations of antibiotics or inducing supplement were added when appropriate: ampicillin (Ap), 100 μg/ml; kanamycin (Km), 50 μg/ml; isopropyl-β-d-thiogalactopyranoside (IPTG), 0.1 or 1.0 mM.
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strain JM109 was used as a host for all plasmid constructions and β-galactosidase analyses. M. tuberculosis strain H37Rv (3) and strain TB233, a recent clinical isolate, were used in this study.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| M. tuberculosis H37Rv | Virulent laboratory strain | ATCCa |
| M. tuberculosis TB233 | Virulent clinical isolate | Laboratory collection |
| E. coli JM109 | e14− (McrA−) recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-proAB) | Stratagene |
| Plasmids | ||
| pJEM15 | E. coli-mycobacterial shuttle vector, Kmr | 31 |
| pACYC184 | E. coli low-copy-number p15A origin cloning vector, Cmr Tcr | 2 |
| pSEH100 | pJEM15 with p15A origin from pACYC184 | This study |
| pSH93 | pSEH100 with trcR promoter cloned upstream of the lacZ gene | This study |
| pGEM-T | E. coli cloning vector, Apr | Promega |
| pYH4 | pZErO/Kmr plasmid containing trcR and 1,039 bp of trcS | 9 |
| pSH33 | His6-tagged recombinant TrcR expression plasmid | 9 |
| pSH234 | pGEM-T with 210 bp of the trcR promoter/intergenic region | This study |
| pSH237 | pGEM-T with 195 bp of the trcR promoter and coding region | This study |
| pSH266 | pSH93 with the 36-bp mutated trcR promoter | This study |
| pSH337 | pGEM-T with 1,311 bp of clpC (Rv3596c) | This study |
ATCC, American Type Culture Collection.
The pJEM15 E. coli-mycobacterial shuttle plasmid (31) was used for M. tuberculosis promoter-lacZ fusions after it was altered to reduce the plasmid copy number by exchanging the E. coli pBR322 origin of replication and a portion of the mycobacterial origin of replication for the p15A origin of replication from pACYC184 (2). The resulting plasmid, pSEH100, had 10 to 12 copies per cell (2) and was used to generate pSH93, which consists of a 300-bp fragment containing the trcR promoter and 60 bp of the trcR coding region cloned upstream of the lacZ gene in pSEH100. pSH93 is compatible with the TrcR expression plasmid, pSH33 (9), which has a ColE1 origin of replication. The pSH227 plasmid was constructed by PCR amplification of the full-length trcR and trcS genes from M. tuberculosis H37Rv with primers SH36 (5′-AACGCGGCTACGGCTGGGAC-3′) and SH22 (5′-CAATATCTCGATAAGGCAGG-3′) and by subsequent cloning into the pGEM-T vector (Promega Corporation, Madison, Wis.). pSH337 contains a portion of the M. tuberculosis clpC (Rv3596c) gene amplified by using primers clpCF (5′-ACATTCCGTTTACCCCCCG-3′) and clpCR (5′-GCTCTTCTTCCATCCGCAACAG-3′) and cloned into pGEM-T (Promega Corporation).
RNA isolation.
Total RNA was isolated from M. tuberculosis (H37Rv and TB233) cultures grown as described above with glycerol by using the FastRNA Blue Kit (BIO 101, Inc., Vista, Calif.) according to the manufacturer's instructions. Extracted RNA was resuspended in diethylpyrocarbonate-treated water and treated with DNase (Ambion, Inc., Austin, Tex.). DNA-free RNA samples were stored at −70°C in diethylpyrocarbonate-treated water and RNase inhibitor (Ambion, Inc.).
RT-PCR.
Reverse transcription (RT)-PCR was performed using Ready-To-Go RT-PCR beads as described in the manufacturer's protocol (Amersham Pharmacia Biotech, Piscataway, N.J.). Each bead contains recombinant Moloney murine leukemia virus reverse transcriptase for cDNA synthesis, DNA polymerase for amplification, RNase inhibitor, buffer, and deoxynucleoside triphosphates in a glass matrix. One to two micrograms of DNA-free total RNA was added to the RT-PCR beads, reverse transcribed at 42°C with the appropriate reverse trcR or trcS primer, and PCR amplified for 30 cycles at an annealing temperature of 55°C. RNA samples were concurrently analyzed in PCR mixtures without reverse transcriptase to verify the absence of contaminating genomic DNA. PCR mixtures were analyzed on a 2% agarose gel. Primers used for trcR were SH29 (5′-ACGACGATGTCGGGGTACACG-3′) and SH20 (5′-CCCCGTCGAGCGTAAGGTC-3′); for trcS, they were SH42 (5′-GACCTACCGCCCACCACCGA-3′) and SH21 (5′-CAATATCTCGATAAGGCAGG-3′); for the trcR and trcS intergenic region, they were SH23 (5′-CCCCGACGAGCACTGAGCCG-3′) and SH16 (5′-GCGTGCAGGGATTCGACTAGC-3′).
SCOTS analysis.
The pYH4 plasmid (9), containing the trcR gene, 1,039 bp of the trcS gene, and upstream flanking DNA, and the pSH337 plasmid, containing a portion of the M. tuberculosis clpC (Rv3596c) gene, were analyzed by using previously described selective capture of transcribed sequences (SCOTS) cDNA probes (8). The clpC gene was expressed during mycobacterial growth in broth and in macrophages and was used as a positive control in all hybridization experiments. Plasmid DNA was digested with appropriate restriction enzymes, separated by agarose gel electrophoresis, and transferred onto GeneScreen Plus membranes (NEN Life Science, Boston, Mass.). Transferred DNAs were then hybridized with the various SCOTS cDNA digoxigenin (DIG)-labeled probes and detected by anti-DIG-alkaline phosphatase and the CDP-Star chemiluminescent alkaline phosphatase substrate as described by the manufacturer (Roche, Indianapolis, Ind.). Membranes were exposed to X-ray film at room temperature for 15 to 60 min.
The M. tuberculosis SCOTS cDNA probes were developed from mRNAs specifically expressed during growth in Middlebrook 7H9 broth and during intracellular growth within human macrophages for 18, 48, or 110 h (8). Briefly, for the generation of SCOTS cDNA probes, total RNA was isolated from M. tuberculosis cultures and converted to cDNA by RT. Terminal linker sequences were added to the cDNA mixtures, and the cDNA was captured by hybridization with biotinylated H37Rv chromosomal DNA that had been prehybridized with rrnA DNA. The cDNA-chromosomal DNA hybrids were then separated from nonhybridized cDNAs by reacting with streptavidin-coated magnetic beads. After elution from chromosomal DNA, the cDNA molecules were amplified by PCR. Three rounds of selective capture of cDNAs (SCOTS) were performed before preparing DIG-labeled probes from each cDNA mixture. DIG-labeled SCOTS probes were generated by PCR or random-primed labeling with DIG-dUTP as described by the manufacturer (Roche).
Mycobacterial protein extracts.
M. tuberculosis H37Rv cultures were grown as described above without glycerol and harvested by centrifugation at 4°C. Cells were washed three times in cold phosphate-buffered saline (PBS) and resuspended in cold PBS containing 1 mM phenylmethylsulfonyl fluoride. The cell suspension was transferred to a 2-ml Sarstedt screw-cap tube containing 0.6 ml of 0.1-mm-diameter zirconia/silica beads (Biospec Products, Bartlesville, Okla.) and was subjected to three 1-min pulses in a Mini-BeadBeater (Biospec Products) with at least 1-min rests on ice. Lysed cells were centrifuged for 10 min at 13,000 × g, and the cell supernatant containing cell-free protein extract was collected. Protein concentrations were determined by using the Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Hercules, Calif.).
TrcR antibody generation and immunoblot analysis.
Recombinant TrcR protein was produced in E. coli and purified as previously described (9). Purified TrcR protein (∼100 μg) was emulsified in Freund's incomplete adjuvant and used to immunize two New Zealand White rabbits by subcutaneous injection. Rabbits were given booster injections of TrcR antigen at 4-week intervals, and antisera were collected after at least two booster injections.
M. tuberculosis protein extracts (20 μg) were mixed with 0.05% sodium dodecyl sulfate (final concentration) and native polyacrylamide gel electrophoresis (PAGE) sample buffer, separated by native 10% PAGE, and electrotransferred onto Protran nitrocellulose membranes (Schleicher & Schuell Inc., Keene, N.H.). For control reactions in E. coli, the pQE40 vector was induced with 1 mM IPTG to express His-tagged dihydrofolate reductase, and E. coli protein extracts were processed as described in the manufacturer's protocol (QIAexpressionist; Qiagen, Inc., Valencia, Calif.). Membranes were blocked with 2.5% milk proteins in PBS containing Tween 20 (0.05%), incubated with anti-TrcR rabbit polyclonal antiserum, and developed with a horseradish peroxidase-labeled goat anti-rabbit antiserum (Bio-Rad Laboratories or Sigma Chemical Co., St. Louis, Mo.) and SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.).
Assay of β-galactosidase activity.
TrcR autoregulatory experiments were performed in E. coli with a trcR promoter-lacZ transcriptional fusion and recombinant His6-tagged TrcR produced from the expression plasmid pSH33 (Apr) (9). The trcR promoter-lacZ fusion plasmid, pSH93, and pSH33 were transformed into E. coli JM109 and selected on LB/Ap/Km plates. Overnight cultures were grown and diluted in fresh LB medium. Fresh cultures were grown to optical densities at 600 nm of between 0.3 and 0.5 and were then induced with 0.1 mM IPTG for 60 min. β-Galactosidase measurements were performed as previously described and expressed as Miller units (18).
Protein purification and treatment of TrcR with acetyl phosphate.
The TrcR and TrcRN-term (N-terminal portion of TrcR) proteins were expressed and purified by nickel-nitrilotriacetic acid agarose columns as previously described (9). In vitro phosphorylation of TrcR by acetyl phosphate was performed as described by McCleary (15). Briefly, TrcR was treated with 50 mM acetyl phosphate in phosphorylation buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, and 3 mM dithiothreitol (DTT) for 75 min at 37°C. Following phosphorylation with acetyl phosphate, TrcR was added to gel mobility shift assay reaction mixtures (described below).
Gel mobility shift assays.
Promoter DNA probes for gel mobility shift assays were amplified by PCR from M. tuberculosis and 3′ end labeled with DIG as described by the DIG Gel Shift Kit manufacturer (Roche). The primers used for PCR amplification were synthesized by IDT, Inc. (Coralville, Iowa) or Oligos, Etc. (Bethel, Maine). The trcR fragments, PCR primer pairs, and their respective product sizes used in this study are as follows: trcR1, SH36 and SH45 (5′-CCAGGATGGCTTGCCGCGGACGTTGACTGC-3′), 352 bp; trcR2, SH35 (5′-GATGACGGGATTGTGCTGGA-3′) and SH58 (5′-CAGGTCCGGGTGCCTTCGAG-3′), 359 bp; trcR3, SH36 and SH51 (5′-GTCGTCATGCTCCCGTATCC-3′), 305 bp; trcR4, SH36 and SH56 (5′-GAAAGCGGATCTCGGTTTCC-3′), 163 bp; trcR5, SH57 (5′-ACAACCTGGTCACGATCAGC-3′) and SH53 (5′-TTCGGCCTGAAAAGGTGACC-3′), 129 bp; trcR6, SH55 (5′-CACCGCCGAAGCCCGACATC-3′) and SH51, 141 bp; trcR7, SH55 and SH52 (5′-ATGCAGTTGTCACGGGATTC-3′), 98 bp; trcR8, SH55 and SH53, 46 bp; trcR9, SH54 (5′-AGGCCGAAGTAACTACATAAG-3′) and SH51, 103 bp; trcR10, SH54 and SH61 (5′-CAAGCGCACATGCAGTTGTC-3′), 69 bp; trcR11, SH54 and SH52, 60 bp. PCR products were electrophoresed through agarose gels, and amplified bands were removed from the gels and purified by using the Elu-Quik kit (Schleicher & Schuell). Purified PCR products were quantitated by using GeneQuant (Amersham Pharmacia Biotech), and approximately 100 to 150 ng of each product was end labeled with DIG-11-ddUTP. Primers SH63 (5′-ATAAGCGACATTTGAAAAATTTATGAAT-3′) and SH64 (5′-ATTCATAAATTTTTCAAATGTCGCTTAT-3′) were synthesized and 3′ end labeled with DIG by IDT, Inc. The SH63 and SH64 complementary primers were heated to 95°C for 10 min in TEN buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 100 mM NaCl) and allowed to anneal slowly by gradual cooling at room temperature to create fragment trcR12. Binding reaction mixtures contained 1 μg of TrcR protein and approximately 1 to 10 ng of DIG-labeled promoter PCR fragments. Binding reactions were performed as described in the manufacturer's protocol (Roche) in binding reaction buffer [20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% (wt/vol) Tween 20, 30 mM KCl] and were supplemented with poly(dI-dC), poly(dA-dT), and 50 mM acetyl phosphate, when appropriate. Incubations were carried out for 30 min at room temperature, and the reaction mixtures were loaded onto 8% nondenaturing polyacrylamide gels containing 0.25× Tris-borate-EDTA. Electrophoresis was performed at 4°C at 80 to 100 V for 1.5 to 2 h. The DNA-protein complexes were then electroblotted onto a positively charged Hybond-N+ nylon membrane (Amersham Pharmacia Biotech) and detected by anti-DIG-alkaline phosphatase and the CSPD chemiluminescent alkaline phosphatase substrate as described by the manufacturer (Roche). Membranes were exposed to X-ray film at room temperature for 15 to 60 min.
DNase I footprinting of the trcR promoter.
A 364-bp DNA fragment comprising the trcR promoter region was PCR amplified by using forward primer SH57 and reverse primer SH27 (5′-CCATCTTGACCAGATTGGTCA-3′). For generating single, end-labeled probes, either SH57 or SH27 was end labeled with T4 polynucleotide kinase (Epicentre Technologies, Madison, Wis.) and [γ-32P]ATP (specific activity, >7,000 Ci/mmol; NEN Life Science) and used in a PCR with plasmid pYH4 (9) serving as a template. Radioactive trcR probes were purified by using QIAquick spin columns (Qiagen Inc.) and quantitated with a Wallac 1410 scintillation counter (Amersham Pharmacia Biotech). Binding reaction mixtures in a total volume of 50 μl consisted of approximately 50,000 cpm of labeled DNA probe and various concentrations of TrcR in binding buffer (50 mM Tris-HCl [pH 7.6], 50 mM KCl, 10 mM MgCl2, 1 mM CaCl2, 1 mM DTT, 0.1 mM EDTA) containing 100 ng of poly(dI-dC) as a nonspecific competitor. After a 30-min incubation at room temperature, binding reactions were subjected to DNase I digestion for 1 min and then terminated by the addition of 100 μl of stop buffer (20 mM EDTA, 1% sodium dodecyl sulfate, 200 mM NaCl, 125 μg of Saccharomyces cerevisiae tRNA per ml). Phosphorylated TrcR (TrcR∼P) was generated by preincubating TrcR with 50 mM acetyl phosphate (final concentration) in binding buffer for 30 min at room temperature before addition to the binding reaction. Samples were extracted once with phenol-chloroform, and nucleic acids were precipitated with ethanol. The DNA was resuspended in formamide dye and separated on a 6% polyacrylamide sequencing gel. Dideoxynucleotide sequencing reactions were performed using the SequiTherm EXCEL II DNA Sequencing Kit (Epicentre Technologies) with the same end-labeled primer as the one used in the footprinting reaction, and the products were electrophoresed in parallel with the TrcR footprinting reactions.
Construction of a trcR promoter mutant.
The trcR promoter mutant in which 36 bp of the wild-type trcR promoter AT-rich sequence was replaced with a random 36-bp sequence was constructed by using various other trcR promoter plasmids. Plasmid pSH234 consists of a 94-bp distal portion of the trcR promoter and 116 bp of the upstream flanking DNA amplified with primers SH36 and SH53 and cloned into the pGEM-T vector. Plasmid pSH237 consists of a 51-bp proximal portion of the trcR promoter and 144 bp of the trcR coding sequence amplified with primers SH75 (5′-CCCGTGACAACTGCATGTGC-3′) and RHtrcR1 (5′-CTTGACCAGATTGGTCAGCGCTGG-3′) and cloned into pGEM-T. Plasmid pSH234 was digested with PstI and EagI and treated with Klenow DNA polymerase to obtain a partially blunt-end fragment containing the distal trcR promoter and 21 bp of the pGEM-T vector. This blunt-end fragment was then ligated to NcoI-restricted and blunt-ended pSH237 plasmid. A portion of the ligation reaction mixture was used as a template for PCR using primers SH36 and RHtrcR1. The amplified DNA which replaces the original trcR 36-bp AT-rich sequence with 36 bp of sequence from the pGEM-T vector was then digested with PshAI and RsaI for ligation into the ScaI-digested pSEH100. The resulting plasmid, pSH266, was cotransformed with pSH33 into E. coli JM109 and selected on LB/Ap/Km plates. Cultures were grown and β-galactosidase assays were performed as described above with the pSH93 plasmid containing the wild-type trcR promoter sequence.
RESULTS
trcR and trcS transcription.
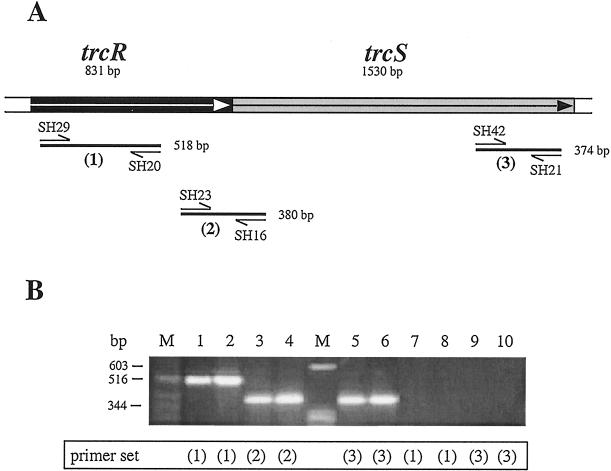
The trcR and trcS genes are adjacent to one another on the M. tuberculosis chromosome, with the response regulator located upstream of the sensor (Fig. 1A). The nucleotide sequence upstream of the apparent translation initiation codon of trcS is separated from the stop codon of trcR by 7 bp. To determine if trcR and trcS are transcribed in the M. tuberculosis H37Rv strain and a clinical isolate strain (TB233), RT-PCR was performed. Primer annealing sites within the coding regions of trcR and trcS and the respective PCR products are depicted in Fig. 1A. The RT-PCR products shown in Fig. 1B indicate that trcR (lanes 1 and 2), trcS (lanes 5 and 6), and the intergenic region between trcR and trcS (lanes 3 and 4) are transcribed. To verify that these RNA samples were not contaminated with genomic DNA, PCRs were performed without the RT step. The lack of amplification products for trcR or trcS (Fig. 1B, lanes 7 to 10) verifies that the RT-PCR products were amplified from RNA that had been reverse transcribed into cDNA. These results indicate that trcR, trcS, and the intergenic region between trcR and trcS are transcribed in both M. tuberculosis H37Rv and a virulent clinical isolate, TB233, and that the two genes are possibly cotranscribed as an operon.
FIG. 1.
RT-PCR analysis of trcR and trcS from M. tuberculosis. (A) Schematic diagram of the M. tuberculosis trcR and trcS genes and the gene-specific primers used in the RT-PCRs. The direction of transcription for trcR and trcS is represented by arrows. PCR primers are indicated by single-headed arrows. Numbers within parentheses below each indicated PCR product refer to the PCR primer sets. (B) Total M. tuberculosis RNA (1 to 2 μg) from two different isolates (H37Rv [odd-numbered lanes] and TB233 [even-numbered lanes]) was reverse transcribed and amplified with the trcR- and/or trcS-specific primers indicated in panel A. The numbers below each set of lanes refer to the primer pairs depicted in panel A. Lanes: 1 and 2, RT-PCR of trcR coding region; 3 and 4, RT-PCR of trcR-trcS intergenic region; 5 and 6, RT-PCR of trcS coding region; 7 to 10, control PCR amplifications without RT using primer sets 1 and 3, respectively, of the same RNA samples used in the RT-PCRs.
Analysis of trcR and trcS gene expression in broth-grown M. tuberculosis cultures and macrophage-infected bacilli.
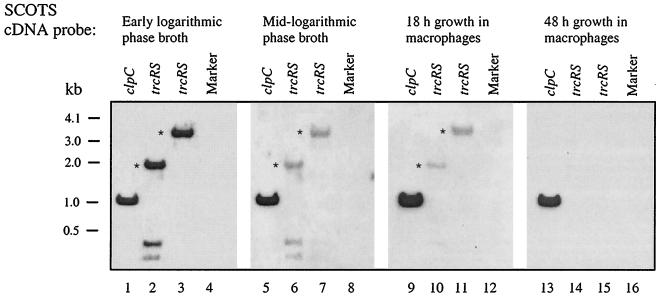
The SCOTS procedure is a technique that can be used to evaluate microbial gene expression in a given environment (8). This technique allows for the capture of cDNAs corresponding to mRNAs that are present in a specific culture condition. The captured cDNAs are hybridized with biotinylated genomic DNA, amplified by PCR, labeled, and used as probes for Southern blot hybridizations. Previously, SCOTS cDNA probes were generated from broth-grown M. tuberculosis and from M. tuberculosis recovered at specific times after infection of macrophages (8). Considering that the above RT-PCR experiments demonstrated that trcR and trcS were transcribed in broth-grown cultures, SCOTS (8) analysis was performed to verify the above results and to determine if trcR was also expressed when M. tuberculosis was growing within macrophages. Figure 2 shows the Southern blot analysis of the trcR and trcS genes using various SCOTS cDNA probes from M. tuberculosis grown in laboratory media and in macrophages. The trcR and trcS genes hybridized with the SCOTS probes generated from early-logarithmic-phase cultures and mid-logarithmic-phase cultures (Fig. 2, lanes 2, 3, 6, and 7). While hybridization is evident with both early-logarithmic- and mid-logarithmic-phase cultures, the regulatory genes are preferentially expressed during early logarithmic growth (Fig. 2, lanes 2 and 3). To determine if these genes were also expressed during M. tuberculosis intracellular growth, Southern analysis was performed using SCOTS cDNA probes developed after 18 and 48 h of M. tuberculosis growth in macrophages. Hybridization of the SCOTS probe from 18 h of growth in macrophages indicates that trcR and trcS are expressed after initial macrophage infection (Fig. 2, lanes 10 and 11). However, the trcR and trcS genes did not hybridize with SCOTS probes developed after 48 h of growth in macrophages (Fig. 2, lanes 14 and 15) or after 110 h of growth in macrophages (data not shown), indicating that these genes are not expressed after longer periods of macrophage infection. The M. tuberculosis clpC (Rv3596c) gene (partial open reading frame) was previously determined to hybridize with SCOTS probes developed after growth in liquid media and after 18 and 48 h of growth in macrophages (S. E. Haydel and J. E. Clark-Curtiss, unpublished data) and was used as a hybridization control (Fig. 2, lanes 1, 5, 9, and 13).
FIG. 2.
SCOTS analysis of trcR and trcS expression. M. tuberculosis SCOTS cDNA probes (8) from early-logarithmic-phase broth-grown cultures, mid-logarithmic-phase broth-grown cultures, 18-h growth in macrophages, and 48-h growth in macrophages were used in Southern blot analysis of the trcR and trcS genes. Plasmid pYH4 was digested with appropriate restriction enzymes to separate the trcR and trcS genes from the remaining vector sequences (lanes 2 and 3, 6 and 7, 10 and 11, and 14 and 15). The detected trcR and trcS DNA fragments are indicated with asterisks, while the trcR and trcS flanking DNA sequences present on pYH4 and detected by the probes remain unmarked. Plasmid pSH337 harboring the partial open reading frame of clpC was also digested with restriction enzymes to release the clpC fragment and used as a positive hybridization control (lanes 1, 5, 9, and 13). Marker, 1-kb DNA ladder (lanes 4, 8, 12, and 16).
Western blot analysis of TrcR in M. tuberculosis cultures.
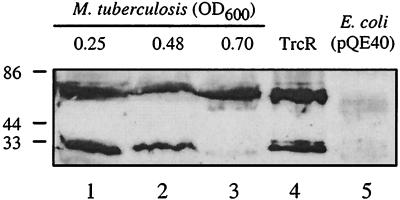
Since RT-PCR and SCOTS analyses established that the trcR and trcS genes are transcribed in broth-grown cultures, M. tuberculosis protein extracts were analyzed by Western blotting using TrcR polyclonal antiserum generated in rabbits. As shown in Fig. 3, TrcR expression was detected during logarithmic growth of M. tuberculosis. These results correlate TrcR protein expression with the trcR RNA data obtained from the aforementioned RT-PCR experiments (Fig. 1B) and trcR SCOTS analysis (Fig. 2). In nondenaturing polyacrylamide gels, purified TrcR migrated as two forms believed to be its monomeric and dimeric forms (Fig. 3, lane 4). In the early- and mid-logarithmic M. tuberculosis protein samples (Fig. 3, lanes 1 and 2), TrcR sera reacted with protein bands that correlate in size with purified TrcR (Fig. 3, lane 4). However, in the late-logarithmic M. tuberculosis protein sample, TrcR sera reacted primarily with a protein antigen corresponding with the larger form of TrcR (Fig. 3, lane 3). These results indicate that the TrcR protein is expressed during logarithmic growth and suggest that TrcR exists in more than one form during logarithmic growth.
FIG. 3.
Western blot analysis of TrcR expression in M. tuberculosis. Whole-cell protein extracts from M. tuberculosis H37Rv were separated by native PAGE and subjected to immunoblot analysis with TrcR polyclonal antiserum. Lanes 1 to 3 represent M. tuberculosis protein samples extracted from cultures grown to optical densities at 600 nm of 0.25, 0.48, and 0.70, respectively. Lane 4 represents purified His-tagged TrcR protein (0.10 μg). As a control, lane 5 contains an E. coli protein extract of strain JM109 expressing the dihydrofolate reductase protein from the pQE40 expression vector. Size standards are indicated in kilodaltons.
TrcR autoregulation.
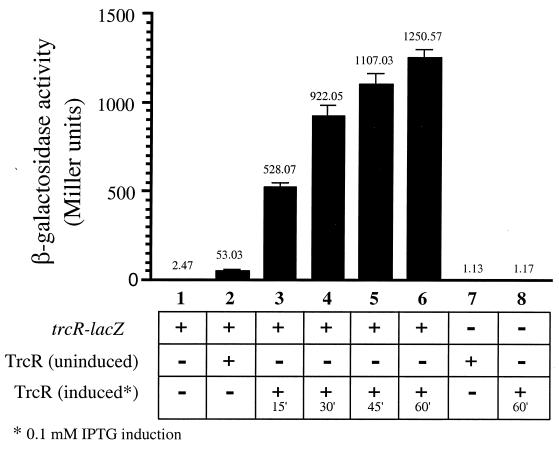
To investigate whether trcR is subject to transcriptional autoregulation, transcriptional fusions were constructed between the trcR promoter and a promoterless lacZ gene. The trcR promoter was fused to the lacZ gene within pSEH100 (described in Materials and Methods), a modified pJEM15 E. coli-mycobacterial shuttle vector (31), to create plasmid pSH93. The pSH33 plasmid with an inducible TrcR protein (9) and pSH93 were cotransformed into E. coli, and β-galactosidase activity was measured to assess the levels of trcR transcription with and without induction of the TrcR response regulator. As shown in Fig. 4, in E. coli cells harboring the trcR-lacZ fusion clone and the IPTG-induced TrcR clone, a steady and significant increase in promoter activity was evident after 15 to 60 min of TrcR induction as indicated by elevated levels of β-galactosidase (columns 3 to 6). After 60 min of TrcR induction, trcR promoter-driven β-galactosidase expression had increased >500-fold compared to cells incubated under identical conditions but without the TrcR expression plasmid. Even in the presence of low levels of TrcR protein (uninduced cells), there was a >20-fold increase in β-galactosidase production from the trcR promoter (Fig. 4, column 2). These experiments demonstrate that the TrcR protein is able to transcriptionally activate expression of the trcR promoter in E. coli.
FIG. 4.
Regulation of the trcR-lacZ transcriptional fusion in the presence of TrcR. Expression of the trcR-lacZ transcriptional fusion in the presence of inducible TrcR. Columns: 1, trcR-lacZ plasmid (pSH93); 2, trcR-lacZ plasmid (pSH93) and uninduced TrcR (pSH33); 3 to 6, trcR-lacZ plasmid (pSH93) and TrcR (pSH33) induced with IPTG for 15, 30, 45, and 60 min, respectively; 7, uninduced TrcR (pSH33); 8, induced TrcR (pSH33). β-Galactosidase activities are expressed as Miller units. Results are the averages (indicated above each column) and standard errors from at least three independent experiments.
Gel shift analyses of the trcR promoter.
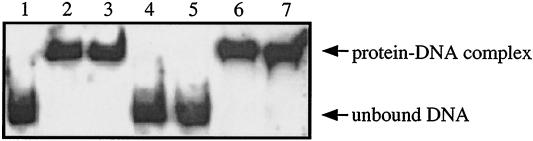
To show that TrcR was regulating trcR expression by directly binding the trcR promoter, DNA gel mobility shift assays were performed. A 352-bp fragment encompassing the entire trcR intergenic region plus flanking DNA was amplified and end labeled with DIG. When recombinant TrcR was incubated with this 352-bp labeled fragment, trcR1, a marked DNA retardation was evident by a complete shift of the labeled DNA (Fig. 5, lane 2). To determine if in vitro phosphorylation of TrcR would alter the binding affinity, TrcR was treated with acetyl phosphate as described in Materials and Methods. TrcR was phosphorylated in vitro via the method of McCleary (15) and was expected to be phosphorylated on the conserved aspartate residue. Since unphosphorylated TrcR efficiently binds the trcR promoter, only a decrease in binding would be detected with phosphorylated TrcR. As shown in Fig. 5 (lane 3), phosphorylated TrcR (TrcR∼P) did not deter DNA retardation of the trcR promoter. (It should be noted that titrations of TrcR and TrcR∼P are performed in the footprinting experiments discussed below to more precisely determine the effect of TrcR phosphorylation.) However, when only the N-terminal portion of the TrcR protein, which contains the phosphoaccepting domain but lacks the DNA binding domain, was added to the trcR promoter fragment, the DNA was not shifted (Fig. 5, lane 4).
FIG. 5.
Gel shift mobility analysis of TrcR binding to the trcR promoter. The full-length trcR promoter is represented by the 352-bp DIG-labeled trcR1 DNA fragment. The trcR1 promoter fragment is shown unbound (DNA only, lane 1), with TrcR (lane 2), with TrcR∼P (lane 3), and with TrcRN-term (lane 4). Lane 5 represents specific competition of the labeled trcR1-TrcR complex with 50-fold excess of unlabeled trcR1. Lanes 6 and 7 represent nonspecific competitions of the labeled trcR1-TrcR complex with 125-fold excess of poly(dI-dC) and 50-fold excess of M. tuberculosis DNA fragment trcR2, respectively. Protein-DNA complexes were separated in 8% native polyacrylamide gels and analyzed by autoradiography. Each reaction mixture contained 1 μg (approximately 1.6 μM) of TrcR or TrcRN-term.
To test if the TrcR binding reaction was specific, unlabeled trcR1 DNA was used in a competition assay. As seen in Fig. 5 (lane 5), with a 50-fold excess of unlabeled trcR1 as a specific DNA competitor, the binding of TrcR is lost. Since the genome of M. tuberculosis is 65% G+C, a competition assay with a 125-fold excess of nonspecific poly(dI-dC) DNA was performed. In assays with poly(dI-dC) DNA (Fig. 5, lane 6) or a 50-fold excess of M. tuberculosis nonspecific DNA competitor, trcR2 (a 359-bp fragment located 240 bp upstream of the trcR coding region) (Fig. 5, lane 7), specific binding activity of TrcR to the trcR1 fragment was retained. These results indicate that TrcR is binding to the trcR promoter region in a sequence-specific manner.
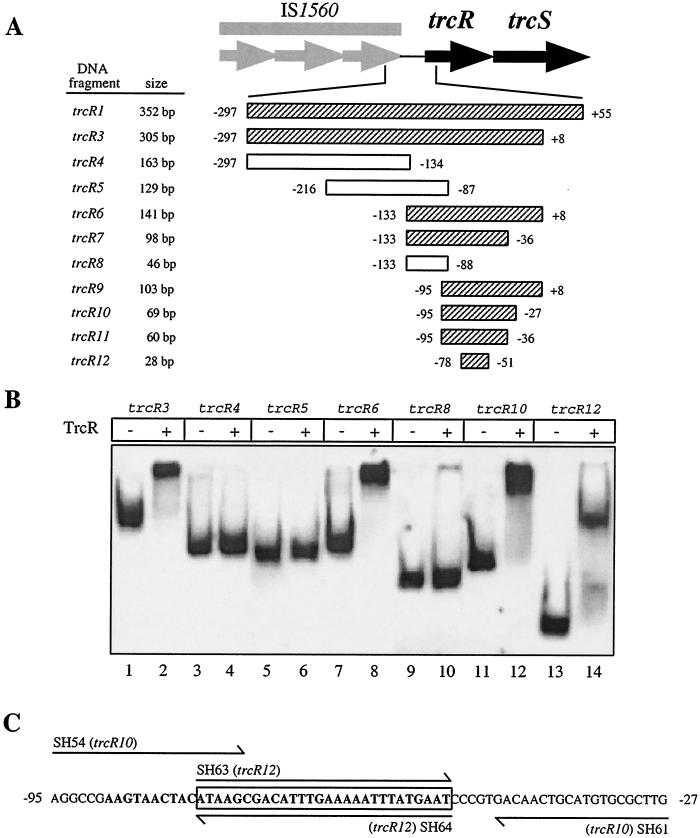
The trcR promoter region important for TrcR binding.
Once it was determined that TrcR was binding directly to the trcR promoter, a more precise characterization of the trcR promoter region important for binding was addressed. To delineate the minimal trcR promoter sequence essential for TrcR binding, various overlapping fragments of the trcR promoter were generated. The trcR promoter regions analyzed are shown schematically in Fig. 6A. As shown in Fig. 6B, a series of gel mobility shift assays were performed with these delimited trcR promoter fragments and 1 μg of unphosphorylated TrcR. As expected, a full-length trcR promoter fragment, trcR3, was shifted in the presence of TrcR (Fig. 6B, lane 2). However, fragments containing the distal upstream regions of the trcR promoter, trcR4 and trcR5, were not shifted upon incubation with TrcR (Fig. 6B, lanes 4 and 6, respectively). Thus, TrcR is apparently binding to a promoter region within 86 bp of the trcR translational start site. Incubation of TrcR with proximal trcR promoter fragments, trcR6 and trcR7, induced complete gel shifts of both fragments (Fig. 6B, lane 8, and data not shown). To further define the TrcR-binding region, five additional and shorter fragments were analyzed. Fragments trcR9, trcR10, and trcR11 were all gel shifted by TrcR (Fig. 6B, lane 12, and data not shown). However, TrcR incubation with fragment trcR8 did not induce a gel shift (Fig. 6B, lane 10). In an attempt to determine a core sequence important for binding, two complementary 28-mer DIG-labeled oligonucleotides, SH63 and SH64, were annealed to create the trcR12 fragment and incubated in gel shift reaction mixtures with TrcR. As shown in Fig. 6B, lane 14, the trcR12 fragment was shifted in the presence of TrcR. Comparing the trcR promoter regions that were bound by TrcR and determining that the smallest fragment was completely shifted by TrcR (trcR12) indicate that TrcR is binding to the trcR promoter within a 28-bp region (Fig. 6C).
FIG. 6.
Schematic representation of the M. tuberculosis IS1560-trcR intergenic region and trcR promoter-TrcR binding specificity. (A) DIG-labeled PCR fragments (trcR1 and trcR3 to trcR11) were used to generate overlapping trcR promoter DNA fragments for TrcR gel mobility shift assays. Fragment trcR12 was created by annealing two DIG-labeled complementary 28-mer primers. The trcR fragment numbering reflects the trcR start codon as +1. Hatched boxes represent PCR fragments that were shifted by TrcR. Open boxes indicate PCR fragments that were not shifted in the presence of TrcR. PCR primers are described in Materials and Methods. (B) Mapping of the TrcR binding site within the trcR promoter using a series of delimiting and overlapping DNA fragments. Fragments encompassing the distal promoter sequences are shown in lanes 3 to 6 and lanes 9 and 10. Proximal promoter regions are shown in lanes 7 and 8 and lanes 11 to 14. All protein-DNA binding reactions (even-numbered lanes) were performed with 1 μg (approximately 1.6 μM) of TrcR. (C) Location of the trcR promoter region important for TrcR binding. The AT-rich sequence is shown in bold type, and the 28-bp sequence bound by TrcR is boxed. PCR primers are indicated by single-headed arrows above and below the sequence.
Sequence analysis of the trcR promoter within a 69-bp region (trcR10) revealed the presence of a 38-bp stretch of DNA that is extremely AT rich (29 of 38, or 76%). The 28-bp trcR12 fragment lies within this 38-bp region and exhibits 78% A+T content (Fig. 6C). It should be noted that this high A+T content is in striking contrast with the 35% A+T content of the M. tuberculosis genome. These delimiting gel shift assays demonstrate that the TrcR protein interacts with the AT-rich region of the trcR promoter.
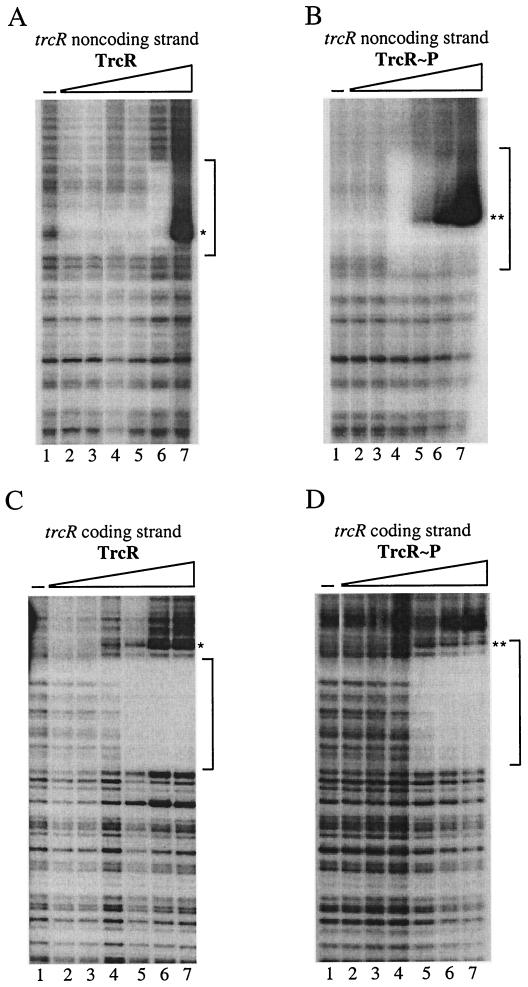
DNase I protection of the trcR promoter by TrcR.
Although gel mobility shift assays revealed that TrcR was interacting with the AT-rich region of the trcR promoter, DNase I protection analyses were also performed on both strands to analyze the effects of TrcR phosphorylation on DNA binding activity (Fig. 7). Using a 364-bp trcR promoter fragment carrying the AT-rich sequence, unphosphorylated TrcR protected 58 bp (from positions −21 to −78 in relation to the translational start site) of the trcR noncoding strand (Fig. 7A) and 72 bp (from −4 to −75) of the trcR coding strand (Fig. 7C). Upon incubation with in vitro-phosphorylated TrcR (TrcR∼P), the TrcR binding site was increased to 68 bp (from −11 to −78) for the trcR noncoding strand (Fig. 7B) and to 86 bp (−4 to −89) for the trcR coding strand (Fig. 7D). With an excess of TrcR (6.5 μM), a marked region of DNase I hypersensitivity was introduced into the footprint of the trcR noncoding strand (Fig. 7A). However, with 10-fold-less protein, TrcR∼P induced a DNase I hypersensitivity site at a slightly different location (Fig. 7B). Protected regions of the trcR coding strand with TrcR and TrcR∼P were similar in that both exhibited the same site of DNase I hypersensitivity; however, TrcR∼P produced a footprint of 86 bp, which was larger than the TrcR footprint of 72 bp (Fig. 7C and D).
FIG. 7.
DNase I footprinting analysis of the trcR promoter with TrcR and TrcR∼P. Binding reaction mixtures containing the trcR promoter noncoding upper strand (A and B) or coding lower strand (C and D) were subjected to nuclease treatment in the absence of TrcR (−) or with increasing amounts of TrcR (lanes 2 to 7). The concentration of TrcR or TrcR∼P present in each reaction mixture is as follows: lane 2, 0.006 μM; lane 3, 0.065 μM; lane 4, 0.325 μM; lane 5, 0.650 μM; lane 6, 3.25 μM; lane 7, 6.50 μM. The protected trcR region is indicated with a notched vertical bar. The single and double asterisks indicate a region of DNase I hypersensitivity for TrcR and TrcR∼P, respectively.
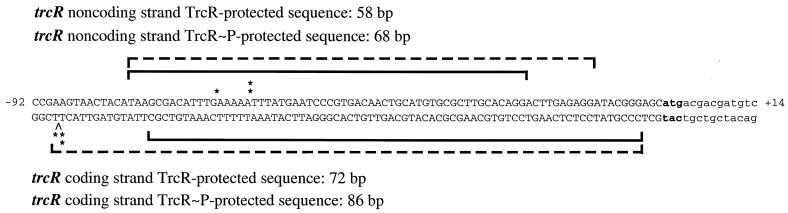
The protected region of the trcR promoter includes the majority of the 28-bp AT-rich sequence deemed important in TrcR binding in gel mobility shift assays (Fig. 8). For the trcR noncoding strand, the TrcR and TrcR∼P nuclease-protected region included 27 of the 28 bp of the AT-rich sequence. For the trcR coding strand, TrcR protected 24 bp of the 28-bp sequence, and TrcR∼P protection encompassed the complete 28-bp AT-rich sequence. The protected regions also extended an additional 31 to 48 bp downstream of this AT-rich region (Fig. 8). Both TrcR and TrcR∼P induced DNase I hypersensitivity sites within the AT-rich region of the trcR noncoding strand (Fig. 8); however, the TrcR∼P hypersensitivity site is evident at a lower protein concentration (Fig. 7B). These data confirm the results of gel shift studies and indicate that TrcR interacts with the AT-rich region of the trcR promoter.
FIG. 8.
Footprinting sequence of the trcR promoter region. The protected regions for TrcR (solid bracket) and TrcR∼P (dashed bracket) are indicated for both strands. The TrcR and TrcR∼P hypersensitivity sites are indicated by single and double asterisks, respectively. The sequence numbering is relative to the trcR translational start site.
Analysis of a trcR promoter mutant in autoregulation experiments.
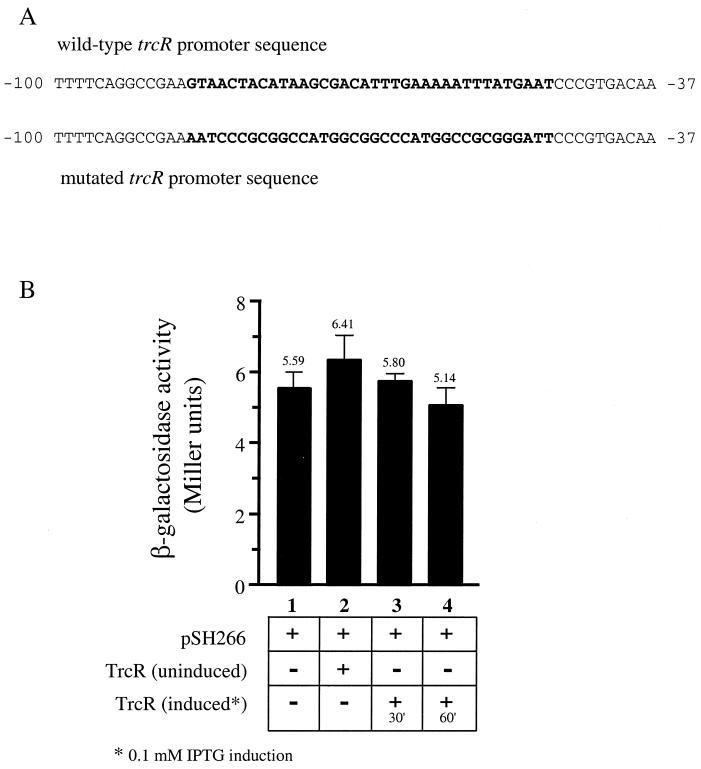
Since previous experiments indicated that TrcR could regulate its own synthesis and bind to the AT-rich sequence within its promoter region, the trcR promoter was mutated in order to determine if the AT-rich sequence was also involved in modulating TrcR autoregulation. A 36-bp region of the trcR promoter which contains the AT-rich sequence was replaced with a nonspecific 36-bp DNA fragment (Fig. 9A). Other than the replacement of the 36-bp stretch of AT-rich sequence, the mutated trcR promoter-lacZ fusion plasmid, pSH266, is identical to the pSH93 plasmid used in the autoregulation studies. The mutated trcR-lacZ fusion plasmid was subjected to experiments identical to the aforementioned TrcR autoregulation experiments (shown in Fig. 4). As shown in Fig. 9B, in comparing E. coli cells harboring the mutated trcR-lacZ fusion clone alone (column 1) and cells harboring both the mutated trcR-lacZ fusion clone and the IPTG-induced TrcR clone (columns 2 to 4), there are no significant changes in β-galactosidase activity produced from the trcR promoter. It should also be noted that β-galactosidase production levels from the wild-type trcR promoter (Fig. 4, lane 1) and from the mutated trcR promoter (Fig. 9B, lane 1) are similar. However, β-galactosidase activity levels driven from the mutated trcR promoter remain unchanged regardless of whether or not TrcR was induced (Fig. 9B, lanes 2 to 4). These results signify that not only does TrcR bind to the AT-rich sequence, but this sequence is also essential in promoting TrcR regulation of trcR transcription.
FIG. 9.
Analysis of the mutated trcR-lacZ fusion in the presence of TrcR. (A) DNA sequence of the wild-type and mutated trcR promoter sequences. A 36-bp region of the wild-type trcR promoter containing the AT-rich sequence was replaced with a nonspecific 36-bp region to create the mutated trcR promoter. (B) Expression of the mutated trcR-lacZ transcriptional fusion in the presence of inducible TrcR. Columns: 1, mutated trcR-lacZ plasmid (pSH266); 2, mutated trcR-lacZ plasmid (pSH266) and uninduced TrcR (pSH33); 3, mutated trcR-lacZ plasmid (pSH266) and TrcR (pSH33) induced for 30 min; 4, mutated trcR-lacZ plasmid (pSH266) and TrcR (pSH33) induced for 60 min. β-Galactosidase activities are expressed as Miller units. Results are the averages (indicated above each column) and standard errors from at least three independent experiments.
DISCUSSION
Two-component system response regulators have been shown to control numerous types of adaptive responses and modulate the expression of genes or activities of many different proteins under a variety of environmental conditions. These regulators can act as DNA binding proteins to activate or repress transcriptional activity of the targeted genes. While it had previously been shown that the M. tuberculosis TrcS and TrcR proteins function as signal-transducing proteins capable of cognate phosphorylation communication (9), the regulated genes of this two-component system had not been identified. The goals of the experiments in this report were to determine if the trcRS genes are expressed in broth-grown cultures and/or in macrophages, to begin identifying the regulated genes of the M. tuberculosis TrcR response regulator, and to biochemically characterize the TrcR binding site.
Based on homology, the TrcR response regulator can be grouped with the OmpR subfamily of response regulators that exhibit carboxy terminus helix-turn-helix DNA binding motifs (22). A number of two-component system genes from this group of response regulators have previously been shown to be autoregulated. The PhoPQ system from S. enterica serovar Typhimurium, the BvgAS system from B. pertussis, and the VanRS system from Enterococcus faecium are all important in regulating virulence pathways and are also involved in autoregulation (12, 27, 29). For Mycobacterium smegmatis, autoregulatory experiments with the regX3 promoter indicate that there is a twofold increase in transcriptional activity when the regX3 and senX3 genes are provided (11). In autoregulation experiments presented here, the TrcR response regulator activates trcR transcription more than 500-fold. Since this substantial activation occurs in E. coli without the TrcS histidine kinase, it is likely that TrcR can regulate its own expression independent of phosphorylation. However, it is currently unknown if TrcR is subjected to low levels of phosphorylation via cross talk with another native E. coli histidine kinase or by a small-molecule phosphodonor, such as acetyl phosphate.
The initial establishment of trcR autoregulation led to the question of how TrcR was activating transcription. In gel mobility shift assays, we have demonstrated that the TrcR response regulator binds to a 28-bp fragment of the trcR promoter. The gel shifts demonstrated with TrcR have also been evident with other two-component response regulators, OmpR (21), ComA (24), BvgA (13, 25), VanR (12), ToxR (10), and their target promoters, and could be the result of oligomerization during DNA binding and/or DNA bending.
Several response regulators in their unphosphorylated state are able to bind DNA. Unphosphorylated BvgA from B. pertussis can efficiently bind to both the fhaB and bvgA promoters (25). The unphosphorylated VanR response regulator from E. faecium is also able to bind the vanH and vanR promoters; however, 10-fold-less-phosphorylated VanR can bind with similar affinity (12). B. subtilis PhoP has been shown to dimerize and bind to the phoB promoter in both phosphorylated and unphosphorylated states (14). However, while 1 μg of PhoP∼P efficiently retarded the phoB promoter, 200 μg of unphosphorylated PhoP was required for binding (14). In contrast, only 1 μg of TrcR or TrcR∼P was able to completely retard the trcR promoter. Response regulator phosphorylation could lead to oligomeric protein states and cooperative binding that might not always be important for binding to certain target promoters (21). The low concentration of TrcR required to induce a complete shift suggests that the phosphorylation state of TrcR is not critical in binding the trcR promoter.
In DNase I footprinting analyses, while similar concentrations of TrcR and TrcR∼P are able to efficiently protect the trcR promoter, TrcR∼P exhibits a 10-fold increase in binding affinity and protects a slightly larger region. This extended footprint observed with TrcR∼P may signify an oligomerization of TrcR or a secondary region for binding by TrcR∼P. Since sites of DNase I hypersensitivity usually indicate tandem protein binding sites (1), symmetric binding of TrcR∼P is suggested by the presence of a hypersensitivity site with low concentrations of protein.
Response regulators often bind to consensus sequences that can be determined by analyzing multiple binding sites. These consensus binding sites are usually found as inverted repeats, direct repeats, or as several sites spaced widely throughout the promoter. In observations of the identified binding sites of B. pertussis BvgA, S. enterica serovar Typhimurium PhoP, E. faecium VanR, and E. coli OmpR, the consensus target DNA for each response regulator is AT rich (12, 25, 29, 32). Common to all of these binding sites is the presence of a TTT sequence within an AT-rich stretch of DNA. Similar to these response regulator binding sites, the 28-bp TrcR binding sequence was also determined to be an AT-rich region that contains two TTT sequences (Fig. 8). Through the use of the available M. tuberculosis genome sequence (3), it was determined that this 28-bp AT-rich sequence in the exact form does not exist elsewhere in the genome. However, sequences similar to this 28-bp AT-rich sequence are present and are currently being investigated as possible TrcR-targeted sequences (S. E. Haydel and J. E. Clark-Curtiss, unpublished data). Mutagenesis of the trcR promoter by replacing a 36-bp AT-rich region with nonspecific DNA demonstrated that this sequence is required for TrcR autoregulation. While the biochemical studies demonstrated the importance of the AT-rich region in TrcR binding, the mutational genetic experiments confirmed that the AT-rich sequence is essential for TrcR autoregulation.
The development of SCOTS to investigate gene expression in various environments (8) has proven to be a very useful laboratory tool. While the initial application identified numerous M. tuberculosis genes that are specifically expressed during growth in human macrophages (8), the SCOTS cDNA probes can be used to analyze the expression of any M. tuberculosis gene under any given (or desired) growth condition. SCOTS analysis of the trcR and trcS genes revealed that these genes are expressed in logarithmic-phase broth-grown cultures as well as after initial M. tuberculosis infection of human blood monocyte-derived macrophages. Expression of trcR and trcS after early infection of macrophages, but not after longer periods of infection indicates that this two-component system may play an important role in allowing the organism to adapt to an intracellular environment. In addition to a possible role in M. tuberculosis intracellular adaptation, the trcR and trcS genes could also be expressed when the tubercle bacilli are replicating extracellularly within the liquefied cavity after breakdown of the walled-off granuloma. Moreover, trcR/trcS expression could occur at the end of the latency stage or at the initiation of reactivation. Thus, the TrcR-TrcS regulatory system may act as a transition regulatory system involved in adapting to an intracellular environment and transitioning from latency to reactivation.
The identification of a gene promoter that the M. tuberculosis TrcR response regulator regulates and the biochemical characterization of its binding interaction are a great advancement in beginning to understand the TrcRS two-component system. While the trcR promoter represents the first TrcR-targeted and TrcR-activated promoter to be identified, efforts are currently under way to identify other TrcR-regulated genes. Elucidation of other promoters targeted by TrcR will lead to the definition of a consensus TrcR-binding site and the possibility of identifying other genes regulated by this system. Identification of the target sites to which TrcR is binding will allow future studies regarding the regulatory features of the TrcR response regulator.
Acknowledgments
We thank Brigitte Gicquel for use of the pJEM15 vector. We gratefully acknowledge Iris Dixon for assistance with gel purification of numerous PCR fragments, Roy Curtiss III for critical review of the manuscript, and Alexander Smith and Randall Harris for helpful suggestions and discussions.
This research was supported by a Heiser Program of the New York Community Trust fellowship (to S.E.H.), NIH grant AI46428 (to J.E.C.-C.), and CDC contract C90117123 (to N.E.D.) from the Alabama Department of Public Health.
REFERENCES
- 1.Boucher, P. E., and S. Stibitz. 1995. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J. Bacteriol. 177:6486-6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 4.Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 80:141-159. [DOI] [PubMed] [Google Scholar]
- 5.DiRita, V. J., and J. J. Mekalanos. 1989. Genetic regulation of bacterial virulence. Annu. Rev. Genet. 23:455-482. [DOI] [PubMed] [Google Scholar]
- 6.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 7.García-Véscovi, E., F. Soncini, and E. A. Groisman. 1994. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res. Microbiol. 145:473-480. [DOI] [PubMed] [Google Scholar]
- 8.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haydel, S. E., N. E. Dunlap, and W. H. Benjamin, Jr. 1999. In vitro evidence of two-component system phosphorylation between the Mycobacterium tuberculosis TrcR/TrcS proteins. Microb. Pathog. 26:195-206. [DOI] [PubMed] [Google Scholar]
- 10.Higgins, D. E., and V. J. DiRita. 1996. Genetic analysis of the interaction between Vibrio cholerae transcription activator ToxR and toxT promoter DNA. J. Bacteriol. 178:1080-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himpens, S., C. Locht, and P. Supply. 2000. Molecular characterization of the mycobacterial SenX3-RegX3 two-component system: evidence for autoregulation. Microbiology 146:3091-3098. [DOI] [PubMed] [Google Scholar]
- 12.Holman, T. R., Z. Wu, B. L. Wanner, and C. T. Walsh. 1994. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry 33:4625-4631. [DOI] [PubMed] [Google Scholar]
- 13.Karimova, G., and A. Ullmann. 1997. Characterization of DNA binding sites for the BvgA protein of Bordetella pertussis. J. Bacteriol. 179:3790-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCleary, W. R. 1996. The activation of PhoB by acetylphosphate. Mol. Microbiol. 20:1155-1163. [DOI] [PubMed] [Google Scholar]
- 16.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. F., J. J. Mekalanos, and S. Falkow. 1989. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science 243:916-922. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Miller, S. L., and J. J. Mekalanos. 1990. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 172:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray, C. J. L., and J. A. Salomon. 1998. Modeling the impact of global tuberculosis control strategies. Proc. Natl. Acad. Sci. USA 95:13881-13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakashima, K., K. Kanamaru, H. Aiba, and T. Mizuno. 1991. Signal transduction and osmoregulation in Escherichia coli. J. Biol. Chem. 266:10774-10780. [PubMed] [Google Scholar]
- 22.Pao, G. M., and J. M. H. Saier. 1995. Response regulators of bacterial signal transduction systems: selective domain shuffling during evolution. J. Mol. Evol. 40:136-154. [DOI] [PubMed] [Google Scholar]
- 23.Pérez, E., S. Samper, Y. Bordas, C. Guilhot, B. Gicquel, and C. Martín. 2001. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 41:179-187. [DOI] [PubMed] [Google Scholar]
- 24.Roggiani, M., and D. Dubnau. 1993. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter regions of srfA. J. Bacteriol. 175:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy, C. R., and S. Falkow. 1991. Identification of Bordetella pertussis regulatory sequences required for transcriptional activation of the fhaB gene and autoregulation of the bvgAS operon. J. Bacteriol. 173:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy, C. R., J. F. Miller, and S. Falkow. 1989. The bvgA gene of Bordetella pertussis encodes a transcriptional activator required for coordinate regulation of several virulence genes. J. Bacteriol. 171:6338-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scarlato, V., A. Prugnola, B. Aricó, and R. Rappuoli. 1990. Positive transcriptional feedback at the bvg locus controls expression of virulence factors in Bordetella pertussis. Proc. Natl. Acad. Sci. USA 87:6753-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soncini, F. C., E. García Véscovi, and E. A. Groisman. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timm, J., E. M. Lim, and B. Gicquel. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsung, K., R. E. Brissette, and M. Inouye. 1989. Identification of the DNA-binding domain of the OmpR protein required for transcriptional activation of the ompF and ompC genes of Escherichia coli by in vivo DNA footprinting. J. Biol. Chem. 264:10104-10109. [PubMed] [Google Scholar]
- 33.Via, L. E., R. Curcic, M. H. Mudd, S. Dhandayuthapani, R. J. Ulmer, and V. Deretic. 1996. Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J. Bacteriol. 178:3314-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volz, K. 1995. Structural and functional conservation in response regulators, p. 53-64. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 35.Zahrt, T. A., and V. Deretic. 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182:3832-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]