Abstract
The conjugative transfer region 1 (Tra1) of the IncHI1 plasmid R27 was subjected to DNA sequence analysis, mutagenesis, genetic complementation, and an H-pilus-specific phage assay. Analysis of the nucleotide sequence indicated that the Tra1 region contains genes coding for mating pair formation (Mpf) and DNA transfer replication (Dtr) and a coupling protein. Insertional disruptions of 9 of the 14 open reading frames (ORFs) in the Tra1 region resulted in a transfer-deficient phenotype. Conjugative transfer was restored for each transfer mutant by genetic complementation. An intergenic region between traH and trhR was cloned and mobilized by R27, indicating the presence of an origin of transfer (oriT). The five ORFs immediately downstream of the oriT region are involved in H-pilus production, as determined by an H-pilus-specific phage assay. Three of these ORFs encode proteins homologous to Mpf proteins from IncF plasmids. Upstream of the oriT region are four ORFs required for plasmid transfer but not H-pilus production. TraI contains sequence motifs that are characteristic of relaxases from the IncP lineage but share no overall homology to known relaxases. TraJ contains both an Arc repressor motif and a leucine zipper motif. A putative coupling protein, TraG, shares a low level of homology to the TraG family of coupling proteins and contains motifs that are important for DNA transfer. This analysis indicates that the Mpf components of R27 share a common lineage with those of the IncF transfer system, whereas the relaxase of R27 is ancestrally related to that of the IncP transfer system.
Bacterial conjugation is a special type of DNA replication during which one strand of a plasmid molecule is transferred from donor to recipient in the 5′-to-3′ direction while the second strand is retained in the donor (38). The host-encoded replisome is responsible for complementary strand synthesis of both the transferred and retained DNA strands (19, 36). Horizontal DNA transfer is directed by the conjugation apparatus, which consists of three multiprotein complexes: the membrane-associated mating apparatus and the cytoplasmic relaxosome, both of which are coupled together by the inner membrane-associated coupling protein (38).
A working model of conjugation suggests that the mating pair formation (Mpf) proteins form a membrane-associated apparatus that functions in synthesizing and assembling mature conjugative pili on the cell surface. The conjugative pilus facilitates the initial contact with the recipient cell, a prerequisite for the formation of stable mating pairs (11). For IncF plasmids, the transfer proteins are believed to function in pilus extension, pilus retraction, formation of stable mating pairs, and formation of a lumen through which plasmid transfer occurs (12). The exact role of the Mpf proteins still remains poorly understood.
The relaxosome is a specific DNA-protein complex that is composed of a relaxase and accessory protein(s). Relaxases initiate and terminate conjugation by catalyzing site- and strand-specific nicking and rejoining reactions at the plasmid-encoded nic site within the origin of transfer (oriT). Nicking results in a covalent bond between the 5′ end of the cleaved strand and the relaxase, forming a complex known as the transferred strand intermediate. Genetic evidence suggests that the transferring intermediate is connected to the mating apparatus via the plasmid-encoded coupling protein (5, 17). Based on an analysis of the crystal structure of TrwB, the coupling protein from the IncW plasmid R388, it has been proposed that coupling proteins may serve as a conduit through which the single-stranded DNA transfers (14, 15).
Conjugative plasmids belonging to the incompatibility group HI1 (IncHI1) encode multiple-antibiotic resistance in Salmonella enterica serovar Typhi and therefore contribute to the persistence of typhoid fever worldwide (10, 27). IncHI1 plasmids are low-copy-number, self-transmissible plasmids that have an unusual mode of conjugation which is thermosensitive for transfer, with transfer occurring optimally between 22 and 30°C, but which is negligible at 37°C (32). Recent evidence indicates that at least one transfer gene, trhC, is not expressed at nonpermissive transfer temperatures, suggesting that transfer thermosensitivity may be imposed at the transcriptional level (13).
R27, the prototype IncHI1 plasmid, was recently sequenced and analyzed (30). The transfer genes of R27 are contained within two separate regions, designated Tra1 and Tra2. Since mutations in either transfer region abolish transfer, both transfer regions contribute to the R27 conjugative apparatus (28, 30). An analysis of the Tra2 region has indicated that this region codes for mating pair formation and H-pilus synthesis phenotypes. Protein homology led us to conclude that the R27 Tra2 region is ancestrally related to the transfer region of the F factor (28).
The objective of this work was to characterize the Tra1 region of R27 using DNA sequence analysis, mutagenesis, genetic complementation, and an H-pilus-specific phage assay. The ultimate goal was to identify IncHI1 conjugal transfer genes and to determine any evolutionary relationships to other type IV secretion systems, especially conjugative transfer systems.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
Escherichia coli strains and plasmids used in this study are listed in Table 1. E. coli was grown at 27 or 37°C in Luria-Bertani broth (Difco Laboratories, Detroit, Mich.) with shaking or on Luria-Bertani agar plates. Antibiotics were added at the following concentration when appropriate: ampicillin, 100 mg/liter; tetracycline, 10 mg/liter; nalidixic acid, 50 mg/liter; rifampin, 50 mg/liter; chloramphenicol, 16 mg/liter. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside and isopropyl-β-d-thiogalactosidewere each used at 50 mg/liter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype, phenotype, or characteristic(s)a | Cloning or gene disruption primers (5′ to 3′)b | Source or reference |
|---|---|---|---|
| E. coli | |||
| DH5α | supE44 lacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 16 | |
| DY330 | W3110 ΔlacU169 gal-490 λcI857 Δ(cro-bioA) | 6 | |
| DY330N | Temperature-resistant revertant; Nalr | This study | |
| DY330R | Temperature-resistant revertant; Rifr | This study | |
| DT1942 | RG192 (Rifr) with pDT1942 | 34 | |
| RG192 | ara leu lac | 2 | |
| Plasmids | |||
| pAT22 | traI cloned into pMS119EH; Ampr | F: TGGATCCATGAATTTCAGGGCACTCTTC | This study |
| R: TCTGCAGTTAATGGTGATGGTGATGGTGCGCCAGATTCCCGACCTCTTTTAG | |||
| pBAD24 | PBAD expression vector; Ampr | 13 | |
| pBAD30 | PBAD expression vector; Ampr | 13 | |
| pJEG31 | R27 with cat in orf115; Tcr Cmr | F: GATGGTGTGGCGAGTGATAGGAATCAGCATTATTGTCCTACTGTGACGGAAGATCACTTC | This study |
| R: CGCTCAGAAGCCCTTCTGCATCATCAGGCAGGTATATTTCTTATTCAGGCGTAGCACCAG | |||
| pJEG36 | orf122 cloned into pMS119HE; Ampr | F: GGATCCATGTCTAAATCGAAGCTATTAAAG | This study |
| R: GAATTCTTAATGGTGATGGTGATGGTGTTTGTCCCACCTCTGATTTTCAG | |||
| pJEG51 | pDT1942 with cat in orf117; Tcr Kmr Cmr | F: TGCTTTTACTAGGGATGATGCTTTATCTTATTTATAGTCTCTGTGACGGAAGATCACTTC | This study |
| R: AGTAGCCTCACTATTTGGCAATAACAGATAACCACCGGTCATTATTCAGGCGTAGCACCAG | |||
| pJEG52 | pDT1942 with cat in orf118; Tcr Kmr Cmr | F: CAGTGCATCTCTGAAAGTAAGTAAAAGCCGCTCGCTCGTTCGTTCTGTGACGGAAGATCACTTC | This study |
| R: TGTCATGGCAAAATTGCCCCACACATTTCAAAGCACCTTTTTATTCAGGCGTAGCACCAG | |||
| pJEG85 | orf128 cloned into pMS119EH; Ampr | F: TATATGAATTCAATGGATTACAACATCTATAC | This study |
| R: TATATGGATCCTTAATGGTGATGGTGATGGTGCGCCTTCTTATTTCATTCAGCCAG | |||
| pJEG94 | pDT1942 with cat in orf123; Tcr Kmr Cmr | F: TCACGCGTAATTTACTCGGTAAACTATTACGTACCGAGATCTGTGACGGAAGATCACTTC | This study |
| R: TCCAAATCCATTACCAAATACTCGGCGTTGGAGGAGTGGTTTATTCAGGCGTAGCACCAG | |||
| pJEG104 | pDT1942 with cat in orf122; Tcr Kmr Cmr | F: CTAAATCGAAGCTATTAAAGTCATTGGAGATTTCACGTACCTGTGACGGAAGATCACTTC | This study |
| R: GCTATCCCTGGGTAAATTCCTCTCAATGTGATTCCGTTCTTTATTCAGGCGTAGCACCAG | |||
| pJEG105 | pDT1942 with cat in orf125; Tcr Kmr Cmr | F: TTCAACAGCCCTTTCAGGGTCTTTCTAGTAGTCACATTTACTGTGACGGAAGATCACTTC | This study |
| R: GGTATTAACGTTAATCGAGTCATTTACCGATCTCCTCATGTTATTCAGGCGTAGCACCAG | |||
| pJEG115 | pDT1942 with cat in orf124; Tcr Kmr Cmr | F: CGCTACAAAAACGATGTCTCTTGGTGCCAATAAAAACTCCCTGTGACGGAAGATCACTTC | This study |
| R: CCCTGCGGTTATCCGTGCGGCGACTCCACCCACGAATCCCTTATTCAGGCGTAGCACCAG | |||
| pJEG123 | orf124 cloned into pMS119EH; Ampr | F: TGAATTCAATGGCTAAAGTCGATCAAACTAAAG | This study |
| R: TGGATCCTTAATGGTGATGGTGATGGTGATCTAACTCTTCCTGGGAATAC | |||
| pLJS4 | orf123 cloned into pMS119HE; Ampr | F: GGATCCATGTCAGAATCAGATAACATC | This study |
| R: GAATTCTTAATGGTGATGGTGATGGTGATAATCCTTCAAGAGCACTTTA | |||
| pLJS33 | orf117 cloned into pMS119EH; Ampr | F: TATATGAATTCAATGGCTGCGGATAATTCTGCTC | This study |
| R: TATATGGATCCTTAATGGTGATGGTGATGGTGCGCCATAAGTTTTTGAAAGTTAG | |||
| pLJS36 | orf119 cloned into pMS119EH; Ampr | F: TATATGAATTCAATGACAAAATCAAAAAGAAC | This study |
| R: TATATGGATCCTTAATGGTGATGGTGATGGTGCGCATGCAATTTCCTTAGATATT | |||
| pLJS42 | orf127 cloned into pMS119EH; Ampr | F: TATATGAATTCAATGCGCGCTGATTTCCATGTTTG | This study |
| R: TATATGGATCCTTAATGGTGATGGTGATGGTGCGCGTTACCTGTGCCACGAAGAC | |||
| pDT1942 | Derepressed R27; R27::TnlacZ | 34 | |
| pDT2956 | pDT1942 with mini::Tn10 inserted into traG | This study | |
| pDT2984 | pDT1942 with mini::Tn10 inserted into orf127 | This study | |
| pDT2987 | pDT1942 with mini::Tn10 inserted into orf126 | This study | |
| pDT2991 | pDT1942 with mini::Tn10 inserted into orf128 | This study | |
| pDT2995 | pDT1942 with mini::Tn10 inserted into traI; Tcr Kmr Cmr | This study | |
| pGEM | Cloning vector; Ampr | Promega | |
| pMS119 | IPTG inducible expression | 8 | |
| HE&EH | vector; Ampr | ||
| pMWG13 | pDT1942 with mini::Tn10 inserted into orf121 | F: GGATCCATGACCGATGTAACAATGAATG | This study |
| R: GAATTCTTAATGGTGATGGTGATGGTGCCGGCGTTTACCAGACAGTTC | |||
| pMWG18 | orf126 cloned into pBAD24; Ampr | F: TATATGAATTCAATGACTCGATTAACGTTAATAC | This study |
| R: TATATGGATCCTTAATGGTGATGGTGATGGTGCGCGCGCGCATATGCACCTCCTAAAC | |||
| pTL138 | 1.3-kbp oriT KpnI-SnaBI fragment into pBAD30; Ampr | This study | |
| pTL139 | 1.6-kbp oriT SnaBI-XbaI fragment into pBAD30; Ampr | This study | |
| R27 | IncHI1 plasmid; Tcr | 18 | |
| pOriT1.1 | 794-bp subclone of oriT region from pTL138 | F: ATATGGTACCTACGGCTATTATCGGACG | This study |
| R: ATATAAGCTTTCAGATCACGATCTCAGC | |||
| pOriT1.2 | 977-bp subclone of oriT region from pTL138 | F: ATATGGTACCTTGCTGAGATCGTGATCTG | This study |
| R: ATATAAGCTTTCTACACACTTAGAACGT | |||
| pOriT2.1 | 360-bp subclone of oriT region from pOriT1.2 | F: ATATGGTACCTTGCTGAGATCGTGATCTG | This study |
| R: ATATAAGCTTAAACCCTGACCTGCTAACG | |||
| pOriT2.2 | 285-bp subclone of oriT region from pOriT1.2 | F: ATATGGTACCGGTTATTGCTACTTAATGCCGA | This study |
| R: ATATAAGCTTATCTGATTCTGACATGTGCG |
Abbreviations: Nalr, nalidixic acid resistance; Rifr, rifampin resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance; Ampr, ampicillin resistance.
Underlined regions of cloning primers designate restriction endonuclease cleavage sites; bold, italicized bases designate a His tag and an engineered stop codon. Underlined regions of disruption primers designate cat-specific bases. F, forward; R, reverse.
DNA manipulations.
R27 DNA was isolated using either ultracentrifugation in a CsCl-ethidium bromide gradient (35) or with a Qiagen (Mississauga, Ont.) Large-Construct Kit. Expression and cloning vectors were purified using Qiagen Midi-preps (Qiagen Inc.). Standard recombinant DNA methods were performed as described by Sambrook et al. (29). Restriction endonucleases were used according to the manufacturer's instructions, and digested DNA was analyzed by agarose gel electrophoresis.
Computer analysis.
Laser gene software (DNASTAR Inc., Madison, Wis.) was used for nucleotide sequence analysis. Repeated nucleotide sequences were identified with GeneQuest. The predicted protein sequence for each open reading frame (ORF) was compared to the GenBank nonredundant database using PSI-BLAST. We identified conserved motifs manually or with ScanProsite (http://ca.expasy.org/tools/scnpsite.html) and obtained predictions for molecular weight and pI values with Compute pI/Mw (http://ca.expasy.org/tools/pi_tool.html).
Mutagenesis.
Mutants of traG, traI, trhF, trhH, and trhG were created prior to this study using random mini-Tn10 mutagenesis (25) as described previously (28). Mini-Tn10 consists of a chloramphenicol resistance cassette flanked by Tn10 inverted repeats (20). To identify the insertion position for each mutant, we sequenced the region flanking the mini-Tn10 insertions using the dideoxy method (Sequenase version 2; United States Biochemical Co., Cleveland, Ohio) with primers PNE11 (5′TATTCTGCCTCCCAGAGCCT) and PNE12 (5′TGGTGCGTAACGGCAAAAGC). Sequence results were compared to the complete nucleotide sequence of R27 (GenBank accession no. NC_002305). All remaining Tra1 genes (orf115, orf116, traJ, orf118, orf121, traH, trhR, trhY, and trhX) were mutated by gene disruption using the E. coli recombination system recently described by Yu and colleagues (37). This method utilizes E. coli strain DY330, containing a defective lambda prophage that serves to protect and recombine an electroporated linear DNA substrate in vivo. Gene disruptions were created by insertion of a chloramphenicol resistance cassette (cat from mini-Tn10) into each of the above-mentioned ORFs in a sequence-specific fashion. DNA substrates were generated through PCR with primers (∼60 nt) that produced a linear cat cassette with 40-bp terminal arms homologous to the desired target site (Table 1). DNA substrates were introduced by electroporation into DY330 harboring R27 that was grown according to the method of Yu et al. (37). Cells were plated on agar plates containing both tetracycline, to select for R27, and chloramphenicol, to select for the desired insertions. To screen presumptive colonies, the target gene was PCR amplified (using cloning primers as described below) and analyzed with 1% agarose gel electrophoresis. An increase in the size of the ORF by ∼900 bp demonstrated that the cat cassette had been inserted into the target gene. The insertion location of the cat cassette in traJ was confirmed by sequencing with primers GIL86 (5′CAGTTATTGGTGCCCTTAAAC) and GIL87 (5′GGTATCAACAGGGACACCAG), verifying the precision of this method.
Cloning of transfer genes.
The primer sequences used for cloning Tra1 transfer genes are listed in Table 1. PCR products, which included terminal BamHI and EcoRI (BamHI and PstI for traI) restriction endonuclease sites, were cloned into pGEM-T (Promega) and then subcloned into either pMS119EH, pMS119HE, or pBAD24. A His6 tag was engineered into the C terminus of each protein to allow for the detection of the recombinant protein by immunoblot analysis with anti-His6 antibodies.
Conjugation assay.
Conjugal transfer of R27 was performed as previously described (32). For complementation experiments, E. coli donors (DY330R) which contained the R27 Tra1 mutant and an expression vector encoding the appropriate transfer protein were mated with recipients (DY330N). To determine the effect of overexpressing transfer proteins on the transfer frequency, each transfer protein was expressed in trans within a donor containing R27 during conjugation experiments. Transfer frequencies were expressed as transconjugants per donor.
Phage plaque assays.
Hgal RNA bacteriophage was prepared as previously described (23). Phage spot tests were performed to determine Hgal infectivity of Tra1 mutants. A 50-μl sample of mid-logarithmic-phase cultures was suspended in 8 ml of brain heart infusion broth (Difco Laboratories) containing 0.6% agar that was incubated at 48°C. The suspension was overlaid onto a fresh brain heart infusion plate. In the case of cultures containing Tra1 gene expression constructs, ampicillin and isopropyl-β-d-thiogalactoside were included in the overlay. After solidification of the overlay, 10 μl of Hgal stock (109 PFU/ml) was spotted on the overlay. Plates were incubated at 27°C overnight and analyzed for zones of clearing in the Hgal-spotted regions.
Cloning of origin of transfer.
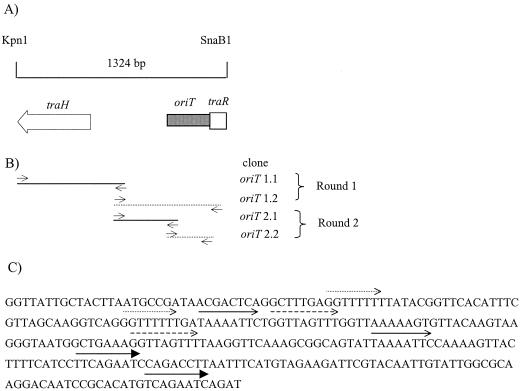
Two intergenic regions from the Tra1 region were considered likely to harbor the origin of transfer (oriT), a 757-bp region between traH and trhR and a 400-bp region between trhY and trhX. Since the origin of transfer is the only element required in cis on a DNA molecule for transfer (21), we cloned each of these regions to determine if either could be mobilized by R27. A 1.3-kbp KpnI-SnaBI region containing the 757-bp intergenic region was cloned into the KpnI and SmaI sites of pBAD30 (16), and a 1.5-kbp HindIII-XbaI fragment containing the 400-bp intergenic region was cloned into the same sites of pBAD30. The 1.3-kbp fragment was mobilized by R27 at a frequency of 10−3 transconjugants/donor, whereas both the 1.5-kbp HindIII-XbaI clone and pBAD30 were not mobilized.
To systematically identify the minimal oriT, regions of the 1.3-kbp clone were PCR amplified, cloned, and tested in mobilization experiments. The strategy is outlined in Fig. 3. Two overlapping regions of the 1.3-kbp fragment were PCR amplified using primers which incorporated KpnI and HindIII sites into the ends. These products were cloned into pBAD30 using the KpnI and HindIII sites, resulting in clones of oriT 1.1 (977 bp) and oriT 1.2 (794 bp). The primers for amplifying oriT 1.1 were oritclone1 and nicprimer3, and the primers for amplifying oriT 1.2 were orit2 and nicprimer5 (Table 1). The oriT 1.2 clone was mobilized by R27, whereas the oriT 1.1 clone and pBAD30 alone were not mobilized under the same conditions. Two overlapping regions of the oriT 1.2 clone were subcloned as described above, resulting in clones of oriT 2.1 (360 bp) and oriT 2.2 (285 bp). The primers for amplifying oriT 2.1 were Trev3000 and Trev3002, and the primers for amplifying oriT 2.2 were Trev3001 and Trev3004 (Table 1). The oriT 2.2 clone was mobilized by R27 at a frequency of 10−3 transconjugants/donor, whereas the oriT 2.1 clone and pBAD30 alone were not mobilized.
FIG. 3.
Origin of transfer. (A) Genetic organization of the 1.3-kbp KpnI-SnaBI fragment containing oriT activity. Shown is the location of the 285-bp oriT between traH and trhR. (B) Cloning strategy used to identify the 285-bp oriT. PCR primers were used to amplify regions and incorporate KpnI and HindIII restriction sites that were used to clone fragments into the same sites of pBAD30 (see Materials and Methods for details). Horizontal lines represent subclones of the 1.3-kbp KpnI-SnaBI fragment and are aligned with the above schematic (panel A). Broken lines represent clones that were mobilized by R27, whereas solid lines represent clones that were not mobilized by R27. Round 1 identified a 977-bp region that was mobilized by R27 and was designated oriT 1.2. Round 2 identified a 285-bp region that was mobilized by R27 and was designated oriT 2.2. (C) Nucleotide analysis of a 285-bp region with oriT activity. Three 8-bp direct repeats and one 9-bp direct repeat were identified and are represented by arrows. No matches to any of the five nic site families were identified.
RESULTS
Nucleotide analysis of Tra1 region.
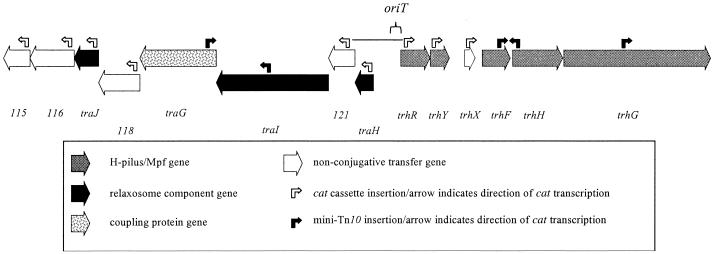
The location of the Tra1 region in R27 was originally identified by using random transposon mutagenesis and screening for transfer-deficient mutants (25). The completion of the nucleotide sequence of R27 allowed us to precisely map the insertion location of 15 mini-Tn10 transfer mutants into five genes: traG, traI, trhF, trhH, and trhG (Fig. 1; Table 2) (30). The genetic organization of this region suggests that these genes are contained within transfer operons, an organization that is commonly seen in other transfer systems, such as those from IncF (12) and IncP plasmids (26, 33). The ends of these putative operons were used to define the Tra1 region as a starting point for this study. Therefore, the Tra1 region was defined as being between the coordinates 98 and 117 kb on R27 and contains 14 ORFs. The ORFs are organized into three operon units (using the operon structure defined in reference 8), consisting of eight, two, and four ORFs, which are separated by two intergenic regions of 757 and 400 bp. The putative transcriptional units are transcribed away from the 757-bp intergenic region (Fig. 1).
FIG. 1.
Open reading frame map of transfer region 1 of R27. Black ORFs indicate genes required for conjugative transfer but not Hgal infection. Cross-hatched ORFs indicate genes required for conjugative transfer and Hgal infection. White ORFs are not essential for conjugative transfer. Black bent arrows represent the insertion location of mini-Tn10 and the direction of cat transcription. White bent arrows represent the insertion location of the cat cassette and the direction of transcription. oriT indicates a 285-bp region of Tra1 that has been cloned and is mobilized by R27. The black horizontal line indicates a KpnI-SnaBI fragment used to identify oriT. The ORF map corresponds to coordinates 98 to 117 kbp and ORF115 to ORF128 on R27 (30).
TABLE 2.
Summary of computer analysis of nucleotide sequence of transfer region 1 from R27
| ORFa | Genea | Lengthb | Molc wt | pId | PSI BLAST resultse (protein/source/length of homolog) | Degree of relatednesse (identical residues/range of identity = % identity) | Accession no. of homologe | Motifsf |
|---|---|---|---|---|---|---|---|---|
| orf115 | 242 | 27.9 | 9.64 | ORF8 from Bacteriodes uniformis | 46/210 = 21% | AF238367 | None | |
| NBU1 element (246 aa) | ||||||||
| orf116 | 393 | 42.9 | 7.54 | Proteinase IV from Vibrio cholerae (616 aa) | 69/343 = 20% | H82130 | Type IV protease (191-325) | |
| orf117 | traJ | 220 | 25.4 | 9.61 | None | AraC transcriptional regulator (8-48) and leucine zipper (182-202) | ||
| orf118 | 367 | 42.2 | 8.15 | None | None | |||
| orf119 | traG | 694 | 80.0 | 8.61 | TraG from R751 (637 aa) | 70/539 = 12% (all in C-terminal end) | S22992 | Walker A (205-212) and Walker B (404-414) |
| orf120 | traI | 1,011 | 113.0 | 4.68 | None | Zinc carboxypeptidase (588-598) and relaxase motifs I-III | ||
| orf121 | 240 | 27.9 | 9.10 | None | None | |||
| orf122 | traH | 161 | 19.2 | 9.50 | None | None | ||
| orf123 | trhR | 266 | 30.3 | 5.15 | None | None | ||
| orf124 | trhY | 170 | 19.4 | 8.57 | None | None | ||
| orf125 | trhX | 95 | 11.0 | 4.91 | None | None | ||
| orf126 | trhF | 348 | 39.6 | 5.20 | TraF from R100 (247 aa) | 46/231 = 19% | AAB61943 | None |
| orf127 | trhH | 470 | 48.1 | 5.02 | TraH from R100 (460 aa) | 108/419 = 25% | BAA78879 | None |
| orf128 | trhG | 1,329 | 140.9 | 5.34 | TraG from F factor (938 aa) | 121/682 = 17% (all in N-terminal end) | AAC44184 | None |
Gene/ORF designation according to Sherburne et al. (30) or from this analysis.
Length in amino acid residues according to Sherburne et al. (30) or from this analysis.
Molecular (Mol) weight was predicted using the Compute pI/Mw tool (http://ca.expasy.org/tools/pi_tool.html).
pI values were predicted using the Compute pI/Mw tool (http://ca.expasy.org/tools/pi_tool.html).
Homologous proteins were identified with PSI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Motifs were identified using Prosite (http://ca.expasy.org/tools/scnpsite.html) or alignment (Fig. 2).
To identify potential transfer genes and to determine any evolutionary relationship to other conjugative systems, the predicted protein sequence from each Tra1 ORF was compared with the GenBank database using PSI-BLAST. Only six of the protein sequences had significant hits in the database (Table 2). ORF115 shares identity with the product of orf8 from the Bacteroides uniformis mobile element NBU1 (31). ORF116 is homologous to putative proteinase IV from Vibrio cholerae (18). TraG shares a low identity with TraG from the IncPβ plasmid R751 (33) but also shares homology to coupling proteins from several other conjugative systems, such as IncW and IncF. TrhF, TrhH, and TrhG share identity with TraF, TraH, and TraG, respectively, from IncF plasmids (2).
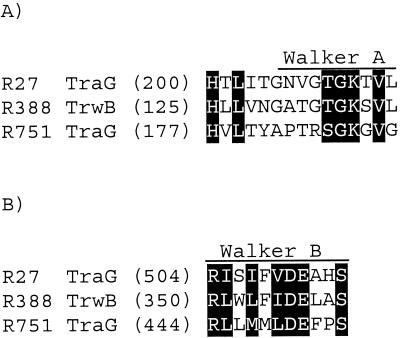
To identify conserved sequence motifs, the predicted amino acid sequence from each ORF from Tra1 was compared to the Prosite database. The ORF116 sequence contains a motif characteristic of type IV proteases, which is consistent with the BLAST analysis. TraJ contains a helix-turn-helix motif at residues 8 to 48, which is characteristic of transcriptional regulators belonging to the AraC family, and a leucine zipper motif at 182 to 202, which is a common dimerization domain. TraG contains a Walker A ATP binding motif at the 205-212 position (Fig. 2). An alignment of TraG (IncHI1) with TrwB (IncW) and TraG (IncPβ) has identified a conserved Walker B ATP binding domain at the 504-514 position (Fig. 2), although the importance of this motif has yet to be demonstrated (15). TraI, which has previously been shown to contain all three relaxase domains (30), contains a zinc carboxypeptidase motif at the 588-598 position, although the significance of this is unknown.
FIG. 2.
Alignment of Walker A (A) and Walker B (B) boxes from coupling proteins from R27 (TraG; GenBank accession no. NP_058332), R388 (TrwB; GenBank accession no. CAA44852), and R751 (TraG; GenBank accession no. S22992). Each number represents the number of amino acid residues preceding the motif. Motifs were aligned in MegAlign (DNASTAR Inc.) and shaded in GeneDoc using the conservative mode and two levels.
Gene disruptions and identification of transfer mutants.
To supplement the mini-Tn10 transfer mutants, and to identify a role for ORFs orf115, orf116, traJ, orf118, orf121, traH, trhR, trhY, and trhX in the conjugative transfer of R27, we systematically created gene disruptions of each of these ORFs. A cat cassette (chloramphenicol resistance gene) was inserted within the first 120 bp of the predicted start codon of each ORF using the methods of Yu et al. (37). Each plasmid mutant was then tested for its ability to transfer (Table 3). Gene disruptions in traJ, traH, trhR, and trhY abolished conjugative transfer of R27. A disruption of trhX reduced the conjugation frequency of R27 10-fold, whereas disruptions of orf115, orf116, orf118, and orf121 had no effect on the conjugation frequency. These data, combined with that from the random transposon mutagenesis experiments, indicate that 9 of the 14 genes within the Tra1 region are essential for conjugative transfer of R27 (Table 3; Fig. 1).
TABLE 3.
Effects of mutations in Tra1 on R27 conjugation
| R27 mutanta in the donor strain | Transferb phenotype of mutantc | Relative conjugation frequency of R27 mutant (%)d | Relative conjugation frequency during complementation (%)e | Relative conjugation frequency of R27 with overexpressed transfer proteinf (%) | Plaque with Hgalg |
|---|---|---|---|---|---|
| orf115 | + | 120 | N/D | N/D | + |
| orf116 | + | 130 | N/D | N/D | + |
| traI | − | 0 | 24 | 23 | + |
| orf118 | + | 100 | N/D | N/D | + |
| traG | − | 0 | 5 | 30 | + |
| traI | − | 0 | 5 | 100 | + |
| orf121 | + | 14 | N/D | N/D | + |
| traH | − | 0 | 83 | 97 | + |
| trhR | − | 0 | 221 | 177 | − |
| trhY | − | 0 | 0.2 | 217 | − |
| trhX | + | 16 | N/D | N/D | + |
| trhF | − | 0 | 0.5 | 65 | − |
| traH | − | 0 | 0.3 | 79 | − |
| trhG | − | 0 | 136 | 77 | − |
Gene designation according to complete sequence of R27 (30) or this study. Donor strain was DY330R.
Ability of mutant to transfer via conjugation. Detectable matings scored as “+”, and nondetectable (<10−8 transconjugants/donor) scored as “−.”
cat cassette introduced into gene by either random transposon mutagenesis or homologous recombination as described in Materials and Methods.
Transfer frequency of R27 mutant relative to R27 [(R27 mutant frequency/R27 frequency) × 100%]. Due to the inherent variation of R27 transfer frequency, relative frequency was determined using the R27 transfer frequency for each experiment. R27 transferred with an average frequency of 5 × 10−1 transconjugants/donor. Values represent the average values of two independent experiments.
Restoration of conjugation by complementation in trans using a cloned gene (see Table 1); N/D, not determined. Transconjugants/donor cell after 18-h liquid mating with recipient DY330N, as described in Materials and Methods.
Designated transfer protein was overexpressed in donor containing R27 during conjugation assay. Relative transfer frequency of R27 with transfer protein/R27 for each experiment. Values represent the average value from two independent experiments.
Ability of H-pilus-specific RNA bacteriophage to form plaques with E. coli harboring designated mutant of R27 (agar spot test). E. coli containing R27 forms plaques when infected with Hgal.
Genetic complementation of transfer-deficient mutants.
The wild-type gene corresponding to each of the transfer-deficient mutants was cloned into the expression vector pMS119EH, pMS119HE, or pBAD24 (Table 1). Each clone was transformed into the strain containing the appropriate R27 mutant. When the wild-type version was expressed in trans, conjugative transfer of R27 was restored for each of the nine transfer-deficient mutants (Table 3), demonstrating that each of these genes is essential for conjugative transfer.
During complementation experiments, the conjugation frequency varied between 221 (trhR) and 0.2% of wild-type levels (trhY) (Table 3). Two explanations for the variation are that it could be due to transcriptional polar effects and/or that the overexpressed transfer proteins affect wild-type conjugation frequencies. To address these possibilities, we compared the complementation frequencies for each mutant to the transfer frequency of R27 when the corresponding transfer protein was overexpressed in donors during conjugation experiments (Table 3). Any difference in the conjugation frequency between the complementation and the overexpression experiments may reflect polar effects on downstream genes.
Overexpression of either TraI or TraH had no effect on the transfer frequency of R27, while the traI and traH mutants were complemented to 5 and 83% of wild-type levels. This indicates that the traI and traH mutants are partially polar. When either TrhR or TrhY was overexpressed, the transfer frequency increased to 177 and 152% of wild-type levels, respectively. The trhR mutant complemented to 221% of wild-type levels, whereas the trhY mutant was complemented to 0.2% of wild-type levels. This indicates that the trhY mutant is partially polar. When TraJ, TraG, TrhF, TrhH, and TrhG were overexpressed individually in donors, the transfer frequencies were reduced to 23, 30, 65, 79, and 77% of wild-type levels, respectively. In the case of both TraJ and TrhG, these frequencies were comparable to their respective complementation frequencies (Table 3). This suggests that the reduced transfer frequency of traJ and trhG during complementation experiments is due to the overexpression of these transfer proteins and not because of polarity, as would be expected for mutants at the ends of transfer operons. Since overexpression of TraG, TrhF, or TrhH cannot account for the low levels of complementation, these mutants are likely partially polar.
These results suggest that both mini-Tn10 and cat cassette insertions resulted in partially polar transfer mutants of traG, traI, traH, trhY, trhF, and trhH. Nevertheless, these mutants allowed the identification of nine essential transfer genes within the Tra1 region of R27.
Hgal plaque assay of R27 transfer-deficient mutants.
Hgal is an H-pilus-specific bacteriophage that lyses E. coli harboring R27 to form distinctive plaques. Hgal binds uniformly along the shaft of H-pili (23); this property of Hgal has been used to document the kinetics of H-pilus assembly (24). We utilized an Hgal plaque assay to determine which of the Tra1 transfer mutants were resistant or sensitive to Hgal lysis (Table 3). E. coli cells containing transfer mutants trhR, trhY, trhF, trhH, and trhG were resistant to Hgal, suggesting that these transfer genes play a role in H-pilus biosynthesis. This is consistent with the roles predicted for trhF, trhH, and trhG using BLAST analysis of these genes. Cells containing plasmids with mutants of traJ, traG, traI, and traH remained sensitive to Hgal, suggesting that their role(s) in conjugation is not related to H-pilus biosynthesis. These results would be expected for the relaxosome components and the coupling protein, since RP4 traG remained sensitive to phage (34).
Identification and cloning of the R27 origin of transfer.
The oriT contains the nic site and is typically located next to the DNA processing genes within conjugative systems. It was therefore anticipated that the oriT of R27 would be present within the Tra1 region. A 757-bp intergenic region between orf122 and orf123 contains several hallmark features of oriT regions, including the following: (i) being located between diverging ORFs, (ii) a high %A+T content (62.5 versus 52% for R27), and (iii) a number of inverted and direct repeat sequences.
We localized a 285-bp minimal oriT region from the 757-bp intergenic region that was mobilized by R27 at a frequency of 10−3 transconjugants/donor (Fig. 3). The oriT is located adjacent to traR and contains three 8-bp direct repeats and one 9-bp direct repeat. A comparison of the oriT nucleotide sequence to the five nic site families (38) revealed no matches. Additional studies are required to precisely identify the nic site within this region.
DISCUSSION
The conjugative transfer genes of R27 are contained within two separate transfer regions, designated Tra1 and Tra2, which are separated by 63 kbp. Each region contributes to the conjugative transfer function of R27, since mutations in either region abolish transfer. The Tra2 region contains Mpf/H-pilus synthesis genes and is interrupted with the insertion of two partitioning modules (28, 30). Nine genes within the Tra2 region encode proteins that share a very low level of homology to F factor transfer proteins; six of these are essential for transfer as determined by mini-Tn10 mutagenesis (28). The Tra2 Mpf genes are trhL, trhE, trhK, trhB, trhV, trhC, trhW, trhU, and trhN, which were named to reflect their homology with the IncF Mpf counterparts (trh designates R27 Mpf genes). In addition, a putative pilin gene is located at the beginning of Tra2. Interestingly, the trhP gene encodes for a peptidase, a protein family essential for IncP pilin processing (7) but not found in IncF plasmids. In this study, genetic and sequence analysis of the Tra1 region has demonstrated that this region contains genes that encode mating pair formation proteins and H-pilus synthesis, a coupling protein, and a DNA transfer replication protein(s), as well as a functional origin of transfer.
There are five Mpf genes within the Tra1 region, and all are transcribed in the same direction and away from the oriT. Three genes, trhF, trhH, and trhG, specify proteins that are homologous to the TraF, TraG, and TraH transfer proteins from IncF plasmids. In the IncF system, these proteins are required for Mpf/F-pilus synthesis, with TraG also being important for stabilizing mating pairs during conjugation (2, 11, 12). These 3 genes and the 10 Mpf genes from the Tra2 region encode the core Mpf complex of R27. It therefore appears that the H-pilus synthesis and Mpf functions of IncHI1 plasmids and IncF plasmids share a common ancestry. It is notable that R27 lacks a homolog of a VirB11 superfamily of nucleoside triphosphate hydrolases (NTPase) (30). VirB11-type NTPases are present in the majority of type IV secretion systems, including IncP and Ti plasmids, but are lacking in the IncF conjugative systems (6).
The other Mpf genes of Tra1 are trhR and trhY, which share no homology to any component of known conjugative systems. These genes may represent type IV secretion system components that are unique to IncHI1 plasmids. Alternatively, since genes that regulate conjugative transfer are generally present around oriT and are the least well-conserved components of transfer systems (38), these ORFs may play a role in regulating conjugative transfer. Consistent with the latter idea, overexpression of TrhR and TrhY increased the transfer frequency of R27 (Table 3).
The major component of the relaxosome is TraI. The relaxase motifs of TraI align best with relaxases from the IncP family (30). Besides the three relaxase motifs, no homology exists between TraI and any of the known relaxases, including the IncP specified relaxases (Table 2). It therefore appears that TraI is distantly related to previously characterized relaxases. Relaxase families have been assigned based on the presence or absence of a helicase domain in their C-terminal region that is responsible for the DNA unwinding activity during conjugative transfer. Relaxases from the IncF/IncW plasmid groups contain a helicase motif, although these motifs and functions are absent in relaxases from the IncP plasmid group (4). TraI from R27 does not contain a helicase domain, suggesting that TraI is from the IncP relaxase lineage; this is consistent with the alignment of the relaxase motifs. From our analysis, the best candidates for relaxosome accessory components are encoded by traJ and traH. Both are essential for transfer but not for H-pilus synthesis, consistent with the characteristics of relaxosome accessory proteins.
The coupling protein of R27, TraG, shares only a low level of identity with characterized coupling proteins. The conserved regions are predominantly at or around the Walker A and Walker B motifs, indicating the functional importance of these motifs. It therefore appears that the coupling protein from IncHI1 plasmids is distantly related to known coupling proteins.
A 285-bp region between traH and trhR was cloned and mobilized by R27. This region is present within a 757-bp intergenic region that contains hallmark features of oriTs. We have compared the nucleotide sequence of the 285-bp oriT to the five nic site families (38) and found no matches. Precisely identifying the nic site of R27 will allow us to determine if it is distantly related to one of the five nic site families or defines its own family.
In addition to the transfer genes present in the Tra1 region, four genes appear to play no role in conjugation. Other transfer systems, such as IncF and IncP, also contain genes within their transfer operons that are not essential for conjugation under laboratory conditions (12, 22). The nonconjugative genes are dispersed among the transfer genes, resulting in an intermittent transfer gene organization. This arrangement creates difficulty in defining the precise boundaries for the Tra1 region. Our results suggest that the Tra1 region is between genes traJ and trhG, thereby eliminating orf115 and orf116 from the original boundaries. This does not, however, exclude genes adjacent to the Tra1 region from being involved in conjugative transfer. For example, ORF130, which is outside the Tra1 region, shares homology to a family of muramidase enzymes found in other conjugative systems (3). Therefore, it is possible that other transfer genes could be present in the vicinity of Tra1.
Our analysis of the Tra1 region indicates that the conjugative transfer system of IncHI1 plasmids shares a common ancestry with multiple transfer systems. The Tra1 region maintains a mosaic-like structure, as indicated by the presence of nontransfer genes, highlighting the complex evolutionary history of IncHI1 plasmids. The transfer systems (trb and vir) of the Ti plasmid are also believed to have evolved by a similar method (1, 9). These observations imply a modular nature for the various transfer system components (Dtr, Mpf , and coupling protein). Indeed, chimeric transfer systems consisting of Dtr (IncQ), coupling protein (IncP), and Mpf (Ti) functions constructed in vivo from different conjugative plasmid systems have been shown to be fully functional for conjugative transfer (17). Therefore, plasmids, especially larger plasmids, appear to evolve conjugative transfer systems by acquiring and combining transfer components from other conjugative transfer systems.
Acknowledgments
This study was supported by grant MOP6200 to D.E.T. from the Canadian Institutes for Health Research (CIHR). T.D.L. is supported by a Studentship from the Alberta Heritage Foundation for Health Research (AHFMR) and a Doctoral Scholarship from the CIHR. M.W.G. is supported by a Studentship from AHFMR. D.E.T. is an AHFMR Scientist.
We thank Andrew Ting for his assistance in cloning traI. We are grateful to Laura Frost for the useful discussions on bacterial conjugation.
REFERENCES
- 1.Alt-Morbe, J., J. L. Stryker, C. Fuqua, P. L. Li, S. K. Farrand, and S. C. Winans. 1996. The conjugal transfer system of Agrobacterium tumefaciens octopine-type Ti plasmids is closely related to the transfer system of an IncP plasmid and distantly related to Ti plasmid vir genes. J. Bacteriol. 178:4248-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, K. G., W. A. Klimke, J. Manchak, and L. S. Frost. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J. Bacteriol. 181:5149-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, M., R. Iberer, K. Bischof, E. Rassi, E. Stabentheiner, G. Zellnig, and G. Koraimann. 2001. Functional and mutational analysis of p19, a DNA transfer protein with muramidase activity. J. Bacteriol. 183:3176-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 25:1011-1022. [DOI] [PubMed] [Google Scholar]
- 5.Cabezon, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 6.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenbrandt, R., M. Kalkum, R. Lurz, and E. Lanka. 2000. Maturation of IncP pilin precursors resembles the catalytic Dyad-like mechanism of leader peptidases. J. Bacteriol. 182:6751-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ermolaeva, M. D., O. White, and S. L. Salzberg. 2001. Prediction of operons in microbial genomes. Nucleic Acids Res. 29:1216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrand, S. K., I. Hwang, and D. M. Cook. 1996. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 178:4233-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fica, A., M. E. Fernandez-Beros, L. Aron-Hott, A. Rivas, K. D'Ottone, J. Chumpitaz, J. M. Guevara, M. Rodriguez, and F. Cabello. 1997. Antibiotic-resistant Salmonella typhi from two outbreaks: few ribotypes and IS200 types harbor Inc HI1 plasmids. Microb. Drug Resist. 3:339-343. [DOI] [PubMed] [Google Scholar]
- 11.Firth, N., K. Ippen-Ihler, and R. A. Skurray. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 12.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmour, M. W., T. D. Lawley, M. M. Rooker, P. J. Newnham, and D. E. Taylor. 2001. Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol. Microbiol. 42:705-715. [DOI] [PubMed] [Google Scholar]
- 14.Gomis-Ruth, F. X., G. Moncalian, R. Perez-Luque, A. Gonzalez, E. Cabezon, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 15.Gomis-Ruth, F. X., G. Moncalian, F. de La Cruz, and M. Coll. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein: detailed structural features and mapping of the active site cleft. J. Biol. Chem., in press. [DOI] [PubMed]
- 16.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton, C. M., H. Lee, P. L. Li, D. M. Cook, K. R. Piper, S. B. von Bodman, E. Lanka, W. Ream, and S. K. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingsman, A., and N. Willetts. 1978. The requirements for conjugal DNA synthesis in the donor strain during flac transfer. J. Mol. Biol. 122:287-300. [DOI] [PubMed] [Google Scholar]
- 20.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 21.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 22.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maher, D., R. Sherburne, and D. E. Taylor. 1991. Bacteriophages for incompatibility group H plasmids: morphological and growth characteristics. Plasmid 26:141-146. [DOI] [PubMed] [Google Scholar]
- 24.Maher, D., R. Sherburne, and D. E. Taylor. 1993. H-pilus assembly kinetics determined by electron microscopy. J. Bacteriol. 175:2175-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newnham, P. J. 1995. Ph.D. dissertation. University of Alberta, Edmonton, Canada.
- 26.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 27.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 28.Rooker, M. M., C. Sherburne, T. D. Lawley, and D. E. Taylor. 1999. Characterization of the Tra2 region of the IncHI1 plasmid R27. Plasmid 41:226-239. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, V. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemaker, N. B., G.-R. Wang, and A. A. Salyers. 2000. Multiple gene products and sequences required for excision of the mobilizable integrated Bacteroides element NBU1. J. Bacteriol. 182:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, D. E., and J. G. Levine. 1980. Studies of temperature-sensitive transfer and maintenance of H incompatibility group plasmids. J. Gen. Microbiol. 116:475-484. [DOI] [PubMed] [Google Scholar]
- 33.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPbeta plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 34.Waters, V. L., B. Strack, W. Pansegrau, E. Lanka, and D. G. Guiney. 1992. Mutational analysis of essential IncP alpha plasmid transfer genes traF and traG and involvement of traF in phage sensitivity. J. Bacteriol. 174:6666-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whelan, K. F., D. Maher, E. Colleran, and D. E. Taylor. 1994. Genetic and nucleotide sequence analysis of the gene htdA, which regulates conjugal transfer of IncHI plasmids. J. Bacteriol. 176:2242-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkins, B. M., and S. E. Hollom. 1974. Conjugational synthesis of F lac+ and Col I DNA in the presence of rifampicin and in Escherichia coli K12 mutants defective in DNA synthesis. Mol. Gen. Genet. 134:143-156. [DOI] [PubMed] [Google Scholar]
- 37.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool. Harwood Academic Publishers, Amsterdam, The Netherlands.