Abstract
Darwin's paradigm holds that the diversity of present-day organisms has arisen via a process of genetic descent with modification, as on a bifurcating tree. Evidence is accumulating that genes are sometimes transferred not along lineages but rather across lineages. To the extent that this is so, Darwin's paradigm can apply only imperfectly to genomes, potentially complicating or perhaps undermining attempts to reconstruct historical relationships among genomes (i.e., a genome tree). Whether most genes in a genome have arisen via treelike (vertical) descent or by lateral transfer across lineages can be tested if enough complete genome sequences are used. We define a phylogenetically discordant sequence (PDS) as an open reading frame (ORF) that exhibits patterns of similarity relationships statistically distinguishable from those of most other ORFs in the same genome. PDSs represent between 6.0 and 16.8% (mean, 10.8%) of the analyzable ORFs in the genomes of 28 bacteria, eight archaea, and one eukaryote (Saccharomyces cerevisiae). In this study we developed and assessed a distance-based approach, based on mean pairwise sequence similarity, for generating genome trees. Exclusion of PDSs improved bootstrap support for basal nodes but altered few topological features, indicating that there is little systematic bias among PDSs. Many but not all features of the genome tree from which PDSs were excluded are consistent with the 16S rRNA tree.
Since the early days of molecular biology, it has been assumed that the historical pattern of genetic relationships among organisms could eventually be reconstructed via statistically based comparisons of the sequences of genes or proteins within individual families (47). In keeping with Darwin's paradigm of treelike descent with modification, these relationships are represented as bifurcating trees. If a gene or protein family has not been affected by gene duplication (i.e., if only orthologs are being compared), its phylogenetic tree shows organismal relationships as well. With the recent rise of genomics, attention has been focused on the genome as an entity that is conceptually distinguishable from both genes and organism (18). As most genes are physically colocated on chromosomes that partition together during meiosis and mitosis, however, the default assumption has been that genetic, genomic, and organismal phylogenies are merely different facets or delineations of the same fundamental historical process. The information content of individual genes may sometimes be insufficient to resolve the complete details of this process, but sequences of multiple genes or proteins—now available from complete genome sequences—can be analyzed jointly to obtain a better (historically more accurate) tree.
However, a potentially serious complication has arisen. It is now appreciated that some genes are sometimes transmitted not vertically (along individual branches of the organismal tree through time) but laterally or horizontally (directly from one branch of the tree to another). The closest relatives (nearest orthologs) of a laterally transferred gene thus occur in genomes in the organismal lineage from which the transfer originated, not in the relatives of its new host; as a consequence, the extrapolated organismal phylogeny is incongruent with the phylogeny inferred from families whose members have been transmitted only vertically. If such lateral gene transfer (LGT) were frequent, the number and diversity of anomalous gene trees might erode our ability to reconstruct correct organismal trees or perhaps even empty this concept of meaning. In the extreme case, what we consider species might have been generated and be maintained not by common ancestry but rather by barriers and firewalls to genetic recombination 38; W. F. Doolittle, personal communication). Many studies have suggested that LGT has occurred frequently in many prokaryotic lineages (7-9, 16, 19-21, 24-26, 31, 33, 38).
The degree to which our understanding of genome evolution may be compromised by LGT depends on the extent to which genetic material has been transferred laterally and, more generally, on our ability to detect and control for incongruent data. We hypothesize that removal of incongruent data should leave sets of genes which, analyzed jointly, should yield a phylogeny that better corresponds with organismal (phenotypic, ultrastructural, and physiological) groupings and with trees inferred from gene families that have undergone little or no lateral transfer (e.g., small-subunit rRNA genes [43]). If the incongruent data are topologically biased (e.g., if they can be identified with one or a few major LGT events), eliminating them from the analysis might yield a topologically different tree. On the other hand, if the incongruent data are unbiased (e.g., if they arose from a large number of dissimilar, quantitatively less-significant events), their removal might improve the statistical support for subtrees but have little effect on tree topology. These competing predictions can be tested directly by inferring whole-genome trees before and after suspect data have been identified and eliminated.
Phylogenetic trees are inferred for individual gene or protein families by maximizing or minimizing an appropriate function over all putatively homologous positions. Trees of organisms are typically derived by parsimony analysis of discrete character states. The first genome trees were constructed by distance analysis and were based on distances derived from proportions of statistically similar open reading frames (ORFs) (presumptive orthologs) or protein folds that could be recognized by pairwise comparisons of genomes (14, 27, 41, 42, 44). Basing these trees on shared ORF or fold contents avoided the perhaps intractable complexities of sequence-based alignment of entire genomes but failed to capture information about the degrees to which sets of ORFs (presumed orthologs) are similar. Grishin et al. (17) used a distance measure based on empirically determined distributions of pairwise interprotein amino acid substitution rates for genomes, while Wolf et al. (44) examined transformations of the pairwise percentages of identity between orthologs. Here we utilized an alternative method of constructing genome trees based instead on mean normalized BLASTP (1) scores, and we compared trees produced by this approach with a content-based genome tree. This study did not constitute a test of the idea that lineages are artifacts of recombinational barriers rather than of shared descent but did explore the extent to which sequences that are incongruent for any reason with the majority signals from their own genomes erode support for a single common phylogenetic tree.
Lateral genetic transfer is not the only process that can obfuscate a phylogeny. An ORF can be phylogenetically discordant (i) if its orthologs have been lost in some but not all other genomes, leaving a patchwork of orthologous and paralogous matches; (ii) because of convergent evolution and nonneutral evolution in general; and (iii) in cases in which certain genes exhibit rates or patterns of sequence change substantially different from those of the other genes in the lineage. To avoid circularity in using phylogenetic analysis to assess phylogenetic incongruity, in this study we instead developed a pairwise statistical approach, correlating patterns of observed genomic similarity among species.
MATERIALS AND METHODS
We established the target database for the initial BLASTP (1) analysis by parsing the GenBank database by species to obtain a large number of “virtual genomes.” Each query database (one for each genome in our analysis) consisted of all of the protein-encoding ORFs recognized for the genome by the respective genome project or community. Each query ORF found a best BLASTP match better than a defined threshold in none, one, or more target species among the species for which there are data in the GenBank database. We ranked these best matches by strength (i.e., improbability of occurrence by chance). Phylogenetic discordance among ORFs in a query genome or between a query ORF and the bulk signal from its own genome was assessed by statistical comparison of the rankings (see below). As explained above, it is the pattern of orthologs that marks an ORF as having arisen by LGT. However, orthology is a tree-based concept (13) that usually cannot be directly established by pairwise comparison. Following established practice (30, 41, 44), we focused instead on reciprocal best matches (RBMs). If a query ORF has no ortholog in a given target genome, BLASTP analysis might nonetheless identify a best match for it (probably a paralog), but the paralog's best match back into the query genome would probably be its own ortholog and not the original query ORF (i.e., the match would be nonreciprocal). This approach is imperfect, as it can be confounded by the absence of a target ORF's true ortholog in the query genome and best BLAST matches do not need to be phylogenetically adjacent sequences (22); nonetheless, it should be unbiased and eliminate many instances of paralogy. Query ORFs involved in fewer than four RBMs were excluded from further analysis, as such patterns contained too little information for rigorous comparison (see below).
If all genes within a genome have the same phylogenetic history, the RBMs for each gene vis-a-vis the target genomes (here, the GenBank virtual genomes) should rank similarly. The rankings should be the same, within statistical variation, for the genome as a whole and for each constituent ORF. If an ORF does not have this common ranking but instead shows a conflicting pattern, it is discordant. Indeed, it is phylogenetically discordant because, by analogy with construction of a tree from a distance matrix, a conflicting pattern of similarity relationships must specify a conflicting tree. Missing orthologs create gaps but not misinformation (incorrectly ordered rankings).
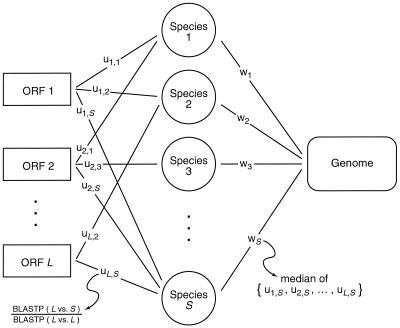
To quantify these relationships, we introduced a normalized similarity score (u) for a query ORF and its RBM in a given target species. We computed this score by dividing the BLASTP-based similarity score (bit score [S′]) by the ORF's self-matching score (Fig. 1). The median of the u values for a target species defined w, a measure of the query genome's overall sequence similarity with sequences from that target species. By correlating u and w for all species in which the ORF found a match at a BLASTP expectation value greater than a defined threshold (here e = 1.0 × 10−10), we could determine if the pattern of relationships was consistent with the ORF having evolved concordantly with the rest of the genome.
FIG. 1.
Statistical strategy. In this schematic diagram, ORF 1 finds RBMs in species 1, 2, and S; ORF 2 finds RBMs in species 1, 3, and S; and ORF L finds RBMs in species 2 and S. Thus, w1 = {u1,1, u2,1, . . .}; w2 = {u1,2, . . ., uL,2}; w3 = {u2,3, . . .}; and wS = {u1,S, u2,S, . . ., uL,S}. In this example, ORF 1's set of w values is {w1, w2, . . ., wS}; ORF 2's set of w values is {w1, w3, . . ., wS}; and ORF L's set of w values is {w2, . . ., wS}. An ORF's set of u values is correlated with its set of w values as described in the text.
The set of w values associated with a given ORF formed the intrinsic hypothesis of the test; that is, it was the relative distribution of w values against which we tested the distribution of u values. Since the distributions of u values from which each w was calculated were not identical, we used an approximate randomization test (32) to determine the significance of the observed correlation between u and w. We chose the Spearman rank correlation (37, 46) as our test statistic because there was no reason to assume that the relationship between u and w was linear. To approximate the distribution of the test statistic, the underlying distribution of u values that made up each w was sampled with replacement to generate random sets of u values that were then correlated with w. The probability of obtaining a correlation (r) value as small as or smaller than the observed r0 value was defined by (C + 1)/(NR + 1), where C is the count of rR ≤ r0 and NR is the total number of randomizations. (NR was the lesser of 199,999 and Πni, ∀ni > 0, where ni is the number of ORFs shared by the query genome and the target genome [i].) For each pair of genomes (the query and each target), the size of each resampled set was equal to the number of RBMs (Table 1).
TABLE 1.
Proportions of ORFs analyzable and proportions of ORFs found to be phylogenetically discordant in each genome
| Genome | Total no. of loci | No. of analyzable ORF (% of total) | No. of PDSs at P < 0.05 (% of analyzable ORFs) |
|---|---|---|---|
| Aquifex aeolicus | 1,522 | 1,063 (70) | 140 (13.2) |
| Bacillus halodurans | 4,066 | 2,187 (54) | 228 (10.4) |
| Bacillus subtilis | 4,099 | 2,133 (52) | 217 (10.2) |
| Borrelia burgdorferi | 1,637 | 543 (33) | 81 (14.9) |
| Buchnera sp. strain APS | 574 | 542 (94) | 41 (7.6) |
| Campylobacter jejuni | 1,634 | 1,052 (64) | 109 (10.4) |
| Chlamydia muridarum | 818 | 470 (57) | 63 (13.4) |
| Chlamydia trachomatis | 877 | 585 (67) | 75 (12.8) |
| Chlamydophila pneumoniae AR39 | 997 | 709 (71) | 75 (10.6) |
| Chlamydophila pneumoniae CWL029 | 1,052 | 602 (57) | 90 (15.0) |
| Deinococcus radiodurans | 3,103 | 1,524 (49) | 166 (10.9) |
| Escherichia coli | 4,289 | 2,410 (56) | 197 (8.2) |
| Haemophilus influenzae | 1,709 | 1,315 (77) | 107 (8.1) |
| Helicobacter pylori 26695 | 1,565 | 851 (54) | 101 (11.9) |
| Helicobacter pylori J99 | 1,491 | 853 (57) | 103 (12.1) |
| Mycobacterium tuberculosis | 3,918 | 1,735 (44) | 201 (11.6) |
| Mycoplasma genitalium | 467 | 317 (68) | 45 (14.2) |
| Mycoplasma pneumoniae | 677 | 331 (49) | 51 (15.4) |
| Neisseria meningitidis MC58 | 1,989 | 1,214 (61) | 117 (9.6) |
| Neisseria meningitidis Z2491 | 2,064 | 1,229 (60) | 128 (10.4) |
| Pseudomonas aeruginosa | 5,565 | 2,980 (54) | 287 (9.6) |
| Rickettsia prowazekii | 834 | 570 (68) | 89 (15.6) |
| Synechocystis sp. strain PCC6803 | 3,169 | 1,570 (50) | 193 (12.3) |
| Thermotoga maritima | 1,846 | 1,180 (64) | 108 (9.2) |
| Treponema pallidum | 1,031 | 558 (54) | 94 (16.8) |
| Ureaplasma urealyticum | 611 | 334 (55) | 52 (15.6) |
| Vibrio cholerae | 3,828 | 2,173 (57) | 180 (8.3) |
| Xylella fastidiosa | 2,831 | 1,270 (45) | 140 (11.0) |
| Aeropyrum pernix | 2,694 | 848 (31) | 52 (6.1) |
| Archaeoglobus fulgidus | 2,407 | 1,183 (49) | 101 (8.5) |
| Halobacterium sp. strain NRC-1 | 2,605 | 1,244 (48) | 104 (8.4) |
| Methanobacterium thermoautotrophicum | 1,869 | 965 (52) | 92 (9.5) |
| Methanococcus jannaschii | 1,770 | 895 (51) | 72 (8.0) |
| Pyrococcus abyssi | 1,765 | 1,065 (60) | 69 (6.5) |
| Pyrococcus horikoshii | 2,064 | 998 (48) | 60 (6.0) |
| Thermoplasma acidophilum | 1,478 | 866 (59) | 64 (7.4) |
| Saccharomyces cerevisiae | 6,245 | 2,561 (41) | 239 (9.3) |
| Mean (n = 37) | 2,194 | (56) | (10.8) |
The 16S rRNA tree was extracted from a much larger (7,322-prokaryotic sequence) maximum-likelihood tree maintained by the Ribosomal Database Project (29).
Genome trees were inferred by Fitch-Margoliash least-squares analysis of distance-type matrices (17), each element of which was an ORF-based measure of pairwise dissimilarity between genomes, either 1.0 minus the proportion of ORFs shared by a given pair of genomes (41) or 1.0 minus the mean of normalized pairwise BLASTP scores. BLASTP scores (42) were normalized (4) by dividing an ORF's score against the target genome by the ORF's score against itself. The target database was the set of ORFs identified for the genome itself, not, as described above, all genes deposited under the species designation in GenBank. Only ORFs with matches better than a defined threshold (BLASTP e = 1.0 × 10−10) were used; the reciprocal best-match criterion was not used to derive genome trees. Distance matrices were generated using the NeuroGadgets Inc. web service (http://www.neurogadgets.com). Distance analysis was carried out using the FITCH program in the PHYLIP software package (11), with global rearrangements and randomized (jumbled) species input order. For bootstrap analysis, samples (n = 100) were taken with replacement from the ORFs shared by each pair of genomes, and from these a mean distance was calculated for that pair of genomes. Distance trees were again generated using FITCH, and the majority rule consensus tree was computed using the CONSENSE program in PHYLIP. Trees were visualized and bootstrap values were added using TREEVIEW (36).
RESULTS
A total of 81,160 ORFs in 37 query genomes representing the Bacteria (28 species or strains), the Archaea (eight species), and Saccharomyces cerevisiae were examined; for 42,925 (52.9%) of these ORFs we found four or more RBMs, and we used these ORFs for analysis (Table 1). The minimum analyzable proportion was found in Aeropyrum pernix (31%), and the maximum analyzable proportion was found in Buchnera sp. strain APS (94%); the unweighted mean for 37 genomes was 56%. Of the 42,925 analyzable ORFs, 4,331 (10.1%) were phylogenetically discordant at P < 0.05. The lowest proportion of phylogenetically discordant sequences (PDSs) was observed in Pyrococcus horikoshi (6.0%), and the highest proportion of PDSs was observed in Treponema pallidum (16.8%); the unweighted mean for these genomes was 10.8%. We suspect that many of the sparsely distributed ORFs excluded from this analysis are phylogenetically discordant as well (39).
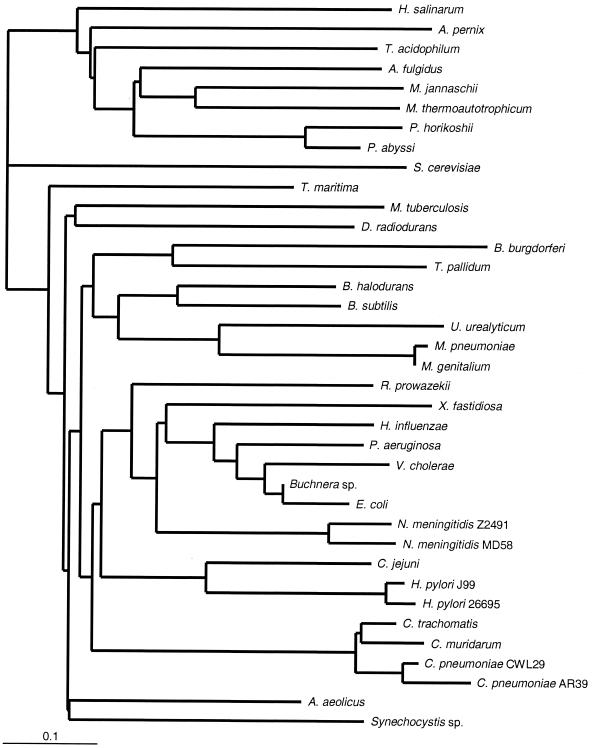
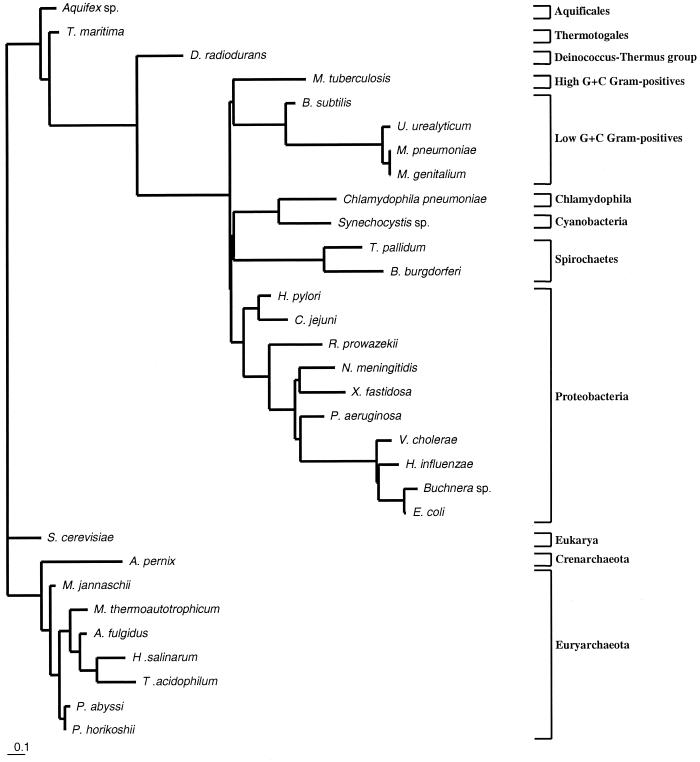
As described above, the first genome phylogenies were based on a distance-type measure derived from the proportions of ORFs shared pairwise by genomes. Figure 2 shows a genome tree for the 37 microbial genomes based on the proportion in each query genome of ORFs that had an initial BLASTP match better than the threshold (e = 1.0 × 10−10) in each other genome. It was not immediately obvious how to assess statistical support (confidence intervals for internal nodes) for such a tree. Snel et al. (41) assessed confidence by the half-delete jackknife method (45), deleting random halves of ORFs in each query genome, reassessing the proportions of shared genes, inferring a new tree for each replicate, and counting the number of times out of 100 that a particular cluster was found. Wolf et al. (44) instead used the nonparametric bootstrap method (12), resampling from among identified orthologs. The proportion-based genome tree for the 37 genomes (Fig. 2) resembles the tree based on 16S rRNA sequences (Fig. 3) in many respects, but there are a number of discrepancies. In the proportion-based genome tree the Thermotoga maritima genome branches basally among bacterial genomes, as the Thermotoga 16S rRNA does. However, in the proportion-based tree, the genome of Aquifex aeolicus does not branch second deepest; instead, it groups with the genome of Synechocystis as the third-deepest branch. The Mycobacterium tuberculosis genome does not group with the genomes of other Firmicutes; instead, it joins the genome of Deinococcus radiodurans on the second-most-basal branch. In addition, the Haemophilus influenzae genome does not group with the genomes of enteric members of the γ subclass of the class Proteobacteria (γ-proteobacteria). Even more discrepant is the archaeal subtree, in which the genomes of Halobacterium salinarum and A. pernix constitute the two most-basal branches (the former does not group with genomes of other euryarchaeotes). The methanogen genomes, together with the genomes of Archaeoglobus fulgidus and the two Pyrococcus species, form a group not seen in 16S rRNA trees or indeed in most other molecular sequence trees.
FIG. 2.
Genome tree based on the proportions of query ORFs that found a match in each target genome better than the threshold BLASTP expectation (e = 1.0 × 10−10). Proportions were computed (http://www.neurogadgets.com) by determining the number of ORFs shared by the query genome (smaller of the pair) and the target genome (larger of the pair) and then dividing this number by the number of ORFs in the query genome. A distance matrix was generated by computing 1.00 minus the proportion of shared loci; this matrix was imported into PHYLIP (12) for analysis by FITCH (see Materials and Methods). The branch lengths reflect distances, as assessed by these criteria, between genomes.
FIG. 3.
16S rRNA gene tree, adapted from the Ribosomal Database Project (29). Where 16S rRNA sequences were not available from the Ribosomal Database Project, close relatives were selected. The major organism classifications are consistent with those described by Olsen et al. (34).
To explore whether the apparent anomalies arose because the underlying similarity matrix was based on proportions of shared ORFs (instead of some other measure of distance), we developed an alternative measure based on mean sequence similarity (see Materials and Methods). Again, only the ORFs having a match better than the threshold (BLASTP e = 1.0 × 10−10) were considered. We assessed confidence in internal nodes by using the nonparametric bootstrap method (11, 44), sampling with replacement from the ORFs shared by each pair of genomes to generate the mean distances used to construct trees. This approach lends itself to filtering (see below) better than the approach based on proportions of shared ORFs does. The genome tree produced by using this alternative method (Fig. 4) differs from the proportion-based tree (Fig. 2) but also exhibits some different discrepancies vis-a-vis the 16S rRNA tree (Fig. 3).
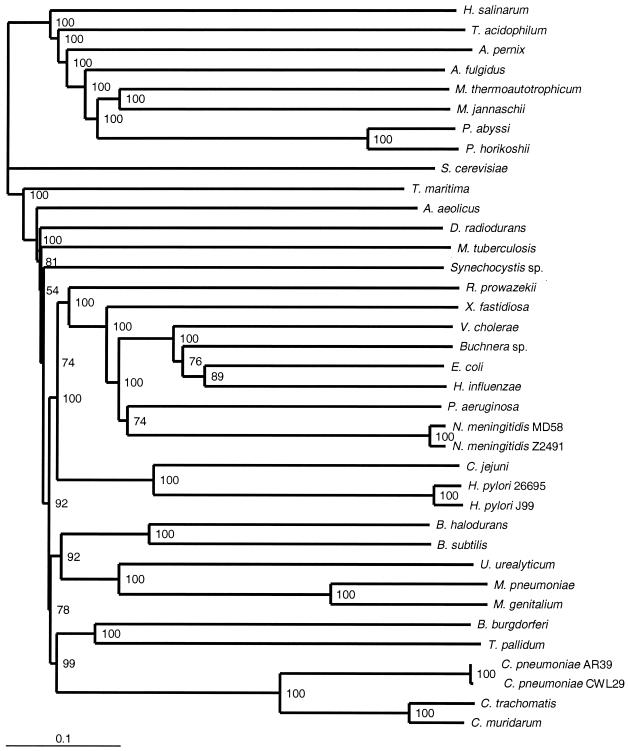
FIG. 4.
Genome tree based on normalized BLASTP scores, constructed by using all query ORFs that found a match in the target genome better than the BLASTP expectation (e = 1.0 × 10−10). The tree was constructed as described in the legend to Fig. 2, but 100 random replicates (pairwise genome scores reconstructed from individual ORF scores, with resampling) were examined. The numbers at the nodes are bootstrap values, which were generated by the CONSENSE program in PHYLIP.
In the genome tree based on mean normalized BLASTP scores (Fig. 4), the T. maritima genome retains its position as the most-basal branch among the bacterial genomes; the A. aeolicus genome is (as it is in the 16S rRNA tree) resolved with good confidence as the second-deepest branch; and the genome of D. radiodurans appears third. Chlamydial genomes form the sister group of spirochete genomes as they do in the 16S rRNA sequence tree, although cyanobacterial genomes are still distant. A large clade of genomes from Proteobacteria, low-G+C-content Firmicutes, and chlamydiae plus spirochetes is well supported, and the genome of the high-G+C-content firmicute M. tuberculosis is completely outside the group. The genome of Xylella fastidiosa is at the base of the γ-proteobacterial genome group, as it is in the proportion tree, although this group includes genomes of β-proteobacteria (represented by the two Neisseria meningitidis genomes). 16S rRNA sequence trees (Fig. 3) (34) also suggest that the β-proteobacterial sequences might be included among the sequences of the γ-proteobacteria. The genome of H. influenzae groups with the genomes of the other enteric γ-proteobacteria, as it does in the 16S rRNA tree, which is consistent with the findings of nonmolecular bacterial systematics. The archaeal portion of the score-based genome tree is rearranged compared to the archaeal portion of the proportion-based genome tree (Fig. 2) and is topologically quite different from the archaeal portion of the 16S rRNA tree (Fig. 3), with genomes of both H. salinarum and Thermoplasma acidophilum branching earlier than the genome of the crenarchaeote A. pernix.
Excluding sequences that could not be analyzed for potential phylogenetic discordance (but retaining the discordant sequences) yielded a tree that was essentially identical to the tree shown in Fig. 4 (data not shown) and differed in only three topological features. The group of five Bacillus, Mycoplasma, and Ureaplasma genomes was resolved as a sister group of the spirochete and chlamydial genomes, albeit with very weak bootstrap support (55%), instead of branching next from the backbone as in Fig. 4. The genome of Pseudomonas aeruginosa branches immediately after the two Neisseria genomes, instead of forming a sister group with them as in Fig. 4, although the bootstrap support is weak in both cases (67 and 62%). Finally, the branching order of the H. influenzae and Buchnera sp. genomes is reversed, again with low bootstrap support. Thus, excluding the 47.1% of the ORFs that had relatively few strong matches in these genomes had an almost negligible effect on the topology of the genome tree based on mean normalized BLAST scores.
Exclusion of PDSs (P < 0.05) from the latter set yielded the tree shown in Fig. 5. The topology of this tree is almost identical to that of the tree described above, differing only in the position of the P. aeruginosa genome, which groups weakly with the two Neisseria genomes, as in Fig. 4. The two other topological changes compared with Fig. 4 resulted from excluding the 38,235 nonanalyzable ORFs, not from removing the 4,331 discordant ORFs. Exclusion of discordant sequences did, however, substantially improve the resolution of some subtrees, as measured by the nonparametric bootstrap method. Within the γ and β-proteobacteria, for example, the mean bootstrap values increased from 86.1% (Fig. 4) and 86.6% (in the tree containing all analyzable ORFs [data not shown]) to 91.3% (Fig. 5).
FIG. 5.
Genome tree based on normalized BLASTP scores better than the threshold (e = 1.0 × 10−10), after removal of PDSs (P < 0.05). Tree construction and bootstrapping were performed as described in the legend to Fig. 4.
DISCUSSION
Our results demonstrate that a biologically reasonable, statistically well-supported genome tree can be produced from a distance type of matrix based on mean normalized BLASTP scores. BLAST (1) is based on pairwise local alignments and consequently does not guarantee that all homologous characters are recognized and scored as such; on the other hand, normalized BLAST similarity (bit) scores are well defined statistically and can be compared and ranked much more naturally than global alignments can. By analogy with algorithms by which trees are derived from distance matrices, we hypothesized that it is possible to define statistically a difference in rankings among elements within individual rows or columns of distance (here, BLASTP-based) matrices such that matrices with anomalously ranked elements would almost always specify an incongruent tree. Our results indicate that despite its inherent approximations, our approach was successful enough to warrant further development and refinement (e.g., iterative comparisons and use of a rescaled distance measure to more accurately represent the weaker pairwise matches). Distance-based genome trees based on much less stringent thresholds have recently been published by Grishin et al. (17) and Wolf et al. (44), although PDSs were not identified or removed.
We identified 4,331 ORFs (10.1% of the 42,925 analyzable ORFs) that have patterns of BLASTP matches which are significantly different from the patterns of their host genomes and would thus probably specify incongruent trees. Removing these ORFs from the analysis affected the topology of the genome tree very little, although it did improve the confidence in a number of subtrees as assessed by the nonparametric bootstrap method. We therefore concluded that both according to the proportions of shared ORFs and according to sequence divergence, genome phylogeny largely agrees with the canonical view of prokaryote evolution based on 16S rRNA gene sequences. It is especially noteworthy that restricting the analysis to ORFs that were found not to be discordant produced so few topological changes; this demonstrates that neither the set of nonanalyzable ORFs (47.1% of the ORFs) nor the 5.3% of the total ORFs identified as PDSs at P < 0.05 is specifically biased toward a preferred alternative topology. Removal of PDSs improved bootstrap support most noticeably in the γ- and β-proteobacterial subtree, although the comparison was imperfect, as there was limited or no room for improvement in several regions of the tree.
Trees based on overall pairwise genomic distances do not, however, agree completely with the 16S rRNA tree. This is especially true for the archaeal subtree, since in all our genome trees the genome of the crenarchaeote A. pernix grouped with the genomes of euryarchaeotes. Euryarchaeota are paraphyletic in other genome trees as well (Fig. 3 and 5 of reference 44) and in a multigene tree based on sequences of ribosomal proteins (Fig. 6 of reference 44), while Crenarchaeota are paraphyletic in trees based on radA (40). Thus, single-gene trees, such as 16S rRNA trees (5, 35), are increasingly isolated in suggesting that Euryarchaeota and Crenarchaeota are monophyletic. The basal position of the genome of Halobacterium sp. (Fig. 2, 4, and 5) is unexpected based on small-subunit ribosomal DNA analysis (Fig. 3) but occurs in the distance-based genome tree of Wolf et al. (Fig. 8 of reference 44) and in a multigene tree based on concatenated ribosomal proteins (Fig. 6 of reference 44). We suspect that this could be an artifact of the high G+C content of this organism and the resultant systematic bias in the amino acid composition of the proteins (15) or an artifact of the elevated content of acidic amino acids that help stabilize proteins in the presence of the high concentrations of intracellular salt characteristic of halobacteria (10). Haloarchaeal genomes may also contain significant numbers of laterally transferred genes (6). It is more difficult to explain why the genome of T. acidophilum branches more basally than expected, although this occurs in other genome trees (Fig. 3 and 5 of reference 44) and in the tree based on ribosomal proteins (Fig. 6 of reference 44). Methanogens are monophyletic in our trees (Fig. 2, 4, and 5) and other genome trees (Fig. 3 of reference 17; Fig. 3 and 5 of reference 44) but not in the trees based on small-subunit ribosomal DNA (5, 40) (Fig. 3), radA (40), or ribosomal proteins (Fig. 6 of reference 44).
Among the bacteria, T. maritima and A. aeolicus appear on the deepest (most basal) branches in the trees based on 16S rRNA (5, 35) (Fig. 3) and ribosomal proteins (Fig. 6 of reference 44) and in some (Fig. 3 and 5 of reference 44) but not all genome trees. In our analyses, the genome of T. maritima always appeared to be the most basal genome, but the genome of A. aeolicus was second deepest only in our distance-based trees (with or without removal of discordant sequences). The numbers of and degrees of similarity between proteins in one or both of these hyperthermophilic bacteria and some archaea have opened a debate about common ancestry versus LGT (2, 3, 23, 28).
Spirochete and chlamydial genomes form a single clade in our distance-based genome trees (Fig. 4 and 5), in the distance-based genomes tree of Grishin et al. (Fig. 3 of reference 17) and Wolf et al. (Fig. 5 of reference 44), and in a multigene ribosomal protein-based tree (Fig. 6 of reference 44). They do not group together in genome trees based on proportions of shared genes (Fig. 2 of reference 42; Fig. 3 of reference 44) (Fig. 2) or in many 16S rRNA-based trees (34) (Fig. 3). No genome trees support monophyletic grouping of all Firmicutes (low-G+C-content and high-G+C-content gram-positive organisms), as some 16S rRNA trees do (Fig. 3). Previously, the low-G+C-content gram-positive bacteria (Bacillus-Clostridium group of Firmicutes) were polyphyletic in genome trees based on proportions of shared genes (Fig. 2 of reference 42; Fig. 3 of reference 44) but monophyletic in distance-based genome trees (Fig. 3 of reference 17; Fig. 5 of reference 44), 16S rRNA-based trees 34) (Fig. 3), and concatenated ribosomal protein gene-based trees (Fig. 6 of reference 44). These organisms were monophyletic in all of our genome trees (Fig. 2, 4, and 5).
Our genome trees are the first trees to resolve the Proteobacteria as a stable monophyletic group, in agreement with 16S rRNA trees (34) (Fig. 3) and concatenated ribosomal protein trees (Fig. 6 of reference 44); the levels of bootstrap support were high (99 and 100% before and after removal of discordant sequences, respectively). Our proportion-based genome tree (Fig. 2) may be unique in resolving the β-proteobacteria as a sister lineage of the γ-proteobacteria; in our distance-based trees (Fig. 4 and 5), as in the trees based on16S rRNA (34) (Fig. 3) and ribosomal proteins (Fig. 6 of reference 44), Neisseria groups with the γ-proteobacteria.
Details aside, many genome trees are substantially similar to the 16S rRNA gene tree and to many other single-gene trees and contain physiologically coherent groups largely consistent with modern prokaryote systematics. This finding is obviously consistent with the Darwinian model of descent along genealogical lineages, as on a bifurcating tree, but in and of itself it does not prove that prokaryotes have evolved mostly by traditional vertical descent. Doolittle (8, 9; personal communication) has suggested a scenario of genome evolution in which LGT plays a predominant role and lineages owe their existence to selective sharing of gene pools that might be constrained by environmental, physiological, and other nongenealogical factors. In this scenario, distances such as those underlying the construction of genome trees might reflect the frequency of LGT, not the time since a common ancestor. The genes that confuse genome phylogenies are a testament to this lateral exchange; we explore them further elsewhere (39). Thus, it remains to be determined whether the topologies in genome trees reflect Darwinian evolutionary lineages or are artifacts of an elaborate network of differential lateral genetic exchange. We strongly urge that our results be interpreted in this context.
Acknowledgments
We are grateful to A. St. Jean for his efforts in developing the NeuroGadgets Inc. Bioinformatics Web Service and to members of the Evolutionary Biology Program of the Canadian Institute for Advanced Research for helpful comments.
This work was funded by the Natural Sciences and Engineering Research Council of Canada and by the Institute for Molecular Bioscience, University of Queensland. We are grateful to G. Drouin for financial support of G.D.P.C.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., R. L. Tatusov, Y. I. Wolf, R. Walker, and E. V. Koonin. 1998. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 14:442-444. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., R. L. Tatusov, Y. I. Wolf, R. Walker, and E. V. Koonin. 1999. Reply. Trends Genet. 15:299-300. [DOI] [PubMed] [Google Scholar]
- 4.Bansal, A. K., P. Bork, and P. J. Stuckey. 1998. Automated pair-wise comparisons of microbial genomes. Math. Modelling Sci. Comput. 9:1-23. [Google Scholar]
- 5.Barns, S. M., C. F. Delwiche, J. D. Palmer, and N. R. Pace. 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA 93:9188-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlebois, R. L. 1999. Evolutionary origins of the haloarchaeal genome, p. 309-317. In A. Oren (ed.), Microbiology and biogeochemistry of hypersaline environments. CRC Press, Boca Raton, Fla.
- 7.Charlebois, R. L. 1999. Archaea: whose sister lineage?, p. 309-317. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. American Society for Microbiology, Washington, D.C.
- 8.Doolittle, W. F. 1999. Phylogenetic classification and the universal tree. Science 284:2124-2128. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle, W. F. 2000. Uprooting the tree of life. Sci. Am. 282(2):90-95. [DOI] [PubMed] [Google Scholar]
- 10.Ebel, C., P. Faou, B. Franzetti, B. Kernel, D. Madern, M. Pascu, C. Pfister, S. Richard, and G. Zaccai. 1999. Molecular interactions in extreme halophiles—the solvation-stabilization hypothesis for halophilic proteins, p. 227-237. In A. Oren (ed.), Microbiology and biogeochemistry of hypersaline environments. CRC Press, Boca Raton, Fla.
- 11.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Fitch, W. M. 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19:99-113. [PubMed] [Google Scholar]
- 14.Fitz-Gibbon, S. T., and C. H. House. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27:4218-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, P. G., and D. A. Hickey. 1999. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol. 48:284-290. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Vallvé, S., A. Romeu, and J. Palau. 2000. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 10:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grishin, N. V., Y. I. Wolf, and E. V. Koonin. 2000. From complete genomes to measures of substitution rate variability within and between proteins. Genome Res. 10:991-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huynen, M. A., and P. Bork. 1998. Measuring genome evolution. Proc. Natl. Acad. Sci. USA 95:5849-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin, E. V., and M. Y. Galperin. 1997. Prokaryotic genomes: the emerging paradigm of genome-based microbiology. Curr. Opin. Genet. Dev. 7:757-763. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V., A. R. Mushegian, M. Y. Galperin, and D. R. Walker. 1997. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol. Microbiol. 25:619-637. [DOI] [PubMed] [Google Scholar]
- 22.Koski, L. B., and G. B. Golding. 2001. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 52:540-542. [DOI] [PubMed] [Google Scholar]
- 23.Kyrpides, N. C., and G. J. Olsen. 1999. Archaeal and bacterial hyperthermophiles. Horizontal gene exchange or common ancestry? Trends Genet. 15:298-299. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence, J. G., and H. Ochman. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383-397. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence, J. G., and H. Ochman. 2002. Reconciling the many faces of lateral gene transfer. Trends Microbiol. 10:1-4. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence, J. G., and J. R. Roth. 1999. Genomic flux: genome evolution by gene loss and acquisition, p. 263-289. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. American Society for Microbiology, Washington, D.C.
- 27.Lin, J., and M. Gerstein. 2000. Whole-genome trees based on the occurrence of folds and orthologs: implications for comparing genomes on different levels. Genome Res. 10:808-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Logdson, J. M., Jr., and D. M. Faguy. 1999. Evolutionary genomics: Thermotoga heats up lateral gene transfer. Curr. Biol. 9:R747-R751. [DOI] [PubMed] [Google Scholar]
- 29.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mushegian, A. R., and E. V. Koonin. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 93:10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed]
- 32.Noreen, E. W. 1989. Computer intensive methods for testing hypotheses: an introduction. John Wiley & Sons, New York, N.Y.
- 33.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 34.Olsen, G. J., C. L. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 176:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 36.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Applic. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 37.Press, W. H., S. A. Teukolsky, W. T. Vetterling, and B. P. Flannery. 1992. Numerical recipes in C: the art of scientific computing, 2nd ed. Cambridge University Press, New York, N.Y.
- 38.Ragan, M. A. 2001. Detection of lateral gene transfer among microbial genomes. Curr. Opin. Genet. Dev. 11:620-626. [DOI] [PubMed] [Google Scholar]
- 39.Ragan, M. A., and R. L. Charlebois. Distributional profiles of homologous open reading frames among bacterial phyla: implications for vertical and lateral transmission. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 40.Sandler, S. J., P. Hugenholtz, C. Schleper, E. F. DeLong, N. R. Pace, and A. J. Clark. 1999. Diversity of radA genes from cultured and uncultured Archaea: comparative analysis of putative RadA proteins and their use as a phylogenetic marker. J. Bacteriol. 181:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snel, B., P. Bork, and M. A. Huynen. 1999. Genome phylogeny based on gene content. Nat. Genet. 21:108-110. [DOI] [PubMed] [Google Scholar]
- 42.Tekaia, F., A. Lazcano, and B. Dujon. 1999. The genomic tree as revealed from whole proteome comparisons. Genome Res. 9:550-557. [PMC free article] [PubMed] [Google Scholar]
- 43.Woese, C. R. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA 97:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf, Y. I., I. B. Rogozin, N. V. Grishin, R. L. Tatusov, and E. V. Koonin. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 1:8. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, C. F. J. 1986. Jackknife, bootstrap and other resampling methods in regression analysis. Ann. Stat. 14:1261-1295. [Google Scholar]
- 46.Zar, J. H. 1996. Biostatistical analysis, 3rd ed. Prentice-Hall, Upper Saddle River, N.J.
- 47.Zuckerkandl, E., and L. Pauling. 1965. Evolutionary divergence and convergence in proteins, p. 97-166. In V. Bryson and H. J. Vogel (ed.), Evolving genes and proteins. Academic Press, New York, N.Y.