Abstract
Many bacteria employ the nonmevalonate pathway for synthesis of isopentenyl diphosphate, the monomer unit for isoprenoid biosynthesis. However, gram-positive cocci exclusively use the mevalonate pathway, which is essential for their growth (E. I. Wilding et al., J. Bacteriol. 182:4319-4327, 2000). Enzymes of the mevalonate pathway are thus potential targets for drug intervention. Uniquely, the enterococci possess a single open reading frame, mvaE, that appears to encode two enzymes of the mevalonate pathway, acetoacetyl-coenzyme A thiolase and 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase. Western blotting revealed that the mvaE gene product is a single polypeptide in Enterococcus faecalis, Enterococcus faecium, and Enterococcus hirae. The mvaE gene was cloned from E. faecalis and was expressed with an N-terminal His tag in Escherichia coli. The gene product was then purified by nickel affinity chromatography. As predicted, the 86.5-kDa mvaE gene product catalyzed both the acetoacetyl-CoA thiolase and HMG-CoA reductase reactions. Temperature optima, ΔHa and Km values, and pH optima were determined for both activities. Kinetic studies of acetoacetyl-CoA thiolase implicated a ping-pong mechanism. CoA acted as an inhibitor competitive with acetyl-CoA. A millimolar Ki for a statin drug confirmed that E. faecalis HMG-CoA reductase is a class II enzyme. The oxidoreductant was NADP(H). A role for an active-site histidine during the first redox step of the HMG-CoA, reductase reaction was suggested by the ability of diethylpyrocarbonate to block formation of mevalonate from HMG-CoA, but not from mevaldehyde. Sequence comparisons with other HMG-CoA reductases suggest that the essential active-site histidine is His756. The mvaE gene product represents the first example of an HMG-CoA reductase fused to another enzyme.
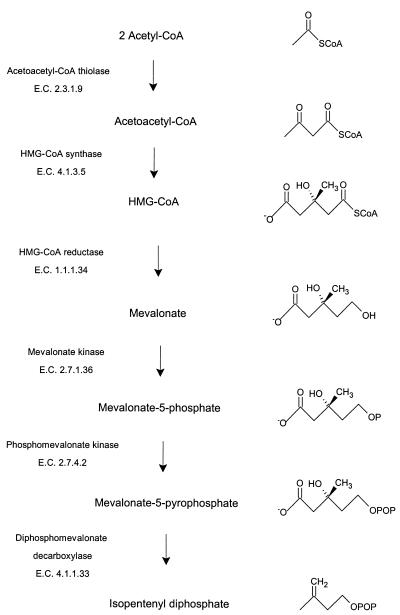
Isoprenoids, lipids synthesized from the 5-carbon isoprene units of isopentenyl diphosphate, serve diverse and numerous functions in all living organisms. Isopentenyl diphosphate is formed by one of two pathways (14, 21), the mevalonate pathway (Fig. 1) or the nonmevalonate pathway. Mammals and archaea appear to use the mevalonate pathway exclusively, whereas plants use both pathways (19).
FIG. 1.
Intermediates and enzymes of the mevalonate pathway for isopentenyl diphosphate biosynthesis.
Until recently, all bacteria were thought to use only the nonmevalonate pathway. However, analysis of microbial genome sequences has revealed that the gram-positive cocci and the spirochete Borrelia burgdorferi possess genes that encode only enzymes of the mevalonate pathway (25), whereas a eubacterial streptomycete strain appears to possess genes that encode both pathways (13, 23). Genetic disruption experiments have shown that the mevalonate pathway enzymes are essential for the growth of gram-positive cocci (25). Genes encoding enzymes of the mevalonate pathway are essential for the growth of Staphylococcus aureus, and mutants auxotrophic for mevalonate were severely attenuated for virulence in mice (26).
In enterococci, the mevalonate pathway genes are clustered at two positions on the genome, and, uniquely, two enzymes of the mevalonate pathway appear to be encoded by a single gene, mvaE (25). One enzyme is acetoacetyl-coenzyme A (CoA) thiolase (acetyl-CoA C-acetyltransferase, EC 2.3.1.9), the catalyst for the first reaction of the mevalonate pathway (Fig. 1).
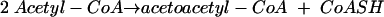 |
(1) |
Reaction 1 is freely reversible, and thiolysis (reaction 2) is favored.
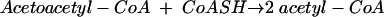 |
(2) |
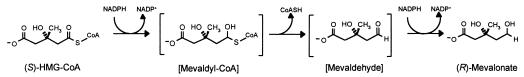
The second enzyme is 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase, the best-characterized enzyme of the mevalonate pathway (3) and the target of the statin drugs that lower human blood cholesterol levels (5). HMG-CoA reductase catalyzes the rate-limiting step of the mevalonate pathway, the reductive deacylation of (S)-HMG-CoA to (R)-mevalonate, reaction 3 (Fig. 2). Reaction 3 involves successive formation of enzyme-bound mevaldyl-CoA and mevaldehyde and proceeds in three stages, the first and third of which are reductive.While mevaldehyde is not released during the course of the reaction, HMG-CoA reductase catalyzes two reactions of free mevaldehyde. Reaction 4 resembles the third stage, and reaction 5 resembles the reverse of stages one and two of the overall reaction. HMG-CoA reductase also catalyzes reaction 6, the oxidative acylation of (R)-mevalonate to (S)-HMG-CoA.
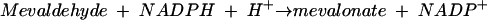 |
(4) |
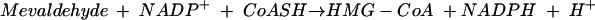 |
(5) |
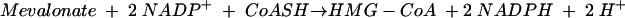 |
(6) |
There are two structural classes of HMG-CoA reductase (2): the class I enzymes of eukarya and several archaea and the class II HMG-CoA reductases of Pseudomonas mevalonii (11), Archaeoglobus fulgidus (12), S. aureus (26), and some streptomycetes (6, 23). Crystal structures of both classes of the enzyme have been solved (10, 15). Relative to the class I HMG-CoA reductases, those of class II are over four orders of magnitude less sensitive to inhibition by statin drugs (1, 12, 26).
FIG. 2.
Substrates and products of the reaction catalyzed by HMG-CoA reductase (reaction 3). The putative enzyme-bound intermediates mevaldyl-CoA and mevaldehyde are shown in brackets.
Enterococci are a major cause of nosocomial infections, and Enterococcus faecalis is responsible for about 85% of all enterococcal infections. Since the mevalonate pathway is essential for the survival of gram-positive cocci, the class II HMG-CoA reductases, and potentially other enzymes of the mevalonate pathway, represent potential targets for development of active-site-directed inhibitors for use as antibiotics against multiple-drug-resistant strains.
We report here the cloning of the mvaE gene of the gram-positive pathogen E. faecalis, its expression in Escherichia coli, and the purification and characterization of the gene product, which exists in the enterococci examined as a true bifunctional protein.
MATERIALS AND METHODS
Reagents.
Purchased materials included deoxynucleoside triphosphates, T4 polynucleotide kinase, Ts-alkaline phosphatase, isopropyl-β-d-thiogalactopyranoside (IPTG) and NuPAGE gels (Invitrogen), T4 DNA ligase and restriction enzymes (New England Biolabs), Pwo polymerase (Roche Biochemicals), lysozyme, mutanolysin, and immunoglobulin G horseradish peroxidase (Sigma). Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) was used for nickel affinity chromatography. Plasmid DNA preparations employed a QIAprep Spin Miniprep Kit (Qiagen), and agarose gel extractions employed a Qiagen Gel Extraction kit. Fluvastatin was a gift from Novartis. Antibodies against the E. faecalis fusion protein were raised in New Zealand white rabbits by Covance Research Products, Denver, Pa. Western blotting employed the NOVEX NuPAGE System (Invitrogen Corp.) and the ECL Western blotting system (Amersham Pharmacia Biotech). Synthetic oligonucleotides were prepared, and automated DNA sequencing was performed in-house at GlaxoSmithKline. Unless otherwise specified, all other reagents were from Sigma.
Plasmids, bacterial strains, and culture media.
Expression vector pET28a(+) (Novagen) was modified to allow blunt cloning by replacing the NdeI site with ScaI to generate pET28-6Hblunt. Bacterial strains used included E. coli DH5α and BL21(DE3) (Invitrogen), Enterococcus hirae (ATCC 8043), E. faecalis strain 41, and Enterococcus faecium H62738 (GlaxoSmithKline culture collection). Genomic DNA from E. faecalis strain 41 was used for amplification of the mvaE open reading frame. Luria-Bertani (LB) medium and agar (20) supplemented with 50 μg of kanamycin per ml served for the growth of E. coli strains. Tryptone soy broth and agar or Todd Hewitt Broth supplemented with 5% (wt/vol) yeast extract served for the growth of enterococcal strains.
Construction of the expression plasmid.
The mvaE gene was PCR amplified from E. faecalis genomic DNA by using a forward primer with the start codon changed from TTG to ATG (5′-ATGAAAACAGTAGTTATTATTGATGC-3′) and a reverse primer (5′-ATGTTATTGTTTTCTTAAATCATTTAAAATAGC-3′). The resulting 2.4-kb fragment was phosphorylated, gel purified, and ligated into ScaI-digested pET28-6Hblunt that had been dephosphorylated and gel purified (20). Plasmid DNA isolated from transformed E. coli DH5α cells was sequenced to confirm the presence of unaltered mvaE. BL21(DE3) cells were transformed with MvaEef-pET28-6Hblunt to express the N-terminally 6-His-tagged protein.
Expression and purification of the gene product.
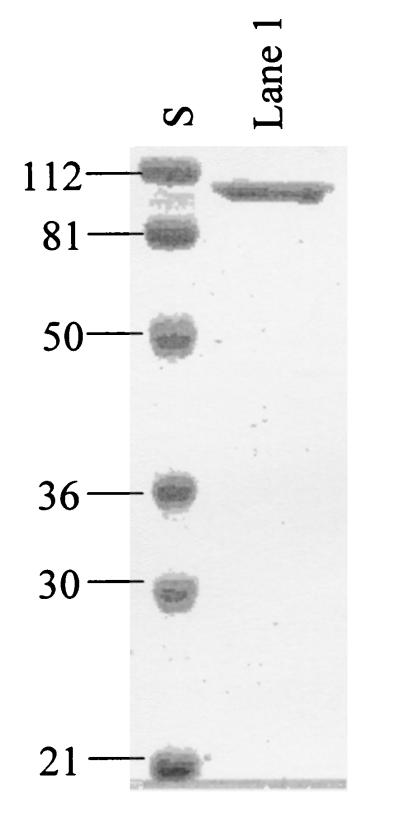
E. coli BL21(DE3) cells transformed with MvaEef-pET28-6Hblunt were grown initially at 37°C. Following addition of 0.5 mM IPTG, growth was continued at 18°C. Harvested cells were washed with 0.9% saline and were suspended in a solution containing 10 mM imidazole, 300 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 20 mM HEPES, pH 8.0 (Buffer A), and then were lysed in a French press. The supernatant liquid obtained by centrifugation of the cell lysate in a Beckman L8-70 ultracentrifuge (30,000 rpm, 60 min, 4°C) was applied to an Ni-NTA column. The column was washed with Buffer A and then was eluted successively with 50 and 100 mM imidazole in Buffer A. Fractions with high activity were combined and stored at −70°C. The most active fractions contained ∼95% homogeneous His-tagged protein (Fig. 3). Overall yields averaged 10 mg/liter.
FIG. 3.
SDS-PAGE of the expressed mvaE gene product purified by nickel affinity chromatography. S, standards of the indicated mass. Lane 1, early fractions of E. faecalis acetoacetyl-CoA thiolase/HMG-CoA reductase. Numbers indicate molecular sizes in kilodaltons.
Preparation of enterococcal cell lysates and Western blotting.
Proteins present in extracts of E. faecalis, E. hirae, and E. faecium prepared with mutanolysin and lysozyme were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were transferred to polyvinylidene difluoride membranes. Following incubation with rabbit antibody raised against the E. faecalis mvaE gene product and incubation with rabbit immunoglobulin G horseradish peroxidase, color was developed with diaminobenzidine tetrahydrochloride and hydrogen peroxide.
Acetoacetyl-CoA thiolase activity.
Determination of acetoacetyl-CoA thiolase activity employed a Hewlett-Packard model 8452 diode array spectrophotometer with the cell compartment maintained at 37°C to monitor the change in absorbance at 302 nm that accompanies the formation or thiolysis of acetoacetyl-CoA (4). Standard assay conditions for each reaction studied follow: for reaction 1, synthesis of acetoacetyl-CoA, 1 mM acetyl-CoA, 10 mM MgCl2, 50 mM Tris, pH 10.5; reaction 2, thiolysis of acetoacetyl-CoA, 30 μM acetoacetyl-CoA, 100 μM CoA, 10 mM MgCl2, 50 mM Tris-HCl, pH 9.5. Both reactions were initiated by addition of enzyme. Assays employed a final volume of 200 μl. For both assays, 1 enzyme unit (eu) represents the synthesis or thiolysis in 1 min of 1 μmol of acetoacetyl-CoA. Reported data represent mean values for at least triplicate determinations.
HMG-CoA reductase activity.
Spectrophotometric assays of HMG-CoA reductase activity at 37°C monitored the appearance or disappearance of NADP(H) at 340 nm. Standard assay conditions for each reaction studied follow: reaction 3, reductive deacylation of HMG-CoA to mevalonate, 0.4 mM NADPH, 1.0 mM (R,S)-HMG-CoA, 100 mM KCl, and 100 mM K xPO4, pH 6.5; reaction 4, reduction of mevaldehyde to mevalonate, 0.4 mM NADPH, 3.8 mM (R,S)-mevaldehyde, 100 mM KCl, and 100 mM KxPO4, pH 7.0; reaction 5, oxidative acylation of mevaldehyde to HMG-CoA, 3.0 mM NADP+, 0.5 mM CoA, 2.5 mM (R,S)-mevaldehyde, 100 mM KCl, 100 mM Tris-HCl, pH 9.3; reaction 6, oxidative acylation of mevalonate to HMG-CoA, 3 mM NADP+, 1.5 mM CoA, 4.5 mM (R,S)-mevalonate, 100 mM KCl, and 100 mM Tris-HCl, pH 8.0. Assays employed a final volume of 200 μl. Reactions were initiated by adding HMG-CoA, mevaldehyde, or mevalonate. For all four assays, 1 eu represents the turnover, in 1 min, of 1 μmol of NADP(H). This corresponds to the turnover of 1 μmol of mevaldehyde or 0.5 μmol of HMG-CoA or mevalonate. Reported data represent mean values for at least duplicate determinations.
RESULTS
The mvaE gene product is a true bifunctional protein.
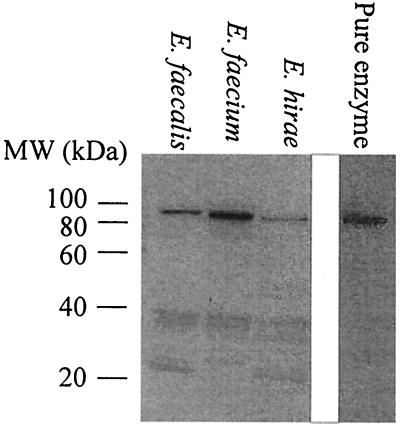
To ascertain whether the mvaE gene product from a representative sample of enterococci undergoes posttranslational cleavage, cell lysates of E. faecalis, E. faecium, and E. hirae were probed with antibodies against the E. faecalis mvaE gene product (Fig. 4). The mvaE gene product was in all instances present as a single peptide in vivo.
FIG. 4.
The mvaE gene product is expressed as a fusion protein in enterococci. Western blots of the indicated enterococcal lysates and of the purified mvaE gene product are shown. Numbers indicate molecular sizes in kilodaltons.
Expression and purification of the E. faecalis mvaE gene product.
Expression in E. coli of E. faecalis mvaE yielded a single soluble polypeptide that was readily purified on a nickel-chelating column. SDS-PAGE indicated that the product was over 95% homogeneous and had a mass of approximately 90 kDa (Fig. 3, lane 1). The smaller peptides more prominent in later fractions were recognized by Ni-NTA antibodies (Qiagen), and thus derive from the N terminus of the expressed protein. The calculated molecular mass for the 803-residue polypeptide encoded by mvaE is 86,496 Da. A mass of 88,712 Da estimated for the expressed protein by matrix-assisted laser desorption ionization mass spectrometry, corresponds to the peptide plus the 6-His tag and minus the N-terminal methionine. Peptide sequencing of the purified protein confirmed that this methionine had been removed and that the N-terminal sequence was otherwise intact.
Described below are the kinetic properties of the two catalytic activities, acetoacetyl-CoA thiolase and HMG-CoA reductase.
Acetoacetyl-CoA thiolase. (i) Optimal temperature.
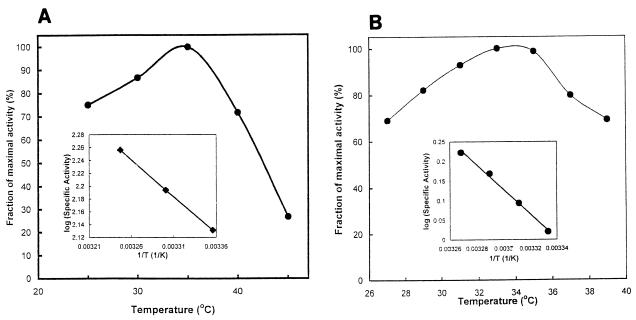
Optimum activity both for acetoacetyl-CoA synthesis and for thiolysis of acetoacetyl-CoA occurred at about 37°C (Fig. 5A). An Arrhenius plot yielded an estimated ΔHa of 5.3 kcal for thiolysis of acetoacetyl-CoA (Fig. 5A, inset).
FIG. 5.
Effect of temperature on activity. Assays were conducted at the indicated temperatures under otherwise standard conditions. (A) Reaction 2, thiolysis of acetoacetyl-CoA; (B) reaction 3, reductive deacylation of HMG-CoA to mevalonate. The insets are selected data shown as Arrhenius plots.
(ii) Optimal pH.
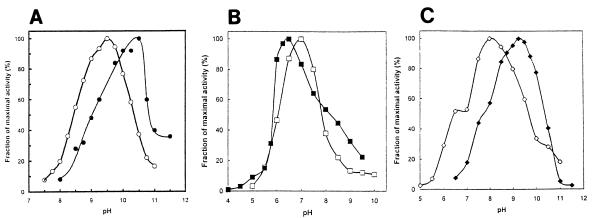
Activity was optimal at pH 10.5 for acetoacetyl-CoA synthesis and at pH 9.5 for acetoacetyl-CoA thiolysis (Fig. 6A).
FIG. 6.
Effect of hydrogen ion concentration on activity. All assays were conducted in a solution containing 50 mM sodium acetate, 50 mM glycine, 50 mM Tris, 50 mM 2-(N-morpholino)ethanesulfonic acid at the indicated pH under otherwise standard conditions. (A) Reaction 1, synthesis of acetoacetyl-CoA (•), and reaction 2, thiolysis of acetoacetyl-CoA (○); (B) reaction 3, reductive deacylation of HMG-CoA to mevalonate (▪), and reaction 4, reduction of mevaldehyde to mevalonate (□); (C) reaction 5, oxidative acylation of mevaldehyde to HMG-CoA (◊), and reaction 6, oxidative acylation of mevalonate to HMG-CoA (⧫).
(iii) Kinetic parameters.
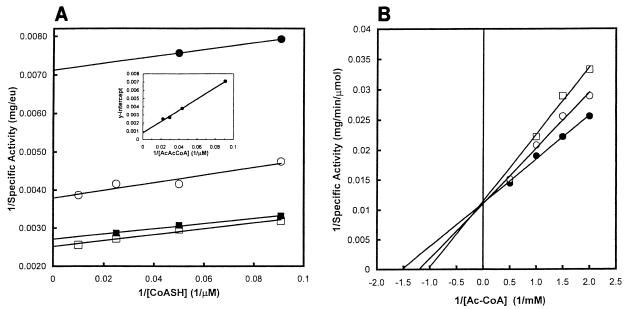
For catalysis of reaction 1, the synthesis of acetoacetyl-CoA, the Km for acetyl-CoA was 0.60 mM (Table 1). For reaction 2, the rate of acetoacetyl-CoA thiolysis was measured at several concentrations of CoA and of acetoacetyl-CoA. A plot of 1/[CoASH] (CoASH is the reduced form of CoA) versus 1/(specific activity) at several concentrations of acetoacetyl-CoA yielded parallel lines, consistent with a ping-pong mechanism (Fig. 7A). For acetoacetyl-CoA thiolysis, the Km for acetoacetyl-CoA was 88 μM and the Km for CoA was 10 μM (Fig. 7A and inset). Table 1 summarizes these kinetic parameters and compares them to those of other acetoacetyl-CoA thiolases.
TABLE 1.
Kinetic parameters for acetoacetyl-CoA thiolasea
| Source and reference | Synthesisb
|
Thiolysis
|
|||
|---|---|---|---|---|---|
| Vmax (eu/mg) | KmAcCoA (μM) | Vmax (eu/mg) | KmAcAcCoA (μM) | KmCoA (μM) | |
| E. faecalis | 85 | 600 | 1,250 | 88 | 10 |
| Z. ramigera (17) | 71 | 1,200 | 810 | 15 | 9.0 |
| Bradyrhizobium japonicum (22) | NR | 100 | NR | 19 | 30 |
| Chicken liver cytosol (4) | NR | 270 | NR | 39 | 6.4 |
Data for the synthesis and thiolysis of acetoacetyl-CoA are shown.
NR, not reported.
FIG. 7.
Acetoacetyl-CoA thiolase proceeds via a ping-pong mechanism. (A) Acetoacetyl-CoA thiolysis (reaction 2). Assays employed the indicated concentrations of CoA and either 11 (•), 23 (○), 34 (▪), or 45 μM (□) acetoacetyl-CoA. Inset: the Y intercepts were plotted versus the reciprocal of acetoacetyl-CoA concentration. (B) CoA competes with acetyl-CoA during synthesis of acetoacetyl-CoA. The reaction employed 0 (•), 30 (○), or 60 mM (□) CoA, the indicated concentrations of acetyl-CoA, and otherwise standard conditions.
(iv) Inhibition by CoA.
CoA inhibited reaction 1 competitive with acetyl-CoA. Ki was 0.12 mM and Ki/KmAcCoA was 0.19 (Fig. 7B).
HMG-CoA reductase. (i) Optimal temperature.
Optimal activity for catalysis of reaction 3 occurred at 37°C. An Arrhenius plot of selected data yielded a ΔHa of 14 kcal (Fig. 5B).
(ii) Coenzyme specificity.
Nucleotide specificity has been established for three class II HMG-CoA reductases. The P. mevalonii enzyme uses NAD(H) exclusively (9), whereas those of S. aureus (26) and A. fulgidus (12) use NAD(H) and NADP(H) equally well. By contrast, E. faecalis HMG-CoA reductase was specific for NADP(H) as the oxidoreductant for all four catalyzed reactions. Activity was undetectable with NAD(H), and even a 10-fold excess of NADH over NADPH did not inhibit reaction 3.
(iii) Optimal pH.
The effect of hydrogen ion concentration on the rates of all four catalyzed reactions was determined. Optimal activity was observed at pH 6.5 and 7.0 for reductive reactions 3 and 4 (Fig. 6B) and at pH 8.0 and 9.5 for oxidative reactions 5 and 6 (Fig. 6C).
(iv) Kinetic parameters.
Kinetic parameters were determined for all four catalyzed reactions. Table 2 compares these data to those for other characterized HMG-CoA reductases.
TABLE 2.
Kinetic parameters for E. faecalis HMG-CoA reductase
| Reaction and substrate | Optimal pH | Vmax (eu/mg) | Km (μM) | Km values (μM) from literaturea |
|---|---|---|---|---|
| HMG-CoA→mevalonate (reaction 3) | 6.5 | 2.0 | ||
| HMG-CoA | 20 | 20-40 | ||
| NADPH | 30 | 80-160b | ||
| Mevaldehyde→mevalonate (reaction 4) | 7.0 | 13 | ||
| Mevaldehyde | 3,800 | 600-8,000 | ||
| NADPH | 250 | 30-1,800b | ||
| Mevaldehyde→HMG-CoA (reaction 5) | 8.0 | 2.4 | ||
| Mevaldehyde | 660 | 100-600 | ||
| CoASH | 220 | 20-500 | ||
| NADP+ | 670 | 40-900b | ||
| Mevalonate→HMG-CoA (reaction 6) | 9.3 | 1.2 | ||
| Mevalonate | 1,000 | 20-1,200 | ||
| CoASH | 230 | 10-1,700 | ||
| NADP+ | 250 | 90-580b |
The ranges of reported Km values for HMG-CoA reductases from different sources are given.
For either NADP(H) or NAD(H).
(v) Inhibition by a statin drug.
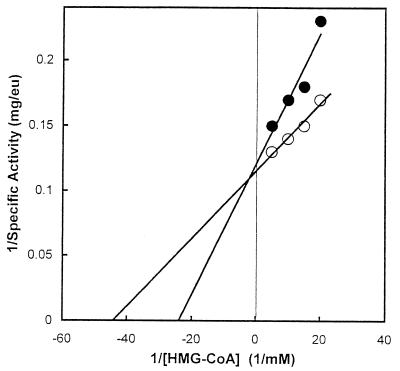
Inspection of sequence alignments suggested that the E. faecalis enzyme is a class II HMG-CoA reductase. A major difference between the two classes of HMG-CoA reductase is their sensitivity to inhibition by statin drugs. Ki values for statins are in the millimolar range for the class II enzymes and in the nanomolar range for the class I enzymes (12, 26). Fluvastatin inhibited reaction 3 competitive with HMG-CoA with a Ki of 0.5 mM (Fig. 8). A Ki of 0.2 to 0.5 mM is diagnostic of a class II HMG-CoA reductase (7, 26).
FIG. 8.
Inhibition by a statin drug of reaction 3, the reductive deacylation of HMG-CoA to mevalonate. Assays were conducted in the presence of 0 (○) or 500 μM (•) fluvastatin at the indicated concentrations of HMG-CoA under otherwise standard conditions. All reactions were initiated by adding NADPH.
(vi) Inactivation by diethylpyrocarbonate and reactivation by hydroxylamine hydrochloride.
In all HMG-CoA reductases studied to date, a histidine plays a key role in catalysis, protonation of the CoA thioanion released during the first stage of the overall reaction (9). The active-site histidine appears to participate exclusively in the first stage of reaction 3 (7, 8). We therefore asked whether the histidine-modifying reagent diethylpyrocarbonate (DEPC) inactivated E. faecalis HMG-CoA reductase. DEPC blocked the reductive deacylation of HMG-CoA to mevalonate, and subsequent treatment with hydroxylamine hydrochloride restored HMG-CoA reductase activity (Fig. 9A). By contrast, under conditions where the catalysis of reaction 3 was severely attenuated, DEPC had no detectable effect on reaction 4, the reduction of mevaldehyde to mevalonate (Fig. 9B).
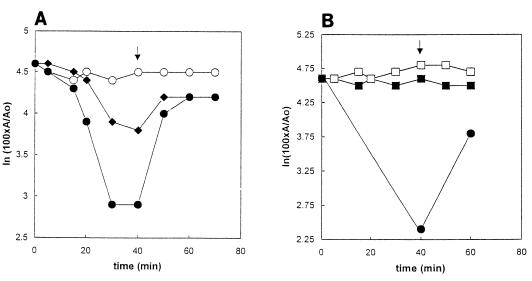
FIG. 9.
Effect of DEPC and subsequent addition of hydroxylamine hydrochloride on HMG-CoA reductase activity. (A) Effect on reaction 3, the reductive deacylation of HMG-CoA to mevalonate. Treatment with DEPC and with hydroxylamine hydrochloride was conduced on ice. Samples contained fusion protein in a solution containing 250 mM KCl, 10% (vol/vol) glycerol, 250 mM K xPO4, pH 6.5. DEPC in ethanol was added to a concentration of 0.8 (⧫) or 8.0 mM (•). A control contained ethanol but no DEPC (○). After 40 min, hydroxylamine hydrochloride, pH 6.5, was added to a concentration of 700 mM (arrow). Ten-microliter portions removed at the indicated times were assayed for the ability to catalyze reaction 3, the reductive deacylation of HMG-CoA to mevalonate. Ao and A are the specific activities at zero time and at the indicated times, respectively. (B) Effect on reaction 4, the reductive deacylation of mevaldehyde to mevalonate. Reaction conditions were as described above but at pH 7.0 and using 8 mM DEPC and an ethanol control (□). Data for reaction 4 (▪) are shown. Also shown are data for reaction 3 (•), included to establish that reaction with DEPC had indeed occurred.
DISCUSSION
Analysis of complete and partial genome sequences for pathogenic eubacteria revealed the presence of genes thought to encode enzymes of the mevalonate pathway of isopentenyl diphosphate biosynthesis. Subsequent inspection of sequence alignments revealed an unusual feature of isopentenyl diphosphate biosynthesis in enterococci, the presence of the mvaE gene that appears to encode, without interruption, both acetoacetyl-CoA thiolase and HMG-CoA reductase on a single polypeptide (25). Western blotting of enterococcal extracts was employed to show that the mvaE gene product is indeed expressed in E. faecalis, E. faecium, and E. hirae as a fusion protein and is not posttranslationally cleaved. Radish seedlings express an acetoacetyl-CoA thiolase/HMG-CoA synthase fusion protein (24). However, the mvaE gene product represents the first bifunctional enzyme of isopentenyl diphosphate biosynthesis in eubacteria and the first example of HMG-CoA reductase fused to another protein.
The kinetic properties of E. faecalis acetoacetyl-CoA thiolase resemble those of previously characterized forms of the enzyme (Table 1). A plot of 1/(specific activity) versus 1/[acetoacetyl-CoA] and inhibition of acetoacetyl-CoA synthesis by CoA suggested that reactions 1 and 2 proceed via a ping-pong mechanism. Prior investigations have established the participation of an acyl-cysteinyl intermediate and roles for a histidine and a second cysteine as proton acceptors and donors during catalysis by acetoacetyl-CoA thiolase (17). Inspection of multiple sequence alignments suggested that the cognates of the acyl-acceptor Cys89Z, His348Z, and Cys278Z of Zoogloea ramigera acetoacetyl-CoA thiolase are Cys88E, His342E, and Cys372E of the E. faecalis enzyme, respectively. (Subscripts on residue numbers refer to the enzymes from E. faecalis [E] and Z. ramigera [Z]).
The sequence-based inference that E. faecalis HMG-CoA reductase is a class II enzyme was confirmed by the 0.5 mM Ki value for inhibition of reaction 3 by the statin drug fluvastatin. This Ki value is diagnostic of a class II enzyme (1, 12). As for both classes of the enzyme, inhibition was competitive with HMG-CoA. As anticipated for an enzyme from a mesophile, the temperature optimum was 37°C. The calculated activation energy ΔHa of 14 kcal is similar to the ΔHa of 13 kcal for A. fulgidus HMG-CoA reductase (12), suggesting that the same step is rate limiting for both enzymes. All four reactions catalyzed by HMG-CoA reductase exclusively employed NADP(H) as the coenzyme. Activity with NAD(H) was below detectable limits, and even a 10-fold molar excess of NAD(H) over NADP(H) did not inhibit reaction 3. The class II HMG-CoA reductase of P. mevalonii uses NAD(H) (11), and those of S. aureus (26) and A. fulgidus (12) use either NAD(H) or NADP(H). The E. faecalis enzyme is thus presently the only known class II HMG-CoA reductase that is specific for NADP(H).
As previously shown for P. mevalonii HMG-CoA reductase (7), exposure of the fusion protein to the histidine-modifying reagent DEPC severely attenuated catalysis of reaction 3, and subsequent treatment with hydroxylamine hydrochloride restored activity. By contrast, DEPC had no effect on reaction 4. Since hydroxylamine hydrochloride removes the ethoxyformyl group from histidyl but not from cysteinyl or lysyl residues (16), no catalytically critical cysteinyl or lysyl residue had been modified. These observations implicate a histidine essential for catalysis of reaction 3 but nonessential for reaction 4. This inference is consistent with the proposed role of a histidine that protonates the CoA thioanion released during the first stage of reaction 3 (7-9). Sequence comparisons suggest that the active-site histidine of E. faecalis HMG-CoA reductase is His756E (Fig. 10).
FIG. 10.
E. faecalis HMG-CoA reductase histidine 756 and its cognates. Amino acid sequences from characterized class II HMG-CoA reductases were aligned with CLUSTALW (www.expasy.ch).
Characterized acetoacetyl-CoA thiolases are either dimers or tetramers (17), whereas HMG-CoA reductases typically are either tetramers (10) or hexamers (15, 18). Gel filtration chromatography and analytical ultracentrifugation suggest that the mvaE gene product exists as a complex mixture of high-molecular-weight species (Hedl et al., unpublished observations). Our future investigations will address the effect of multimeric state on catalytic activity.
Acknowledgments
From GlaxoSmithKline we thank Thomas Mathie for the synthesis of the oligonucleotides, Stephanie van Horn for sequencing the plasmid constructs, and Gilbert Scott for matrix-assisted laser desorption ionization mass spectral and N-terminal sequence analysis.
The Purdue contribution was funded by American Heart Association grant 0150503N (V.W.R.) and by National Institutes of Health grants HL 47113 (V.W.R.) and 52115 (C.V.S.).
Footnotes
Journal paper 16671 from the Purdue Agricultural Experiment Station.
REFERENCES
- 1.Bischoff, K. M., and V. W. Rodwell. 1996. 3-Hydroxy-3-methylglutaryl-CoA reductase from Haloferax volcanii. Purification, characterization, and expression in Escherichia coli. J. Bacteriol. 178:19-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochar, D. A., C. V. Stauffacher, and V. W. Rodwell. 1999. Sequence comparisons reveal two classes of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol. Genet. Metab. 6:122-127. [DOI] [PubMed] [Google Scholar]
- 3.Bochar, D. A., J. A. Friesen, C. V. Stauffacher, and V. W. Rodwell. 1999. Biosynthesis of mevalonic acid from acetyl-CoA, p. 15-44. In David Cane (ed.), Comprehensive natural products chemistry, vol. 2. Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- 4.Clinkenbeard, K. D., T. Sugiyama, and M. D. Lane. 1975. Cystolic acetoacetyl-CoA thiolase from chicken liver. Methods Enzymol. 35:167-173. [DOI] [PubMed] [Google Scholar]
- 5.Corsini, A., F. M. Maggi, and A. L. Catapano. 1995. Pharmacology of competitive inhibitors of HMG-CoA reductase. Pharmacol. Res. 31:9-27. [DOI] [PubMed] [Google Scholar]
- 6.Dairi, T., Y. Motohira, T. Kuzuyama, S. Takahashi, N. Itoh, and H. Seto. 2000. Cloning of the gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from terpenoid antibiotics-producing Streptomyces strains. Mol. Gen. Genet. 262:957-964. [DOI] [PubMed] [Google Scholar]
- 7.Darnay, B. G., Y. Wang, and V. W. Rodwell. 1992. Identification of the catalytically important histidine of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem. 267:15064-15070. [PubMed] [Google Scholar]
- 8.Darnay, B. G., and V. W. Rodwell. 1993. His865 is the catalytically important histidyl residue of Syrian hamster 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem. 268:8429-8435. [PubMed] [Google Scholar]
- 9.Frimpong, K., and V. W. Rodwell. 1994. Catalysis by Syrian hamster 3-hydroxy-3-methylglutaryl-CoA reductase. Proposed roles of histidine 865, glutamate 558, and aspartate 766. J. Biol. Chem. 269:11478-11483. [PubMed] [Google Scholar]
- 10.Istvan, E. S., M. Palnitkar, S. K. Buchanan, and J. Deisenhofer. 2000. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 19:819-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan-Starck, T. C., and V. W. Rodwell. 1989. Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl-CoA reductase characterization and chemical modification. J. Biol. Chem. 264:17913-17918. [PubMed] [Google Scholar]
- 12.Kim, D.-Y., C. V. Stauffacher, and V. W. Rodwell. 2000. Dual coenzyme specificity of Archaeoglobus fulgidus HMG-CoA reductase. Protein Sci. 9:1226-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuzuyama, T., M. Takagi, S. Takahashi, and H. Seto. 2000. Cloning and characterization of 1-deoxy-d-xylulose 5 phosphate synthase from Streptomyces sp. strain CL190, which uses both the mevalonate and nonmevalonate pathways for isopentenyl diphosphate biosynthesis. J. Bacteriol. 182:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange, B. M., T. Rujan, W. Martin, and R. Croteau. 2000. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA 97:13172-13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence, C. M., V. W. Rodwell, and C. V. Stauffacher. 1995. Crystal structure of Pseudomonas mevalonii HMG-CoA reductase at 3.0 angstrom resolution. Science 268:1758-1762. [DOI] [PubMed] [Google Scholar]
- 16.Miles, E. W. 1977. Modification of histidyl residues in proteins by diethylpyrocarbonate. Methods Enzymol. 47:431-442. [DOI] [PubMed] [Google Scholar]
- 17.Modis, Y., and R. K. Wierenga. 2000. Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase. J. Mol. Biol. 297:1171-1182. [DOI] [PubMed] [Google Scholar]
- 18.Rogers, K. S., V. W. Rodwell, and P. Geiger. 1997. Active form of Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochem. Mol. Med. 61:114-120. [DOI] [PubMed] [Google Scholar]
- 19.Rohmer, M. 1999. A mevalonate-independent root to isopentenyl diphosphate, p. 45-67. In David Cane (ed.), Comprehensive natural products chemistry, vol. 2. Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Seto, H., H. Watanabe, and K. Furihata. 1996. Simultaneous operation of the mevalonate and nonmevalonate pathways in the biosynthesis of isopentenyl diphosphate in Streptomyces aeriouvifer. Tetrahedron Lett. 37:7979-7982. [Google Scholar]
- 22.Suzuki, F., W. L. Zahler, and D. W. Emerich. 1987. Acetoacetyl-CoA thiolase of Bradyrhizobium japonicum bacteriods: purification and properties. Arch. Biochem. Biophys. 254:272-281. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi, S., T. Kuzuyama, and H. Seto. 1999. Purification, characterization, and cloning of a eubacterial 3-hydroxy-3-methylglutaryl coenzyme A reductase, a key enzyme involved in biosynthesis of terpenoids. J. Bacteriol. 181:1256-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber, T., and T. J. Bach. 1994. Conversion of acetyl-coenzyme A into 3-hydroxy-3-methylglutaryl-coenzyme A in radish seedlings. Evidence of a single monomeric protein catalyzing a FeII/quinone-stimulated double condensation reaction. Biochim. Biophys. Acta 1211:85-96. [DOI] [PubMed] [Google Scholar]
- 25.Wilding, E. I., J. R. Brown, A. P. Bryant, A. F. Chalker, D. J. Holmes, K. A. Ingraham, S. Iordanescu, C. Y. So, M. Rosenberg, and M. N. Gwynn. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J. Bacteriol. 182:4319-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilding, E. I., D.-Y. Kim, A. P. Bryant, M. N. Gwynn, R. D. Lunsford, D. McDevitt, J. E. Myers, Jr., M. Rosenberg, D. Sylvester, C. V. Stauffacher, and V. W. Rodwell. 2000. Essentiality, expression, and characterization of the class II 3-hydroxy-3-methylglutaryl-coenzyme A reductase of Staphylococcus aureus. J. Bacteriol. 182:5147-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams, A. M., U. M. Rodrigues, and M. D. Collins. 1991. Intrageneric relationships of Enterococci as determined by reverse transcriptase sequencing of the small-subunit rRNA. Res. Microbiol. 142:67-74. [DOI] [PubMed] [Google Scholar]