Abstract
The functional analysis of sequenced genomes will be facilitated by the development of tools for the rapid mapping of mutations. We have developed a systematic approach to genetic mapping in Caulobacter crescentus that is based on bacteriophage-mediated transduction of strategically placed antibiotic resistance markers. The genomic DNA sequence was used to identify sites distributed evenly around the chromosome at which plasmids could be nondisruptively integrated. DNA fragments from these sites were amplified by PCR and cloned into a kanamycin-resistant (Kanr) suicide vector. Delivery of these plasmids into C. crescentus resulted in integration via homologous recombination. A set of 41 strains containing Kanr markers at 100-kb intervals was thereby generated. These strains serve as donors for generalized transduction using bacteriophage φCr30, which can transduce at least 120 kb of DNA. Transductants are selected with kanamycin and screened for loss of the mutant phenotype to assess linkage between the marker and the site of the mutation. The dependence of cotransduction frequency on sequence distance was evaluated using several markers and mutant strains. With these data as a standard, previously unmapped mutations were readily localized to DNA sequence intervals equivalent to less than 1% of the genome. Candidate genes within the interval were then examined further by subcloning and complementation analysis. Mutations resulting in sensitivity to ampicillin, in nutritional auxotrophies, or temperature-sensitive growth were mapped. This approach to genetic mapping should be applicable to other bacteria with sequenced genomes for which generalized transducing phage are available.
Complete genomic DNA sequences have generated fascinating insights into the biology and evolutionary history of many bacterial species. Genome sequences have also enabled powerful new technologies such as microarrays to be applied to understanding the activities of microorganisms (25). Sometimes overlooked is the fact that genomic sequence information can be very productively combined with traditional methods of genetic analysis. We have used the recently completed Caulobacter crescentus genome sequence (17) as a guide to produce a set of genetic mapping strains suitable as donors for generalized transduction. Using these mapping strains, mutations with a variety of phenotypes can be quickly and reliably localized to a small fraction of the genome, identifying candidate genes for further investigation. This technique will significantly speed up genetic analysis of C. crescentus and is likely to be applicable to other bacterial species as well.
C. crescentus is a gram-negative aquatic bacterium that serves as a model system for studying the bacterial cell cycle and development (19). Caulobacter exhibits two distinct appendaged cell types—a motile swarmer cell with a single polar flagellum and a nonmotile cell displaying an adhesive stalk. Swarmer cells can be isolated by density centrifugation to produce synchronized cultures that move through the cell cycle in unison. Swarmer cells differentiate into stalked cells by ejecting their flagellum and subsequently grow a stalk from the same cell pole. Chromosome replication is suppressed in the swarmer cell but initiates upon transition to the stalked form. Synthesis of the new flagellum at the cell pole opposite the stalk depends on ongoing DNA replication. Classical and molecular genetic approaches have generated considerable insight into gene function and regulation during Caulobacter development. For example, the isolation of nonmotile mutants led to the eventual identification of over 40 genes involved in flagellum synthesis and regulation (6, 13). Further genetic analysis uncovered complex connections between flagellar regulation and other cell cycle-dependent processes, including DNA methylation, chromosome replication, and cell division (14, 20, 29).
The first genetic map of the C. crescentus chromosome was developed using generalized transduction by bacteriophage φCr30 and RP4-mediated mobilization of chromosomal DNA (2, 9). Improved versions of the genetic map served as precursors to a physical map of the C. crescentus genome (7, 8) and, ultimately, the genome sequence (17). The C. crescentus strain CB15 genome consists of a single circular chromosome 4,016,942 bp in size, with 3,767 predicted protein-coding genes (17). Less than 10% of these genes had been identified prior to the genome sequence, and only 54% have been assigned possible roles based on similarity to genes from other organisms. Such a high proportion of genes with unknown functions is not unusual among sequenced genomes and is indicative of the need for new tools for elucidating gene function.
Transposon-induced null mutations are easy to generate, and transposon insertion sites can be mapped by a variety of techniques, but such mutations are not generally useful for studying genes involved in critical processes such as the cell cycle. When it is necessary to work with point mutations resulting in subtle or conditional phenotypes, genetic mapping becomes more difficult. Gain-of-function mutations, such as extragenic suppressors that might reveal gene product interactions, are particularly challenging to map. The favored approach for cloning a C. crescentus gene in which a non-transposon-induced mutation resides has been to isolate a complementing cosmid clone from a library of wild-type DNA, followed by the subcloning and sequencing of the complementing gene. Unfortunately, the cosmid complementation approach sometimes fails, due to incomplete representation of the genome in available libraries, or because the mutation in question has a dominant phenotype.
Although a C. crescentus linkage map containing more than 170 genes with identifiable mutant phenotypes has been developed (http://www.cosm.sc.edu/caulobacter), the application of transductional mapping to the study of new mutants has declined in recent years. Establishing transductional linkage of a mutation to a known gene often did not help much for cloning the gene in which the mutation resided, unless the linked gene was present on a known cosmid that might contain the new gene as well. With knowledge of the complete genome sequence, the utility of transductional linkage can be dramatically improved. We reasoned that transductional mapping in C. crescentus could be made both easier and more efficient if certain criteria were met. The genomic locations of markers used for genetic mapping should be known precisely, and the markers must be distributed around the genome so as to ensure complete coverage by transduction. The data obtained in mapping experiments using these markers should be readily convertible into a physical location in the genome. The markers should have selectable phenotypes that do not overlap with those of frequent interest in C. crescentus, such as progression of the cell cycle, cell division, or motility. And finally, the mapping strains containing these genetic markers should be readily available to all researchers. To this end, we developed a set of C. crescentus strains containing a kanamycin resistance gene placed at 100-kb intervals along the genome. φCr30 transduces up to 120 kb of chromosomal DNA at detectable frequencies, so a mutation located anywhere in the C. crescentus genome should be transductionally linked to at least two markers in this collection. We demonstrate here the application of this mapping approach.
MATERIALS AND METHODS
Bacterial strains and media.
C. crescentus and Escherichia coli strains isolated or constructed by others are listed in Table 1. Strains made during the course of this work are listed in Table 2. were routinely grown in PYE broth or agar (5) supplemented with antibiotics as necessary: kanamycin (20 μg/ml in agar, 5 μg/ml in broth), oxytetracycline (2 μg/ml in agar, 1 μg/ml in broth), ampicillin (50 μg/ml in agar), and nalidixic acid (20 μg/ml in agar). Selection for prototrophy was done on M2 minimal salts medium (5) containing glucose (0.2%). E. coli strains were grown in Luria-Bertani (LB) broth or agar (22) supplemented with antibiotics as necessary: kanamycin (50 μg/ml), oxytetracycline (10 μg/ml), or ampicillin (50 μg/ml).
TABLE 1.
Caulobacter crescentus strains used in this worka
| Strain | Relevant genotype | Description |
|---|---|---|
| CB15 | Wild-type C. crescentus | |
| CB15N (NA1000) | Synchronizable derivative of CB15 lacking functional holdfasts | |
| SC111 | CB15 metA101 | Methionine auxotroph, mutation probably in CC2238 (homolog of E. coli metZ) at 2.438 Mb |
| SC117 | CB15 ilvB101 | Isoleucine/leucine auxotroph, mutation probably in CC2100 or CC2101 (acetolactate synthase subunits) at 2.311 Mb |
| SC121 | CB15 hisB101 | Histidine auxotroph, mutation probably in CC2223 (homolog of E. coli hisC) at 2.425 Mb |
| SC141 | CB15 metD104 | Methionine auxotroph, mutation in CC2140 (homolog of E. coli metF) at 2.347 Mb |
| SC268 | CB15 motA102 | Nonmotile, mutation in motA gene (CC0750) at 0.825 Mb |
| SC286 | CB15 motB108 | Nonmotile, mutation in motB gene (CC1573) at 1.739 Mb |
| SC375 | CB15 metB112 | Methionine auxotroph, mutation probably in same gene as metA101 (CC2238, a homolog of E. coli metZ, located at 2.438 Mb) |
| SC415 | CB15 cysE103 | Cysteine auxotroph, mutation probably in CC1596 to CC1598 (sulfate transporter subunits homologous to E. coli cysTWA) at 1.765 Mb |
| SC420 | CB15 aroF110 | Aromatic amino acid auxotroph, mutation probably in CC1882 (homolog of E. coli aroD) at 2.078 Mb |
| SC596 | CB15 ts104 | No growth at 30°C or higher |
| SC1612 | CB15 ts123 | No growth at 37°C |
| SC1615 | CB15 ts129 | No growth at 37°C |
| SC1621 | CB15 ts136 | Very slow growth at 37°C |
| SC1622 | CB15 ts137 | No growth at 37°C |
| SC1623 | CB15 ts138 | No growth at 37°C |
| SC1624 | CB15 ts139 | No growth at 37°C |
| SC1625 | CB15 ts141 | No growth at 37°C |
| LS107 (MRKA208) | CB15N Δbla6 | Amps Kans derivative of SC1107 |
| UJ195 | CB15N pleD::spec | Specr and hypermotile (large swarms) due to omega cassette insertion in pleD (CC2462) at 2.669 Mbb |
TABLE 2.
C. crescentus marker strains
| C. crescentus marker strain | Genome sequence coordinates of targeting fragment (overlapped ORFs) | PCR primers for targeting fragmenta |
|---|---|---|
| CMS0 | 41328-42339 (CC0039-0041) | For: CGGCGTGGGCGGTGGAGGATGT |
| Rev: CGGCGTTCGGTGGGCTGGCTGT | ||
| CMS1 | 135832-137201 (CC0126-0128) | For: GCGTTTTTCCAGCCGGCCCACAG |
| Rev: CCTCGCGGCGCCTTTTTCGTTGG | ||
| CMS2 | 239808-240826 (CC0222-0224) | For: GGCCAGCATGACCCTTCGACCG |
| Rev: CGCCGAGTACAAGCCGACCCCGAC | ||
| CMS3 | 339032-340124 (CC0323-0324) | For: CGGTGTCGGGTCTGGCGTTCG |
| Rev: GCGCGCCGGTGCAGCTGTTCTTCT | ||
| CMS4 | 438585-440195 (CC0424-0426) | For: CCAGCAGGCGGCGGTCGATTTGAT |
| Rev: GCGGCGCGTCCAGAAGCTCACT | ||
| CMS5 | 538835-539841 (CC0512-0513) | For: CCGGCGGCGCTTTCGTCTTC |
| Rev: CCGCGTGGAGGAAAAAATCAGGGG | ||
| CMS6 | 638934-640056 (CC0584-0585) | For: GCGCGCGATCACCCCACAGAC |
| Rev: TGGGCCGCGAAGATCCTGAACAAG | ||
| CMS7 | 741557-742473 (CC0674) | For: CGTCGATCAGGTGCAGGCCCCAGT |
| Rev: GGCGGACGGCAAGACCTCGGT | ||
| CMS8 | 845796-846815 (CC0768-0770) | For: CGCGCATGAAGGGGCAGGACCAG |
| Rev: CGGCTTCACCTCGGGGCGGATCC | ||
| CMS9 | 939587-940578 (CC0843-0844) | For: GCGGGTGCTGTCCAGTTTCGTCGA |
| Rev: CCCATGGCCAGCAGGTCGTCGGTGTC | ||
| CMS10 | 1041014-1042197 (CC0937-0939) | For: GGCAAGCCGCGCACCATCCTGAT |
| Rev: GGCGTGCGCAAGGGTCCGGTCATC | ||
| CMS11 | 1060341-1061762 (CC0955-0957) | For: CAGCCTGGAAACGGTGATGTCGAT |
| Rev: CGCCGAACTGTTGCCCGAGCTC | ||
| CMS12 | 1159257-1160692 (CC1029-1030) | For: CCCCCCCAAGCCGATGCCGATGAC |
| Rev: CCGCCCCACCTGGTTCCGCCTCTC | ||
| CMS13 | 1256079-1257163 (CC1107-1108) | For: CGGTGGGTCTCGCCTTGGATCGG |
| Rev: GCGGCAGGCGGTCCAGCAGCAC | ||
| CMS14 | 1349674-1350629 (CC1189-1190) | For: GGCCGTGAGAGCGAGCGATGGG |
| Rev: GCAGCCCGTCCAGCATCACCCTCAT | ||
| CMS15 | 1462150-1462957 (CC1313-1314) | For: CCGCGGCCCTCACGACATCCAGAT |
| Rev: TCGCGACCGTCACCAAGGCCACAG | ||
| CMS16 | 1559310-1560349 (CC1411-1412) | For: CGCCCGAGATGAGTTGCCGCTGG |
| Rev: CGGCTCAGCGCGTCGGAACCCATA | ||
| CMS17 | 1655260-1656305 (CC1496-1497) | For: GGGTCACTCGGCCGCCTCGCAGAT |
| Rev: GGGGAAGAGGCAGGCGCGGTAGGA | ||
| CMS18 | 1750316-1751345 (CC1583-1585) | For: CGGCCCAAGGTGCGCGTGATCG |
| Rev: CGCGGGCGATGGCTTCCTGGC | ||
| CMS19 | 1855379-1856549 (CC1681-1683) | For: GCCGCCGCGACCTTGCCTAAA |
| Rev: CGGGATGTGCGCGGTCTCGTCG | ||
| CMS20 | 1958583-1959461 (CC1774-1775) | For: TCGCCTATTTCGTCGCCACCCTG |
| Rev: GCGGCCTTGTCCACGTGCGTCTC | ||
| CMS21 | 2049425-2050600 (CC1853-1855) | For: CGGCGCAAGGATGGACATTCAGGACG |
| Rev: CGGGCGGCGATGGTCTTGGTGGTC | ||
| CMS22 | 2153536-2154397 (CC1958) | For: CCGCCGCCTCAACCCGTCTCGATC |
| Rev: CGCAAGCGGGGCAGGAGGGAAG | ||
| CMS23 | 2249405-2250489 (CC2043-2044) | For: GCGCGACAGCAAGAGCAAGCAGACC |
| Rev: TCCGGGCGCAGCGAACCTTCT | ||
| CMS24 | 2353191-2354153 (CC2148-2149) | For: TGGCGTCGTTGGGCGGGCAGC |
| Rev: CGCCGGGCAGCATGTCGATCG | ||
| CMS25 | 2453131-2453919 (CC2256-2257) | For: CCATCCTCGGCCAGCACGTCCT |
| Rev: GCCAGCCCGCCGATCCCAACTT | ||
| CMS26 | 2556531-2557393 (CC2356-2357) | For: CGTCGAGGCCGGTGTTGGTCTG |
| Rev: GCGCCGCCTACATCGTCGAGAAG | ||
| CMS27 | 2670146-2670849 (CC2463-2464) | For: GCTCTAGAGGTGACCTCCAGACCCGAGA |
| Rev: GGAATTCAAGCCGTGGGTCTTCAACC | ||
| CMS28 | 2742248-2743137 (CC2537-2538) | For: AGCGGCGCGAGCGGATGGTTTTTC |
| Rev: GCGCGAGGCCACAAGTTGCATCAG | ||
| CMS29 | 2854644-2856043 (CC2637-2638) | For: CGCGGCGTGACGGACCTGTTCTTC |
| Rev: CCGGGCGGGGTGGCGTTCTTG | ||
| CMS30 | 2950035-2951477 (CC2734) | For: GCCCATCCCCGTGCGCAACATCAG |
| Rev: CGCATCGCCGCCAGCCTAGGAACT | ||
| CMS31 | 3055926-3057204 (CC2830-2832) | For: GGCGGGCGTGCAGGTCGAAGG |
| Rev: GGCGTCGAGGGTCTGGGCGTCA | ||
| CMS32 | 3158908-3159721 (CC2935) | For: GCGTGCTGCTGACCGGCCTCGT |
| Rev: CGCAAGCGGGGCAGGAGGGAAG/PICK>{tt} | ||
| CMS33 | 3256387-3257298 (CC3039-3040) | For: GGACCCACGACCCCCGCATCAAG |
| Rev: GCCCCGGCAGGTGGTGGTGAAGAC | ||
| CMS34 | 3360219-3361164 (CC3123-3125) | For: GGGGTCTCGCCGGGCCTGTTGC |
| Rev: GCTGGCGGATGTTTCGGTGCTGAC | ||
| CMS35 | 3458380-3459584 (CC3203-3204) | For: GCGGTGTAGCGTGATGGGGGGATG |
| Rev: CGGCGAACAGCGGCGACAGGAA | ||
| CMS36 | 3556866-3558327 (CC3307-3308) | For: CCGCCCCTGTCCGCCATCCA |
| Rev: GGGGCGCCATCCTGCTCGACAC | ||
| CMS37 | 3653651-3655404 (CC3411-3413) | For: CGCGGCTCTCTGACGGTCGATGA |
| Rev: CGCCTCGGTGGTGTCGTTCATGA | ||
| CMS38 | 3754553-3755968 (CC3510-3511) | For: CCTGCTGCGCCTGAACGACGAGAT |
| Rev: CTGCGTCGCCTGCTGTCGGTGGT | ||
| CMS39 | 3855274-3856437 (CC3601-3603) | For: CCCGCGCCCGACAGGATCAAC |
| Rev: GGCGCTGCTGACGAATCCGAAGG | ||
| CMS40 | 3950897-3952188 (CC3699-3700) | For: GCCGTGCGCGCCAATCTCGAC |
| Rev: GCGGCTACTTCAAGGGCGGGCTC |
For, forward; Rev, reverse.
Amplification and cloning of DNA segments for marker integration.
Marker sites were chosen at approximately 100-kb intervals along the C. crescentus chromosome. The genome sequence, annotation, and numbering were as described by Nierman et al. (17). The genome sequence and annotation are available online (www.ncbi.nlm.nih.gov/cgi-bin/Entrez/framik?db=Genome&gi=177). In order to minimize the risk of generating a detectable phenotype other than antibiotic resistance in any marker strain, the marker locations were chosen so that plasmid integration in each strain would not disrupt the function of any gene, regardless of whether the function of the gene was known. To accomplish this, the DNA fragments (“targeting fragments”) used to direct integration of the marker plasmids were not wholly contained within any gene or operon, including the presumptive promoter region, which we operationally defined as the 100 bp upstream of the start codon. As no algorithm for identifying operons has been applied to the C. crescentus genome, operons were tentatively identified as sets of closely spaced genes (often with overlapping stop and start codons) arranged in the same orientation and usually containing at least two members predicted to function in the same cellular process.
Once the target site was chosen, primers for PCR were selected using the GeneTools software package (BioTools Inc., Edmonton, Alberta, Canada). Primer sequences and locations are shown in Table 2. Primers were synthesized by Operon Technologies (Alameda, Calif.). All primers were 20 to 24 nucleotides in length, with the melting temperature (Tm) estimated to be in the range of 68 to 73°C. PCRs had the following composition: each primer at 1 μM, 10 ng of CB15N chromosomal DNA, 250 μM deoxynucleotide triphosphates, 5% dimethyl sulfoxide (DMSO), 2.5 mM MgCl2, and 1 U of Taq DNA polymerase (Sigma Chemicals, St. Louis, Mo.). Chromosomal DNA was prepared using the DNEasy kit (Qiagen) according to the manufacturer's instructions. Amplification was done with a thermal cycling regimen as follows: (i) 94°C for 1 min, (ii) 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and (iii) 72°C for 10 min. Reaction products were examined by agarose gel electrophoresis. Of the 41 initial targeting fragments selected, 39 were successfully amplified under these conditions and showed few or no additional nonspecific fragments upon electrophoresis. Modification of magnesium and/or DMSO concentration or annealing temperature allowed the remaining two targeting fragments to be amplified.
PCR products were cloned into the pCR2.1 plasmid vector of the Topo Cloning System (Invitrogen Corp., Carlsbad, Calif.) according to the manufacturer's instructions. Plasmid DNA was purified from colonies that appeared white on LB agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), IPTG (isopropyl-β-d-thiogalactopyranoside), and kanamycin. The presence of inserts was verified by restriction digestion. The identity and orientation of inserts was confirmed by cycle sequencing using flanking primers. Sequencing reactions used the BigDye dideoxyterminator kit (Applied Biosystems, Foster City, Calif.), and reaction products were analyzed on an ABI 310 instrument. Sequence analysis was done with ABI and GeneTools software (BioTools).
Integration of markers into C. crescentus genome.
Targeting fragments were subcloned into the pBGS18T plasmid vector (Kanr), a derivative of pBGS18 that contains the RK2 origin of transfer (oriT) (24, 27). Subcloning of the targeting fragment in most cases was done using the flanking EcoRI sites of the pCR2.1 vector, except where EcoRI restriction sites were present in the amplified segment, in which case alternative restriction sites were used. E. coli strain S17.1 was used as a host for the pBGS18T clones. These plasmids were transferred into C. crescentus CB15N by conjugation, with selection on PYE plates containing kanamycin and nalidixic acid for counterselection against the E. coli donor. Plasmid pBGS18T does not replicate in C. crescentus and thus acts as a suicide vector. The presence of integrated plasmid DNA was verified by PCR. Genomic DNA was isolated from each marker strain, and PCR was performed using combinations of one of the primers initially used to amplify the targeting fragment and one vector-directed primer. When appropriate mutant strains were available, the chromosomal location of the integrated plasmid was verified by cotransduction with genes known to be in the vicinity of the target site.
Phage transduction.
Plate lysates of φCr30 were prepared by infecting 0.1 ml of a saturated PYE culture of each Caulobacter marker strain (CMS) with φCr30 at a multiplicity of infection of approximately 1. Infected cells were incubated at 30°C for 15 min and then mixed with 5 ml of molten PYE top agar (0.3% agar) and poured on a prewarmed PYE plate. After overnight incubation, top agar was scraped off each plate and suspended in 5 ml of PYE. Chloroform (0.1 ml) was added to inactivate viable cells from the donor strain, and the lysate was vortexed vigorously. Lysates were incubated at 4°C for at least 2 h, then vortexed again, and centrifuged for 10 min at 3,400 × g. The supernatant was transferred to a sterile petri dish and subjected to UV irradiation in a UV cross-linker (Hoefer model UVC 1000) at a dose of 100,000 μJ of total energy/cm2. This treatment typically reduced the phage titer (PFU/milliliter) approximately 1,000-fold and was found empirically to result in the largest number of transductant colonies. Lysates were stored at 4°C until use.
Transductions were done by mixing 25 μl of UV-treated phage lysate with 475 μl of PYE broth and 0.5 ml of saturated PYE culture of the intended C. crescentus recipient strain. The infected culture was shaken at 30°C for 1 to 4 h. (The yield of transductant colonies did not appear to vary significantly over this range of incubation times.) Infected cultures were pelleted by centrifugation and resuspended in a small volume of PYE for plating. Cells were spread on PYE agar containing kanamycin, except when a separate phenotype such as nutritional prototrophy was being selected for. Plates were incubated at generally incubated at 30°C, except in the case of temperature-sensitive (ts) mutant strains, in which case incubation was at either 25°C (permissive condition) or 37°C (nonpermissive condition selective for wild-type growth).
The method by which cotransduction rates were determined depended on the recipient strain. In all cases, Kanr was the initial selected phenotype. Restoration of prototrophy to auxotrophic mutants was scored by patching Kanr transductants onto M2/glucose agar. Motility was scored by stabbing transductants into PYE swarm agar (0.3%). Loss of temperature sensitivity was measured by patching onto PYE agar and incubating at 37°C. The pleD mutant strain UJ195 (1) was screened for the loss of spectinomycin resistance. At least 100 colonies were tested for each strain in determining the reported cotransduction frequencies.
Transcomplementation analysis of mutants.
After transduction experiments had identified the region of the genome in which a particular mutation resides, candidate wild-type genes were amplified by PCR for complementation testing, using wild-type CB15N DNA as a template. For LS107 (Amps), the β-lactamase homolog CC2139 (blaA) at bp 2345897 to 2346766 was amplified as a PCR product starting 191 bp 5′ to the coding region and ending 87 bp 3′ to the coding region. Because blaA is suspected to be part of an operon, a series of additional PCR products were generated that all started 237 bp upstream of CC2141, the first gene in the putative operon, and progressively added each gene in the operon (CC2141 to 2137) (see Fig. 3). For temperature-sensitive strain SC1621, seven PCR products were amplified from the chromosomal region around bp 1180000 to 1200000, ranging in size from 2 to 6 kb. Product 1 contained CC1043 and CC1044 (phenylalanyl-tRNA synthetase subunits). Product 2 contained CC1045 and CC1046 (ribosomal subunits L20 and L35). Product 3 contained CC1049 to CC1052. Product 4 contained CC1053 to CC1055 and the two adjacent glycine tRNA genes. Product 5 also contained the glycine tRNA genes, plus CC1056 to CC1059. Product 6 contained CC1061 to CC1063. Product 7 contained CC1070 (peptide chain release factor 3). PCR was done with Accutaq Long-read Taq polymerase (Sigma Chemical Co.) and the supplied reaction buffer, plus 2% DMSO, and then cloned into plasmid pCR2.1 by using the Topo Cloning System (Invitrogen). The amplified fragments were subcloned into plasmid pMR20 in the E. coli S17.1 host and introduced into mutant strains by conjugation. Conjugants were selected on PYE containing tetracycline and nalidixic acid. For LS107, complementation was tested by streaking on PYE agar containing 50 μg of ampicillin/ml. For SC1621, complementation was tested by streaking on PYE agar containing tetracycline and incubating at 37°C. The CC1061 and CC1062 genes were subsequently amplified individually using primers flanking the coding regions, allowing at least 100 bp upstream of each for possible promoter sequences. The PCR products were cloned initially into pCR2.1 and then subcloned into pMR10 (Kanr). (Attempts to clone the products into pMR20 were unsuccessful.) As with pMR20, E. coli S17.1 was used as the cloning host, and plasmids containing inserts were transferred into SC1621 by conjugation. The same flanking primers were used to amplify CC1061 from SC1621 chromosomal DNA. The 0.6-kb PCR product was cloned into the pCR2.1 vector for sequencing.
FIG. 3.
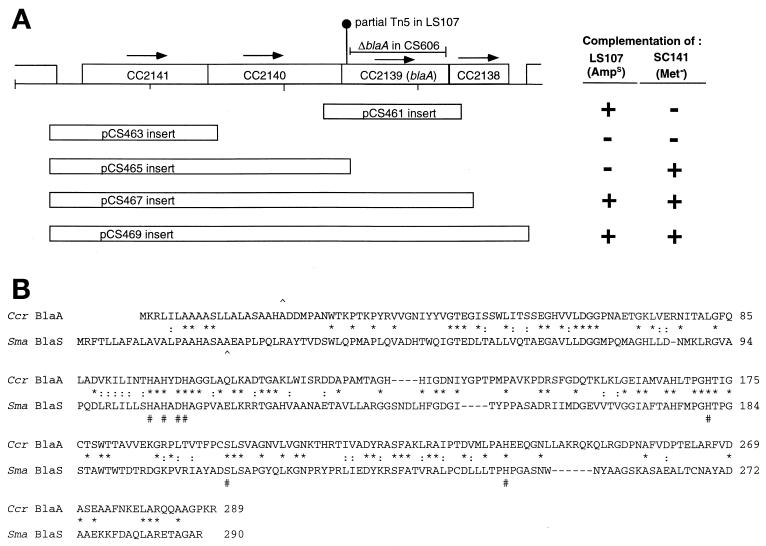
Analysis of blaA region. (A) Schematic of the chromosomal region from approximately bp 2349000 to 2343000. CC2141 encodes a 326-amino-acid polypeptide similar to those of the ArsR family of transcriptional regulators. CC2140 encodes a 315-amino-acid polypeptide homologous to the E. coli 5,10-methylenetetrahydrofolate reductase. CC2139 encodes a 289-amino-acid homologous to L1-type metallo-β-lactamases. CC2138 encodes a 359-amino-acid polypeptide homologous to the amino-terminal domain of 5-methyltetrahydrofolate-homocysteine methyltransferase, while CC2137 encodes a 900-amino-acid polypeptide homologous to the C-terminal domain of the same enzyme. The locations of the Tn5 remnant in LS107 and the extent of the deletion in CS606 are indicated. Boxes beneath the genes indicate fragments amplified by PCR and inserted into the pMR20 vector to generate the pCS plasmids indicated. Complementation of LS107 was scored positive (+) if colonies grew on PYE agar containing 50 μg of ampicillin/ml. Complementation of SC141 was scored positive (+) if cultures grew to saturation in M2/glucose broth containing no amino acid supplements. (B) Alignment of the C. crescentus BlaA polypeptide sequence with the S. maltophilia BlaS sequence (protein identifier g111125376). ∧, known (BlaS) or predicted (BlaA) signal peptide cleavage sites; #, residues known to be involved in zinc coordination or the reaction mechanism of the BlaS protein (28); ∗, residues that are identical in all five polypeptides; :, chemically similar residues.
Construction of ΔblaA strain.
The wild-type blaA gene contains a SacI restriction site at bp 832 to 837, near the 3′ end of the coding region. To construct a deletion of the blaA coding region, PCR-mediated site-directed mutagenesis was used to introduce a new SacI restriction site at bases 72 to 77 of the coding region. A pair of complementary primers was synthesized (BlaSacFor [ACGTCGTCCGAGCTCCATGTGGTG] and BlaSacRev [CACCACATGGAGCTCGGACGACGT]) containing the two-base change (GG to CT at bp 75 and 76) necessary to generate the SacI site. These primers were used in separate amplification reactions with primers located in the upstream (metF) and downstream (metH) genes. The PCR products were isolated and combined together in a second amplification reaction to generate the complete blaA coding region containing the additional SacI site. The mutant gene was subcloned into the pNPTS138 vector, which has no SacI sites. pNPTS138 is a 5.4-kb Kanr plasmid that contains the counterselectable sacB gene, encoding levansucrase (21; M. R. K. Alley, personal communication). Once in pNPTS138, the 760-bp SacI fragment was removed, deleting 90% of the blaA coding region but retaining a continuous open reading frame (ORF) across the deletion to avoid potential polar effects. pNPTS138 was introduced into C. crescentus CB15N by conjugation, with integrants selected for on PYE agar containing kanamycin. Several colonies containing the integrated plasmid were then grown to saturation in PYE broth under nonselective conditions (no antibiotics) and plated on PYE agar containing 3% sucrose to select for plasmid excision. Sucrose-resistant colonies were scored for resistance to kanamycin and ampicillin. Of the kanamycin-sensitive isolates (i.e., those in which most or all of the plasmid was deleted from the chromosome), approximately one-third were also sensitive to ampicillin, suggesting that the blaA gene containing the internal deletion remained. Chromosomal DNA was prepared from four such isolates, and PCR analysis verified the presence of an internal deletion in the sole copy of blaA.
RESULTS
Construction of marker strain collection.
A major goal of this project was to develop a set of C. crescentus strains containing genetic markers whose location in the genome sequence is precisely known. The main factor in deciding how to position Kanr markers was to ensure that every chromosomal gene would be within a transducible distance from a marker. To determine how much chromosomal DNA bacteriophage φCr30 can transfer, published transduction data was reexamined by using the genome sequence to determine the actual sequence lengths separating loci that can be cotransduced. Examples of such data are the argG locus (arginosuccinate synthase; CC0129, bp 137246 to 138472) and the leuA locus (an operon encoding 3-isopropylmalate dehydratase [CC0193] and 3-isopropylmalate dehydrogenase [CC0194], bp 206989 to 210531), which are separated by approximately 70 kb and were reported to cotransduce at a frequency of 8 to 26% (according to Barrett et al. [2]). The hisD locus (histidinol dehydrogenase; CC2346, 2545970 bp) and the argA locus (an operon encoding ornithine carbamoyltransferase [CC2242] and acetylornithine aminotransferase [CC2243], bp 2443239 to 2440969) are separated by 105 kb and cotransduce at a rate of 2% (2). Based on the published data and personal communications from other investigators, it appears that the transducing capacity of bacteriophage φCr30 is limited to 120 to 150 kb of chromosomal DNA. In light of this, we chose to integrate markers at approximately 100-kb intervals, so that any gene on the chromosome could theoretically be cotransduced with at least two markers.
A collection of 41 CMS strains was constructed (Table 2 and Fig. 1). The Kanr markers were inserted at the desired chromosomal locations by using PCR to amplify a small targeting fragment (0.8 to 1.7 kb) representing the insertion site and cloning the targeting fragment into a suicide vector (pBGS18T) to direct integration via homologous recombination. Insertion sites were chosen to minimize the possibility of gene disruption (see Materials and Methods). Integration of the marker plasmids was obtained following conjugal transfer from the E. coli cloning host to C. crescentus CB15N. Integrants were selected on PYE agar containing kanamycin. The integrated plasmids appear to be stable. Three randomly chosen CMS strains were grown for at least 10 generations in PYE broth lacking kanamycin and then were plated on PYE agar with no antibiotic. In all three cases, all 100 of the colonies tested retained kanamycin resistance, suggesting that plasmid excision and loss under nonselective conditions occurs at a rate significantly less than 1% per generation. Excised recircularized plasmid DNA can be detected by PCR in DNA preparations from these strains (data not shown), but its occurrence appears to have no impact on use of the strains for genetic mapping.
FIG. 1.
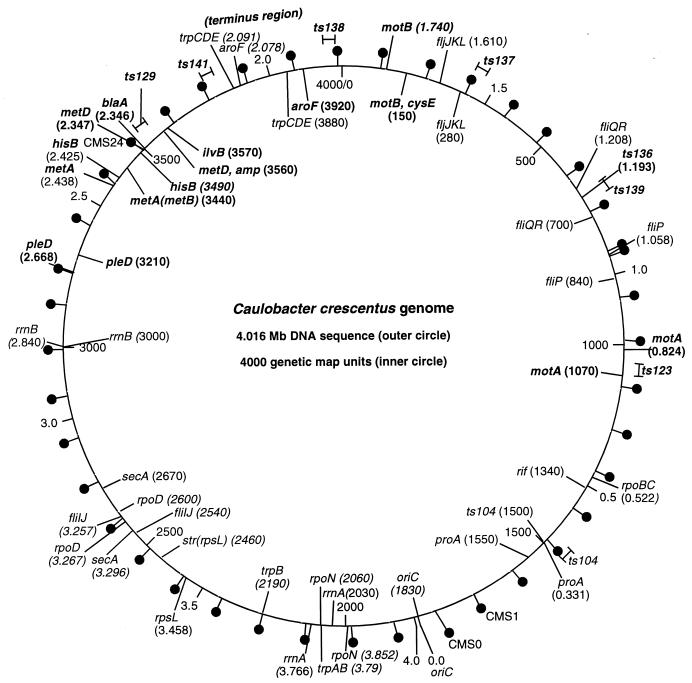
Correlation of genetic and sequence-based maps of C. crescentus and locations of antibiotic resistance markers. The hashmarks and numerical labeling on the inside of the circle indicate genetic map coordinates, as reported by Ely (http://www.cosm.sc.edu/caulobacter/map.html). The hashmarks and numerical labeling on the outside of the circle indicate DNA sequence coordinates, based on the GenBank-deposited genome sequence and annotation (17). The genome sequence numbering starts at the origin of replication, located at 1830 on the genetic map, and proceeds in the direction opposite that of the genetic map coordinates. Representative loci shown on both the inside and the outside have been mapped genetically and can be associated with specific genes or operons in the DNA sequence with a high degree of confidence. “Lollipops” on the outer circle indicate the locations of Kanr integrated plasmid markers in CMS strains, as described in the text and Table 2. Mutations in the genes indicated in bold were used for the cotransduction frequency analysis. The approximate locations of mutations resulting in temperature-sensitive growth are shown on the outer circle.
It is important that the marker strains are free of additional phenotypes resulting from plasmid integration, as other non-wild-type properties could interfere with the use of a strain in mapping certain mutations. All strains were examined microscopically for obvious morphological defects, but none were observed. The growth of each strain in PYE broth cultures was assayed by monitoring optical density. The resulting growth curves were very similar to control cultures of the parental wild-type strain CB15N (data not shown).
The marker strains all produce plaques at similar efficiencies when infected with φCr30, and produced lysates of similar titers (PFU/milliliter) (data not shown). However, some differences were observed in the numbers of Kanr colonies obtained in transduction experiments using identically prepared φCr30 lysates from each donor strain, with CB15N as the recipient. For most of the markers, the number of Kanr colonies obtained varied over a two- to threefold range from marker to marker and from experiment to experiment. The exception was that lysates prepared from strains containing markers integrated in the chromosomal region between 3.2 and 3.7 Mb consistently yielded only 1 to 10% of the number of Kanr colonies typically obtained with markers outside this region.
Testing the marker strain collection.
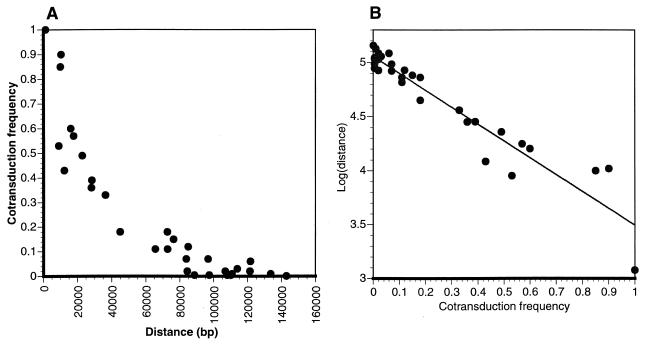
The mapping system was tested with 12 strains containing mutations whose approximate chromosomal locations are known (Table 1 and Fig. 1) (12). These strains, which served as recipients for transductions, were primarily auxotrophs and motility mutants. In some cases, the mutation in the test strain had been experimentally shown to reside in a specific gene. In other cases, the mutation had been mapped relative to other loci, but inspection of the genome sequence in the predicted region (Fig. 1) revealed a strong candidate gene. φCr30 plate lysates were prepared from each Kanr donor strain. Mutant recipient strains were then infected, with selection for Kanr on PYE agar. Transductants were screened for the absence of the mutant phenotype. In all cases, linkage to markers near the previously mapped location was observed. No cotransduction was observed (among the 100 to 200 colonies tested in each experiment) when the distance between sites was 130 kb or greater, suggesting this was a practical upper limit for φCr30. The results of many test transductions are compiled in Fig. 2. Cotransduction frequency declined logarithmically as the sequence length separating the marker and the mutant gene increased (Fig. 2A and B). A plot of log(distance) as a function of cotransduction frequency produced a linear regression as described by the following equation: log(distance) = −1.56(CTF) + 5.06, where CTF is cotransduction frequency. The high correlation coefficient (R2 = 0.903) of the linear regression is indicative of the strong association between the CTF and log(distance) variables. This equation should be useful for estimating distances from φCr30 cotransduction data in future work.
FIG. 2.
Cotransduction frequency as a function of distance from marker. Data were derived from transduction experiments by using combinations of 12 different mutant strains (noted in Fig. 1 and Table 1) and 19 different CMS markers. Cotransduction frequency was calculated as the number of transductants in which the wild-type version of a gene being tested was transferred along with Kanr marker divided by the total number of Kanr transductants tested. (A) Plot of cotransduction frequency as a function of the distance separating the marker and test gene. Distances were corrected for the length of plasmid DNA between insertion site and the kanamycin resistance gene (∼1 or 3 kb, depending on orientation of intergrated plasmid). (B) Plot of log(distance) as a function of cotransduction frequency. The linear regression is described by the following equation: log(distance) = −1.56(CTF) + 5.06, with R2 = 0.903.
Mapping ampicillin sensitivity.
Wild-type C. crescentus is resistant to ampicillin (MIC, 100 μg/ml in PYE broth). The C. crescentus genome sequence contains nine ORFs with homology to the β-lactamase superfamily distributed throughout the genome. Despite this multiplicity of β-lactamase homologs, Barrett et al. (2) isolated strain SC1107 as an ampicillin-sensitive Tn5-induced mutant, suggesting that a single gene is responsible for high-level ampicillin resistance. The amp locus is transductionally linked to the metD and ilvB loci (2). Based on the alignment in Fig. 1, the amp locus is in the 2.3- to 2.4-Mb region of the genome (Fig. 1). An Amps Kans derivative of SC1107 (LS107) in which the Tn5 was thought to have been excised (27) was used to map the amp mutation relative to the CMS markers. LS107 was infected with φCr30 lysates derived from each of the marker strains. Only the marker at 2.35 Mb in CMS24 was tightly linked to Ampr, with an 85% cotransduction rate.
Inspection of genes near the CMS24 marker showed a β-lactamase homolog 8 kb away (ORF CC2139 at bp 2345897 to 2346766). The CC2139 product is a 289-amino-acid polypeptide related to the Stenotrophomonas maltophilia L1 metallo-β-lactamase (Fig. 3a). Several lines of evidence support the identification of this gene, for which we suggest the designation blaA, as responsible for ampicillin resistance in C. crescentus. PCR analysis using primers positioned both upstream from and within the blaA coding region demonstrated the presence of a 2-kb insertion in LS107 near the 5′ side of the coding region. Sequence analysis of a PCR product generated from LS107 showed that a remnant of the original Tn5 is inserted at codon 31 of blaA. The wild-type blaA gene, amplified from CB15N and cloned into the pMR20 plasmid vector, was sufficient to restore ampicillin resistance to LS107. The chromosomal context of the blaA gene suggests that it is part of an operon containing at least three other genes. The blaA start codon overlaps the stop codon of the preceding ORF (CC2140), and its stop codon overlaps the start codon of the subsequent ORF (CC2138) (Fig. 3b). A PCR product containing the upstream genes of the putative operon and the intergenic region expected to contain the operon promoter was also cloned into pMR20. When introduced into LS107, this construct produced colonies that grew more rapidly and to larger size in the presence of ampicillin than the pMR20 construct containing only the region immediately surrounding blaA.
An ampicillin-sensitive strain of C. crescentus could be useful for various genetic applications, including the use of ampicillin-resistant plasmid vectors. SC1107 is not an ideal strain for such applications, as it is in the nonsynchronizable CB15 background and it is also Kanr. LS107 is Kans and synchronizable, but it shows some anomalous behavior, including low conjugation efficiency and poor growth on M2G minimal medium (C. Stephens, unpublished observations). The growth phenotype may result from a polar effect of the Tn5 remnant on expression of the metH gene (see below). We therefore constructed an in-frame deletion of nearly 90% of the blaA coding sequence (codons 59 to 279) and exchanged it with the wild-type locus to generate strain CS606. This deletion results in sensitivity to ampicillin (MIC, 1 μg/ml) comparable to that of LS107 (MIC, 0.5 μg/ml). CS606 is morphologically indistinguishable from CB15N and is superior to LS107 in that it is wild type with respect to growth on minimal media and as a recipient for conjugal transfer of plasmid DNA (data not shown).
Of the other genes in the putative blaA operon, at least two are predicted to be involved in methionine synthesis. The gene immediately 5′ to blaA (CC2140) is homologous to the E. coli metF gene, encoding 5,10-methylenetetrahydrofolate reductase, while the gene immediately 3′ to blaA (CC2138) is homologous to the 5′ portion of the E. coli metH gene, encoding the N-terminal domain of 5-methyltetrahydrofolate-homocysteine methyltransferase. Application of the transductional mapping method to C. crescentus strain SC141, a methionine auxotroph with a mutation at a locus designated metD, indicated that the metD gene is also closely linked to the CMS24 marker (53% cotransduction). The plasmids constructed for mapping amp were tested for complementation of SC141. The results indicated that the metF homolog (CC2140) was necessary to complement the SC141 auxotrophy, suggesting that the mutation in SC141 resides in this gene (Fig. 3).
Mapping mutations responsible for temperature-sensitive growth.
Johnson and Ely (12) isolated a number of spontaneous mutant strains exhibiting temperature-sensitive growth. Such mutations are generally thought to be indicative of genes involved in critical cellular processes. Transductional mapping with the CMS marker collection was used to localize several of these ts mutations. Mutant strains were infected with φCr30 lysates prepared from the marker strains and spread on PYE/Kan agar. For each marker, one infection plate was incubated at room temperature (∼24°C) and one at 37°C. Any 37°C plates that produced Kanr colonies indicated that the marker in that donor strain was linked to the wild-type version of the desired mutant gene. Transductants were then patched from the corresponding permissive temperature plate onto PYE/Kan and incubated at 37°C, to determine the frequency of cotransduction of the wild-type gene with the markers.
C. crescentus strain SC1625 cells grow very slowly at 24 and 30°C, form colonies significantly smaller than CB15N, and do not form colonies at 37°C. The ts141 mutation in SC1625 was in a gene tightly linked to the marker in strain CMS22 at 2.15 Mb (94% cotransduction) and was weakly linked to the marker in CMS21 at 2.05 Mb (15% cotransduction). Based on Fig. 2, the gene was estimated to be located at approximately 2.135 to 2.145 Mb. A large operon or set of operons encoding 15 subunits of the NADH dehydrogenase I complex is located at bp 2135337 to 2151496, suggesting that the mutation in SC1625 affects a subunit of this enzyme. C. crescentus is obligately aerobic, and the small-colony phenotype of this strain is consistent with a mutation causing a defect in respiratory electron transport. Given the large size of the gene cluster and the likely organization of the genes into one or a small number of operons, we did not attempt to define which gene the mutation resides in by sequencing or complementation.
C. crescentus strain SC1621 forms colonies similar to CB15N at 24 and 30°C but grows very slowly at 37°C. The ts136 mutation in SC1621 resides in a gene linked to the CMS12 marker at 1.16 Mb (33% cotransduction) and the CMS13 marker at 1.26 Mb (11% cotransduction). Based on Fig. 2, the gene was estimated to be located at approximately 1.18 to 1.21 Mb. To identify the actual gene in which this mutation is located, the genomic region from 1.173 to 1.203 Mb was divided into segments containing two to four genes for amplification by PCR. The PCR products were cloned into plasmid pMR20 and then introduced into SC1621. Plasmid-bearing strains were grown in PYE at 37°C to test for complementation. The only plasmid from this set that allowed growth of SC1621 cells at 37°C contained a 3.6-kb insert including CC1061 to CC1063 (Fig. 4A). A smaller insert in which the majority of CC1063 (divJ) was removed also allowed growth at 37°C, suggesting that one of the two remaining ORFs contained the mutation in strain SC1621. CC1061 is a small gene starting at bp 1193102 that encodes a 141-amino-acid polypeptide whose sequence is homologous to those of ORFs found in diverse bacteria (Fig. 4b) but whose functions are unknown. CC1062 encodes a sensor kinase with a high degree of similarity to the adjacent divJ gene. Although CC1061 and CC1062 are in the same orientation, the spacing between the coding regions (43 bp) suggests they do not form an operon. The two wild-type genes were amplified separately, but we were unable to subclone them into pMR20 for unknown reasons. They were, however, successfully subcloned into pMR10, a low-copy-number Kanr plasmid vector. Only CC1061 restored growth to SC1621 at 37°C (Fig. 4a). CC1061 was amplified by PCR from the SC1621 chromosome for sequencing, and a single base change (T to C) was identified that changes the Ser 114 codon (TCC) to a proline codon (CCC).
FIG. 4.
(A) Comparison of C. crescentus ORF CC1061 product to homologs from other species. Alignment was done using CLUSTALW (26). (B) The aligned polypeptides are C. crescentus CC1061 (protein identifier [PID] g16125313), Mesorhizobium loti (PID 14027113), Sinorhizobium meliloti (PID g7404510), Agrobacterium tumifaciens (PID g15155913), and Pectobacterium (Erwinia) chrysanthemi (PID 11342550). ∗, residues that are identical in all five polypeptides; :, chemically similar residues. The underlined regions are the predicted membrane-spanning regions. Serine 114, the site of the ts136 mutation in SC1621, is highlighted in bold in the C. crescentus sequence. The mutational change to proline is indicated above S114.
Several additional mutations resulting in temperature-sensitive growth were mapped but have not yet been attributed to individual genes. The ts104 mutation in strain SC596 maps to the 0.34- to 0.38-Mb region of the genome, consistent with its previously established linkage to the proA locus (2), which is suggested by the genome sequence to be the glutamate kinase gene (CC0314) at 0.331 Mb. The ts123 mutation in strain SC1612 maps to 0.76 to 0.80 Mb. The ts129 mutation in strain SC1615 maps to the 2.29- to 2.33-Mb region of the genome. The ts137 mutation in strain SC1622 maps to the 1.52- to 1.56-Mb region of the genome. The ts138 mutation in strain SC1623 maps to the 1.85- to 1.89-Mb region of the genome. The ts139 mutation in strain SC1624 maps to the 1.16- to 1.20-Mb region of the genome, which is very close to the ts136 mutant allele in SC1621. However, none of the plasmid clones tested for complementation of SC1621 restored growth at 37°C to SC1624, suggesting that the mutation either is not in one of the genes tested or is dominant. The ts121 mutation in SC1610 could not be mapped because this strain failed to yield Kanr transductant colonies even at the permissive temperature, despite being sensitive to infection by φCr30, as indicated by plaque formation.
DISCUSSION
We have described a method for rapid and efficient genetic mapping using a collection of markers strategically positioned in the C. crescentus genome and used this method to localize several mutations with interesting phenotypes. Linking phenotype to genotype is the essence of genetic analysis in any organism, and tools that allow this connection to be made as quickly as possible by taking advantage of newly available genomic DNA sequences will be quite valuable. The C. crescentus chromosome is comprised of slightly more than 4 million base pairs of DNA and approximately 3,800 genes (17). The collection of C. crescentus strains described here can be used to localize a mutant gene to a sequence interval equivalent to less than 1% of the genome. After the initial set of transducing lysates is made from the donor strains, mapping any mutation to this level of resolution can be completed in less than 1 week and multiple mutant strains can easily be analyzed in parallel. Transductional mapping is also attractive in that it is inexpensive (particularly since these marker strains are freely available to research labs working with Caulobacter), requiring little more than agar plates, centrifuge tubes, and toothpicks.
The mapping of a mutation resulting in ampicillin sensitivity demonstrated that ampicillin resistance in wild-type C. crescentus is due to the β-lactamase gene (blaA) located at 2.35 Mb in the genome. The dependence on blaA for high-level resistance to ampicillin in PYE medium suggests that the remaining eight β-lactamase genes identified in the C. crescentus genome annotation either are not expressed under these conditions, encode inactive enzymes, or encode enzymes active against β-lactams other than ampicillin. Seven of these sequences lack a functional signal peptide, based on analysis with the SignalP algorithm (16), and thus may not be exportable to the periplasm even if expressed. Other than BlaA, only the CC1540 gene product is predicted to have a functional signal peptide; whether this product is expressed and contributes to the residual low-level ampicillin resistance seen in the absence of blaA in LS107 and CS606 is not known. The blaA gene was probably acquired by horizontal transfer from another organism, as the protein sequence is most similar to class B metallo-β-lactamases from four phylogenetically diverse species: S. maltophilia (Proteobacteria, gamma subdivision) (aligned in Fig. 4), Fluoribacter gormanii (Proteobacteria, gamma subdivision), Janthinobacterium lividum (Proteobacteria, beta subdivision), and Chryseobacterium meningosepticum (Flavibacteria group). The C. crescentus BlaA sequence contains the critical zinc coordinating residues and active site aspartate identified in the S. maltophilia L1 β-lactamase crystal structure (28). Interestingly, despite its apparent acquisition from another organism, the G+C content of the blaA coding region (67%) and its codon usage pattern are quite typical of C. crescentus genes. blaA is seamlessly integrated into an operon involved in methionine synthesis, including homologs of the E. coli metF and metH genes that overlap the blaA start and stop codons, respectively.
Mutations resulting in temperature-sensitive growth were also mapped. One of these mutations (ts141) is likely to affect NADH dehydrogenase I, a complex enzyme made up of at least 14 subunits. This integral membrane complex carries out the initial electron transfer and proton pumping events of respiratory electron transport, which are critical in an obligately aerobic organism, such as C. crescentus. NADH dehydrogenase complex I gene expression and protein levels are cell cycle-regulated in C. crescentus, suggesting that the enzyme may be preferentially active in the stalked and predivisional cells (10, 14). A second mutation causing temperature sensitivity (ts136) resides in a conserved gene whose function is unknown. The CC1061 product is a 15-kDa polypeptide whose most notable properties are that it is quite acidic (pI = 4.66) and it is predicted to have a transmembrane domain spanning residues 80 to 92 (Fig. 4). Homologs of this gene product are found in related gram-negative proteobacteria from the alpha subdivision (e.g., Sinorhizobium meliloti and Agrobacterium tumifaciens) and in more distantly related proteobacteria (e.g., E. coli, Yersinia pestis, and Xylella fastidiosa). One of these homologs (Erwinia chrysanthemi SufE) has been postulated to have a role in insertion of Fe-S centers in proteins (15). The mutation in strain SC1621 (Ser114 to Pro) is predicted to disrupt a conserved α helix in the cytoplasmic domain of the CC1061 protein. It is not known at this time how this results in temperature sensitivity. Further experiments to define the role of this gene product in C. crescentus metabolism and growth are under way.
Before this work, transductional mapping could be used to determine the position of a mutation in the C. crescentus genome relative to other genes (markers) that had been previously mapped. With the genome sequence now known, data on cotransduction frequency become much more useful for locating a mutation, assuming the precise location of the markers used for mapping is known. An alignment of the C. crescentus genetic map and the genome sequence is shown in Fig. 1, with the positions of the CMS markers indicated. There is generally good agreement between the genetic map and actual gene locations derived from the genome sequence. A few discrepancies in gene order are apparent, such as the inversion of the secA-rpoD-fliI gene order on the genetic map relative to the actual sequence, and the incorrect order of rrnA, trpB, and rpoN. Perhaps not coincidentally, both of these problematic segments occur in or near a region of the genome (3.2 to 3.7 Mb) that is difficult to map genetically.
How well do relative distances determined by genetic analysis (especially φCr30 transduction) correlate with actual DNA sequence lengths? This is a critical question, as maximally effective use of transductional mapping data requires cotransduction frequency data to be reliably convertible into DNA sequence distances so that the investigator can determine what section of the genome sequence to focus on. In Fig. 1, distances between loci on the genetic map compared to the DNA sequence are generally consistent, with 1 map unit approximately equal to 1 kb, but there are regions where genetically determined distances appear to not match well with sequence distances, leading to an offset in the two maps. Our own cotransduction data from several markers and mutant strains exhibit a reasonably consistent mathematical relationship between cotransduction frequency and distance (Fig. 2), though some variation is apparent. How much of this is due to experimental factors versus intrinsic differences between sites on the genome is not clear, but investigators using this method should be aware that the relationship between cotransduction frequency and distance may not be constant for all parts of the genome. Local variations in recombination probability that affect the incorporation of transduced or conjugally transferred DNA into the host chromosome may at least partially explain such discrepancies. For example, early versions of the C. crescentus genetic map could not be closed into a circle, as loci at the ends could not be linked by either transduction or RP4-mediated chromosome transfer. This was later attributed to an exceptionally high frequency of recombination in the region in which the terminus of replication is predicted to lie (1.8 to 2.0 Mb; Fig. 1) (7).
Factors unique to transduction can also affect mapping results. It is not known to what extent φCr30 has cleavage preferences for different DNA sequences or what the distribution of preferred sites is on the genome. We observed that transductants containing markers in the chromosomal region from 3.2 to 3.7 Mb are recovered at a much lower rate than markers elsewhere. Barrett et al. (2) noted this as well and suggested that it might be due to a low frequency of phage packaging start sites in this region. Whatever the reason, poor recovery of transductants makes it more difficult to map genes in this region, as extra effort is required to recover enough transductants to accurately assess linkage.
During the course of this work we encountered a few mutant strains that could not be transduced by φCr30 and were thus not amenable to this mapping approach. Some strains cannot be infected by φCr30, as indicated by a lack of plaque formation on top agar. Such strains may contain mutations affecting the expression or assembly of the proteinaceous surface array that serves as the receptor for φCr30 (4). Other strains, such as SC1610, yield plaques on top agar, indicating that there are no problems with phage infection, but nevertheless yield no transductants. Mutations in recA have this phenotype, presumably because the exchange of incoming DNA fragments with the host chromosome is blocked (18). Of course, the mutation responsible for transduction problems in a given strain might not also cause the original phenotype(s) of interest, as secondary mutations causing resistance to φCr30 are not uncommon (12).
The transductional mapping approach described here avoids potential problems associated with cosmid complementation. Dominant mutations are not a problem to map, since the mutant gene is replaced by a transduced wild-type version. Furthermore, no region of the genome is left uncovered. With selectable markers distributed at 100-kb intervals throughout the genome, no gene can be located farther than ∼50 kb from a marker in at least one of the strains in this collection. Depending on the phenotype and location of the mutation being investigated, other mutant genes (Fig. 1), including many spontaneous or Tn5 mutants with easily scored auxotrophies (12), might be used as additional genetic markers to potentially increase the resolution of transductional mapping. Transductional mapping does not, however, eliminate the utility of cosmids and smaller plasmids for testing complementation. Indeed, combining this method with an enhanced cosmid library, particularly an ordered set of cosmid clones with inserts spanning the genome, would further accelerate genetic research. Using a combined approach, a 20- to 40-kb interval of the genome identified by transduction to contain a mutation (assuming it is recessive) could immediately be tested for complementation by appropriate cosmid clones to confirm and narrow the location of the mutation. Efforts to make an ordered set of cosmid clones spanning the C. crescentus genome, as has recently been done for Pseudomonas aeruginosa (11), are under way.
A genetic mapping system similar to that described here could be developed for any bacterium whose genome has been sequenced and for which a mechanism for chromosomal transfer from one strain to another exists. With respect to marker production, usable plasmid-contained antibiotic resistance genes are available for most species. Plasmid pBGS18T is an especially useful delivery vector, since the RK2 oriT sequence allows it to be transferred from E. coli into a variety of bacteria, but it is not necessary to use this plasmid. Any plasmid that can be forced to stably integrate via a cloned targeting sequence should be acceptable. With respect to chromosome transfer, transducing phages have been isolated for many different bacterial species; for a recent example, see work by Burke et al. (3). Other transfer mechanisms could be used to deduce relative distances from markers. Chromosomal DNA can be transferred via conjugation, as with E. coli Hfr strains (23) or RP4-mediated mobilization of the C. crescentus chromosome (2). At this point, however, it appears that transduction may be the best approach for genetic mapping in bacteria in conjunction with a genomic DNA sequence, as phages transfer consistent lengths of chromosomal DNA between cells, yielding reproducible quantitative results for inferring sequence distances between genes.
Acknowledgments
We gratefully acknowledge the contributions of the students in the Spring 2001 “Bio 176: Recombinant DNA Technology” course at Santa Clara University (Lynae Green, Anna Wallebach, Trisha Palafox, Doran Navarro, Carly McLean, Madeline Miller, Rena Kibblewhite, Leah Lacgapan, Amber Bailey, and Jackie Araiza), who constructed many of the marker strains used herein. We thank Yves Brun, Urs Jenal, Dickon Alley, and Lucy Shapiro for sharing plasmids and bacterial strains and for helpful discussions. We also thank Bert Ely for sharing bacterial strains, and Janice Edgerly-Rooks and Michelle Marvier for advice on graphical and statistical analysis.
This work was made possible by research grant MCB-9808798 from the National Science Foundation. Desiree Yang was supported by a summer research fellowship from the “Community of Science Scholars Initiative” program at SCU, made possible by a grant from the Howard Hughes Medical Institute to support undergraduate research.
REFERENCES
- 1.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32:379-391. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, J., C. Rhodes, D. Ferber, B. Jenkins, S. Kuhl, and B. Ely. 1982. Construction of a genetic map for Caulobacter crescentus. J. Bacteriol. 149:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke, J., D. Schneider, and J. Westpheling. 2001. Generalized transduction in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 98:6289-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards, P., and J. Smit. 1991. A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface layer protein as a receptor. J. Bacteriol. 173:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 6.Ely, B., and R. Croft. 1982. Transposon mutagenesis in Caulobacter crescentus. J. Bacteriol. 149:620-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ely, B., T. W. Ely, C. J. Gerardot, and A. Dingwall. 1990. Circularity of the Caulobacter crescentus chromosome determined by pulsed-field gel electrophoresis. J. Bacteriol. 172:1262-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ely, B., and C. J. Gerardot. 1988. Use of pulsed-field-gradient gel electrophoresis to construct a physical map of the Caulobacter crescentus genome. Gene 68:323-333. [DOI] [PubMed] [Google Scholar]
- 9.Ely, B., and R. Johnson. 1977. Generalized transduction in Caulobacter crescentus. Genetics 87:391-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunenfelder, B., G. Rummel, J. Vohradsky, D. Roder, H. Langen, and U. Jenal. 2001. Proteomic analysis of the bacterial cell cycle. Proc. Natl. Acad. Sci. USA 98:4681-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, B., C. B. Whitchurch, L. Croft, S. A. Beatson, and J. S. Mattick. 2000. A minimal tiling path cosmid library for functional analysis of the Pseudomonas aeruginosa PAO1 genome. Microb. Comp. Genomics 5:189-203. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, R. C., and B. Ely. 1977. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics 86:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, R. C., and B. Ely. 1979. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J. Bacteriol. 137:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 15.Nachin, L., M. El Hassouni, L. Loiseau, D. Expert, and F. Barras. 2001. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39:960-972. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 17.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Neill, E. A., R. H. Hynes, and R. A. Bender. 1985. Recombination deficient mutant of Caulobacter crescentus. Mol. Gen. Genet. 198:275-278. [DOI] [PubMed] [Google Scholar]
- 19.Osteras, M., and U. Jenal. 2000. Regulatory circuits in Caulobacter. Curr. Opin. Microbiol. 3:171-176. [DOI] [PubMed] [Google Scholar]
- 20.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 21.Ried, J. L., and A. Collmer. 1987. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spratt, B. G., P. J. Hedge, S. Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 25.Stephens, C. 2001. Bacterial cell cycle: seeing the big picture with microarrays. Curr. Biol. 11:R222-R225. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai, J. W., and M. R. K. Alley. 2000. Proteolysis of the McpA chemoreceptor does not require the Caulobacter major chemotaxis operon. J. Bacteriol. 182:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ullah, J. H., T. R. Walsh, I. A. Taylor, D. C. Emery, C. S. Verma, S. J. Gamblin, and J. Spencer. 1998. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution. J. Mol. Biol. 284:125-136. [DOI] [PubMed] [Google Scholar]
- 29.Wu, J., N. Ohta, and A. Newton. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. USA 95:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]