Abstract
Low-cost and rescue treatments for Helicobacter pylori infections involve combinations of several drugs including tetracycline. Resistance to tetracycline has recently emerged in H. pylori. The 16S rRNA gene sequences of two tetracycline-resistant clinical isolates (MIC = 64 μg/ml) were determined and compared to the consensus H. pylori 16S rRNA sequence. One isolate had four nucleotide substitutions, and the other had four substitutions and two deletions. Natural transformation with the 16S rRNA genes from the resistant organisms conferred tetracycline resistance on susceptible strains. 16S rRNA genes containing the individual mutations were constructed and tested for the ability to confer resistance. Only the 16S rRNA gene containing the triple mutation, AGA965-967TTC, was able to confer tetracycline resistance on H. pylori 26695. The MICs of tetracycline for the transformed strains were equivalent to those for the original clinical isolates. The two original isolates were also metronidazole resistant, but this trait was not linked to the tetracycline resistance phenotype. Serial passage of several H. pylori strains on increasing concentrations of tetracycline yielded mutants with only a very modest increase in tetracycline resistance to a MIC of 4 to 8 μg/ml. These mutants all had a deletion of G942 in the 16S rRNA genes. The mutations in the 16S rRNA are clearly responsible for tetracycline resistance in H. pylori.
Helicobacter pylori is a gram-negative, microaerophilic bacterium that colonizes the human stomach (16). Once acquired, H. pylori may persist for decades unless eradicated with antibiotic treatment. Colonization with H. pylori may lead to a chromic inflammatory response resulting in peptic ulcers, gastritis, mucosa-associated lymphoid tissue lymphomas, and gastric cancer (3, 27). Present first-line treatments for eradication of H. pylori consist of combinations of three drugs and achieve clinical cure rates of >80%. The most common combinations are a proton pump inhibitor or ranitidine bismuth citrate and two antibiotics, clarithromycin and either amoxicillin or metronidazole (17, 27). An older method (standard triple therapy) of bismuth, tetracycline, and metronidazole is also used when cost is a factor or as a second-line treatment in developed countries (17). Quadruple therapy, standard therapy plus a proton pump inhibitor, is also used as a rescue treatment when the first treatment has not been successful.
Tetracycline is a protein synthesis inhibitor with a broad spectrum of activity, active against gram-negative and -positive bacteria and mycoplasmas (24). Its safety and low cost have led to its widespread use in human and veterinary medicine and in the food production industry (5). The drug is actively taken up by bacteria and accumulates in the cytoplasm, where its target is the ribosome. Tetracycline binds to the 30S subunit of the ribosome and blocks the binding of aminoacyl-tRNA, thus stalling the synthesis of nascent peptide chains. Two recent crystal structures of the 30S ribosome have identified two and six tetracycline binding sites (4, 23); the reason for the discrepancy in the number of sites may be related to the fact that multiple low-affinity binding sites (potentially hundreds) have been identified by many biochemical studies over the last 3 decades.
With the increased use of tetracyclines has come the emerging problem of microbial resistance. There are four main mechanisms by which bacteria become resistant to tetracycline (for a recent review see reference 5). There are several classes of tetracycline efflux pumps that are widely disseminated in bacteria and located on transmissible plasmids and transposons. These pumps are single subunits containing multiple transmembrane segments and are specific for the active transport of tetracycline. A putative tetracycline efflux pump of this kind was identified in the genome of H. pylori 26695 (34). The gene HP1165 from this organism was 27% identical to the gene for the tetracycline efflux protein tetA(P) from Clostridium spp. (29). This putative transporter was also present in the H. pylori J99 genome sequence (1), but neither J99 nor 26695 is resistant to tetracycline. In addition to the tetracycline-specific transporters, chromosomally encoded multidrug efflux pumps, such as the MexAB-OprM system of Pseudomonas aeruginosa, confer resistance to several drugs including tetracycline, chloramphenicol, and quinolones (5). Overexpression of the efflux pumps increases multidrug resistance, while mutants with deletions of these operons are hypersensitive to antibiotics (15, 20).
Ribosomal protection by a soluble resistance protein, such as Tet(O), is another common mechanism in which the ribosome is able to continue protein synthesis in the presence of tetracycline (35). Tetracycline is chased from the ribosome by the protection protein, and protein synthesis can continue. The third mechanism is the unknown chemical modification of tetracycline by an oxidoreductase that requires NADP (31).
The most recent mechanism of resistance to be described is a mutation in the 16S rRNA. Several clinical isolates of Propionibacterium acnes (the causative agent of acne vulgaris) were found to contain a G-to-C mutation at position 1058 (Escherichia coli numbering) in their 16S rRNA genes (25). This mutation gave a variable level of tetracycline resistance, with the MIC of tetracycline ranging from 2 to 64 μg/ml (the MICs of tetracycline for wild-type P. acnes were 0.12 to 1 μg/ml). There are three copies of the 16S rRNA genes in this organism, but all of the copies were apparently homogeneous. Nucleotide G1058 is located in helix 34 (h34) of the 16S rRNA adjacent to the primary (Tet-1) binding site (4, 23), suggesting that a disturbance in the base pairing of h34 may interfere with the binding of tetracycline and thus prevent its inhibitory effect. This mutation was reproduced in an E. coli 16S rRNA gene and expressed from a plasmid, but the MIC of tetracycline for the recombinant strain increased only twofold (25). The very slight increase in tetracycline resistance is probably due to the presence of the seven chromosomal rRNA operons in E. coli competing with the mutant plasmid copy.
Tetracycline resistance in H. pylori is relatively rare, with the vast majority of studies reporting no tetracycline resistance in clinical isolates. However, with the use of tetracycline in therapy, tetracycline-resistant isolates have started to emerge at low levels, on average 5 to 7% of isolates (12, 14). In one recent study of clinical isolates from Shanghai, China, the level of tetracycline resistance was an astonishing 59% (39). An early case of tetracycline resistance in Australia saw the isolation of two H. pylori strains from biopsy samples of the antrum and corpus of a patient who had failed triple therapy with colloidal bismuth subcitrate, metronidazole, and tetracycline (18). The patient had failed two previous attempts to eradicate H. pylori by treatment with omeprazole and amoxicillin and with omeprazole, amoxicillin, and metronidazole. These isolates were highly resistant to tetracycline, with the MICs of tetracycline for the strains being ≥256 μg/ml (antrum isolate) and 32 μg/ml (corpus isolate) as determined by E test. They were also resistant to metronidazole (E-test MIC, ≥32 μg/ml). This cross-resistance to tetracycline and metronidazole was also identified in a study of tetracycline-resistant H. pylori isolates from Korea and Japan (14). Furthermore, the resistance to both tetracycline and metronidazole was transferred to susceptible strains by transformation with the genomic DNA of the resistant organisms. Attempts to characterize the tetracycline-resistant H. pylori from the Australian study by shotgun cloning, DNA-DNA hybridizations, and PCR screening turned up no evidence of the known tetracycline resistance determinants (J. Rood, P. Crellin, D. Lyras, and P. Midolo, personal communication), suggesting that perhaps a mutation in the 16S rRNA may be responsible for resistance. The complete genome sequences of two H. pylori strains, 26695 and J99 (1, 34), have been published elsewhere and demonstrate that this organism contains only two rRNA operons, increasing the likelihood that rRNA mutations could arise. The goal of this study was to determine the sequence of the 16S rRNA from two tetracycline-resistant H. pylori strains and test the ability of mutations in the 16S rRNA genes to mediate tetracycline resistance in this organism.
MATERIALS AND METHODS
Strains and growth conditions.
Two strains of tetracycline-resistant H. pylori isolated from biopsy samples of one patient, 108 (from the antrum) and 109 (from the corpus), were obtained from P. Midolo (Monash Medical Center, Melbourne, Australia). H. pylori strains 26695, sequenced by Tomb et al. (34), and NCTC 11639 and A12 (a metronidazole-resistant derivative of 11639 [37]) were also used. Strains were grown on brain heart infusion-yeast extract (BHI-YE) agar (37 g of BHI [Difco, Detroit, Mich.]/liter, 5 g of YE [Difco]/liter, 50 ml of equine serum [HyClone, Logan, Utah]/liter, 15 g of Select agar [Invitrogen, Carlsbad, Calif.]/liter) supplemented with 10 μg of amphotericin B (Sigma-Aldrich, Oakville, Ontario, Canada)/liter and 10 μg of vancomycin (Eli Lilly Canada, Toronto, Ontario, Canada)/liter. Plates were incubated in anaerobic jars at 37°C under an atmosphere of 10% CO2-5% O2-85% N2. Chloramphenicol (Sigma) was used at a concentration of 15 μg/ml while the amounts of tetracycline (Sigma) and metronidazole (Sigma) used varied and are specified below. When chloramphenicol, tetracycline, or metronidazole was added to plates, the amphotericin B and vancomycin were omitted. E. coli DH5α (Invitrogen) was used as the host strain for plasmids created in this study.
MIC determinations.
BHI-YE plates were spread with 50 μl of a frozen stock of H. pylori and allowed to grow for 3 to 4 days. A small amount of cells was scraped and suspended in 500 μl of BHI broth. The cell concentration was determined by readings of optical density at 600 nm and adjusted to 106 cells/ml by dilution in BHI-YE broth. A 4-μl aliquot of each cell suspension was spotted onto BHI-YE plates containing twofold serial dilutions of tetracycline from 128 to 0.25 μg/ml. Plates were incubated for up to 5 days and scored for growth at 2 to 3 days and 5 days. When it was difficult to tell if a culture had grown, the spot was scraped and plated onto a fresh BHI-YE agar plate with no tetracycline to test the viability of the bacteria. Determination of MICs of metronidazole (0.25 to 256 μg/ml) was carried out using the same technique, but plates were incubated for 3 days. Plates were used within 24 h to ensure that the drug was not degraded. The MICs were confirmed by using the alternate method of MIC determination developed by Jeong et al. (11). The quality control strain ATCC 43504 was used as a control, and the MIC for it was found to be 64 μg/ml, in agreement with literature values. Both methods gave consistent results within one MIC level.
Molecular biology techniques.
DNA manipulations were carried out essentially as described in the work of Sambrook et al. (26). Restriction enzymes, polymerases, and oligonucleotides were obtained from Invitrogen and used according to the manufacturer's recommendations. Chromosomal DNA was isolated from H. pylori as described in the work of Ge and Taylor (8). Sequencing of the H. pylori 16S rRNA genes was carried out using oligonucleotides Hp1 through Hp9 (sequences published in reference 7), and only those used in PCR are included in Table 1 along with the oligonucleotides used for mutagenesis. The oligonucleotides used to sequence the rdxA genes, rdxAP1 and rdxAP12, are described in the work of Wang et al. (37). PCR amplification of DNA for sequencing was carried out using Taq polymerase as specified by the manufacturer. DNA sequencing was carried out using the Thermo Sequenase Cycle Sequencing kit (Amersham Pharmacia Biotech) or performed using a CEQ2000 sequencer (Beckman Coulter, Inc., Mississauga, Ontario, Canada) in the DNA Services Laboratory, Department of Biochemistry, University of Alberta. DNA sequence analysis was performed using the LASERGENE software package (DNASTAR, Inc., Madison, Wis.), and Blast searches were carried out using the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
TABLE 1.
Oligonucleotides
| Use and name | Sequence | Positiona | Purpose or 16S rRNA mutation |
|---|---|---|---|
| Sequencing, cloning and PCR | |||
| Hp12 | 5′ATATGCATGCGAAAACCAAGAAAGTCTGG3′ | −603 to −585 | PCR, forward, SphI |
| Hp13 | 5′ATATGGATCCAAGCAAAATTTAAAACAAAACAC3′ | 1766 to 1788 | PCR, reverse, BamHI |
| Hp1 | 5′AGAGTTTGATCCTGGCTCAG3′ | 8 to 27 | PCR, forward |
| Hp3 | 5′CGACCTGCTGGAACATT3′ | 693 to 709 | PCR, forward |
| Hp7 | 5′ATCCTAAAACCTTCATCCTC3′ | 412 to 393 | PCR, reverse |
| Hp8 | 5′TCGTTGCGGGACTTAACCCAA3′ | 1073 to 1053 | PCR, reverse |
| Hp9 | 5′AAGGAGGTGATCCAGCCGCA3′ | 1503 to 1484 | PCR, reverse |
| Hp14 | 5′AGACGCATGCATTTTCCCAAACATTCCCTA3′ | −528 to −507 | PCR, forward, SphI |
| Hp15 | 5′ACCTGGATCCAACAAAGACAAAAAGCTAA3′ | 1688 to 1712 | PCR, reverse, BamHI |
| Mutagenesis | |||
| Hpmut1 | 5′AGGCAGCAGTAAGGAATA3′ | 335 to 352 | G360A, forward |
| Hpmut2 | 5′TATTCCTTACTGCTGCCT3′ | 351 to 334 | G360A, reverse |
| Hpmut3 | 5′GCGAAAGCΔTGGGGAG3′ | 724 to 740 | Deletion G771, forward |
| Hpmut4 | 5′CTCCCCAΔGCTTTCGC3′ | 740 to 724 | Deletion G771, reverse |
| Hpmut5 | 5′CACAAGCGΔTGGAGCA3′ | 897 to 912 | Deletion G942, forward |
| Hpmut6 | 5′ATGCTCCAΔCGCTTGT3′ | 913 to 898 | Deletion G942, reverse |
| Hpmut7 | 5′AGCATGTGGTTTAATTCGATTCTACAC3′ | 909 to 935 | AGA965-967TTC, forward |
| Hpmut8 | 5′TTCTTCGTGTAGAATCGAATTAAACCAC3′ | 941 to 913 | AGA965-967TTC, reverse |
| Hpmut9 | 5′GTGGTTTAATTCGATGATACAC3′ | 914 to 935 | A965T, forward |
| Hpmut10 | 5′GTGTATCATCGAATTAAACCAG3′ | 935 to 913 | A965T, reverse |
| Hpmut11 | 5′GTGGTTTAATTCGAATATACAC3′ | 914 to 935 | G966T, forward |
| Hpmut12 | 5′GTGTATATTCGAATTAAACCAC3′ | 935 to 913 | G966T, reverse |
| Hpmut13 | 5′GTGGTTTAATTCGAAGCTACAC3′ | 914 to 935 | A967C, forward |
| Hpmut14 | 5′GTGTAGCTTCGAATTAAACCAG3′ | 935 to 913 | A967C, reverse |
| Hpmut15 | 5′GCTGCACGCCTGTCGTCAGC3′ | 1018 to 1037 | G1058C, forward |
| Hpmut16 | 5′GACGACAGGCGTGCAGCAC3′ | 1034 to 1016 | G1058C, reverse |
Cloning of an H. pylori 16S rRNA gene.
With the genome sequence of H. pylori 26695 (34), primers were designed to amplify one of the 16S rRNA genes, including the promoter region, by PCR. The sequences of Hp12 and Hp13 are shown in Table 1. The primers contained restriction sites, SphI (Hp12) and BamHI (Hp13), to generate products that could then be cloned into the pOK12 vector (36). The gene was amplified using the high-fidelity polymerase Platinum Pfx (Invitrogen) to minimize errors. The manufacturer's recommended conditions were followed for 30 cycles of 94°C for 45 s, 50°C for 1 min, and 68°C for 2.5 min. PCR products (2.4 kb) were purified by extraction from agarose gels and cloned into pOK12 to give p16S. Restriction digestion and DNA sequencing were used to confirm that clones were correct.
Mutation of 16S rRNA genes.
Recombinant PCR, as outlined previously (10) but with substitution of Platinum Pfx polymerase for Taq polymerase, was used to individually recreate the mutations identified in H. pylori 109. With the p16S plasmid as template, the Hp12 primer and one reverse Hpmut primer formed the first product, while Hp13 and a forward Hpmut primer produced the second product. The two products were purified from an agarose gel and mixed in equivalent amounts, as judged from agarose gel electrophoresis, to be the template for a third round of PCR with the Hp12 and Hp13 primers. The product of this reaction was then cloned into pOK12 and confirmed to be correct by restriction digestion and DNA sequencing.
Transformation of H. pylori.
The procedure for natural transformation is essentially that described in the work of Ge and Taylor (8). H. pylori 26695 was spread onto BHI-YE plates and grown for 3 days. Cells were scraped onto fresh plates and incubated at 37°C for 6 h, after which 15 μl of aliquots of DNA was spotted onto the cells. The plates were incubated for another 20 h, at which time the spots were scraped and the cells were spread onto plates containing 2 or 4 μg of tetracycline/ml. The spots contained an average of 5 × 107 cells as determined from plating serial dilutions of mock-treated spots. Plates were incubated for 5 days to select for transformants. The DNA samples used in the natural transformation experiments were purified PCR products. The method for electroporation of H. pylori was described previously (8). Bacteria were harvested and washed in 10% glycerol. DNA (approximately 0.5 μg) was mixed with 0.2 ml of cells, transferred to an electroporation cuvette (0.2-cm gap), and placed in a Bio-Rad Gene Pulser (Bio-Rad, Hercules, Calif.). A single pulse of 12.5 ms at 2.5 kV (12.5 kV/cm) with a 25-μF capacitor and a resistance of 600 Ω was used for transformation. Cell suspensions were plated on BHI-YE plates and incubated for 20 h before being transferred to selective plates.
Construction of a 16S rRNA gene deletion strain of H. pylori.
A deletion mutant of the 16S rRNA gene was constructed by cutting p16S with HindIII, leaving only 110 bp of the upstream promoter and about 300 bp downstream of the gene. The ends were filled in using T4 DNA polymerase, and a chloramphenicol acetyltransferase (cat) gene (38) was cloned in, allowing the deletion mutant to be selected by growth on 15 μg of chloramphenicol/ml. The 16S rRNA gene deletion was amplified with Hp12 and Hp13 primers to give a 1.1-kb product that was purified and transformed into 26695 by electroporation. Transformants appeared only after incubation for 10 days. The insertion of the 16S deletion in the chromosome was detected by the PCR amplification of two bands with primers Hp14 and Hp15, one of 2.4 kb (wild type) and one of 1.1 kb (chloramphenicol-disrupted gene). One of the strains, UA1374 (26695 rrs::cat) carrying the 16S deletion, was chosen for further study.
Serial passage on tetracycline.
Tetracycline concentrations in the plates ranged from 0.063 to 16 μg/ml. Metronidazole was added at a concentration of 0.5 or 4 μg/ml. Cells were streaked onto a plate containing the lowest concentration of tetracycline, allowed to grow for 3 days, and then streaked onto a plate with a twofold increase in tetracycline. Four strains, 26695, UA1374, NCTC 11639, and A12, were used. Strains were passaged until no growth occurred.
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA genes (accession no. AY062898 and AY062899) and the rdxA genes (AY063488 and AY063489) of H. pylori strains 108 and 109, respectively, have been deposited in GenBank.
RESULTS
Tetracycline resistance of H. pylori.
The tetracycline-resistant H. pylori isolates 108 and 109 were cultured to determine the MICs of tetracycline for them by the agar dilution method. MICs of tetracycline for susceptible H. pylori strains are usually between 0.125 and 0.5 μg/ml (33), and the MIC for strain 26695 is 0.5 μg/ml (Table 2). Both strain 108 and strain 109 grew very well on agar containing 0.5 to 16 μg of tetracycline/ml after only 2 days of incubation. When plates were incubated for 3 additional days, colonies appeared on the 32-μg/ml tetracycline plates and it appeared that faint growth was present on plates containing 64 to 128 μg/ml. However, when the apparent bacterial growth on these plates was restreaked onto nonselective plates, only the bacteria from the 32-μg/ml tetracycline plate were viable. Thus, the MIC of tetracycline for both 108 and 109 is 64 μg/ml. The reported MICs of tetracycline were >256 μg/ml for 108 and 32 μg/ml for 109, but these had originally been determined by an alternate method, the E test (18). MIC determinations were repeated several times and remained stable.
TABLE 2.
Summary of MIC determinations and 16S rRNA gene mutations in H. pylori
| Strain, transformant, or mutant | MIC (μg/ml) of drug:
|
16S rRNA gene(s) | |
|---|---|---|---|
| Tetra- cycline | Metroni- dazolea | ||
| Strains | |||
| 26695 | 0.5 | 2 (1) | Wild type |
| 108 | 64 | 128 (64) | G360A, AGA965-967TTC |
| 109 | 64 | 128 (64) | G360A, ΔG771, ΔG942, AGA965-967TTC |
| 11639 | 1 | 2 (1) | Wild type |
| 11639rdxA12 | 0.5 | 64 (64) | Wild type |
| Transformants | |||
| 26695 (109 16S) | 4-16 | 2 (2) | Mixed |
| 26695 (16S TTC) | 64 | 2 (2) | AGA965-967TTC |
| 11639 (16S TTC) | 128 | 2 (2) | AGA965-967TTC |
| Mutants | |||
| UA1374 | 0.25 | 1 (2) | Insertion of cat gene into one 16S gene |
| 26695Tcr | 2 | NDb | ΔG942 |
| 6/2Tcr | 2 | ND | ΔG942 |
| 11639Tcr | 8 | ND | ΔG942 |
| 11639rdxA12Tcr | 2 | ND | ΔG942 |
The MICs derived by the method of Jeong et al. (11) are in parentheses.
ND, not done.
The 16S rRNA genes of strains 108 and 109 were amplified by PCR and sequenced. The 16S rRNA genes were amplified with primers based upon the published sequences of H. pylori 16S rRNA genes (7) and did not distinguish between the two copies present on the chromosome. Previous sequencing of these genes found only one sequence per strain, indicating a homogeneity of the 16S rRNA genes in each strain (7), as has also been shown previously for the 23S rRNA genes in H. pylori (32). A single sequence was obtained for the 16S rRNA genes of each tetracycline-resistant strain, indicating that the genes in these strains were also homogeneous.
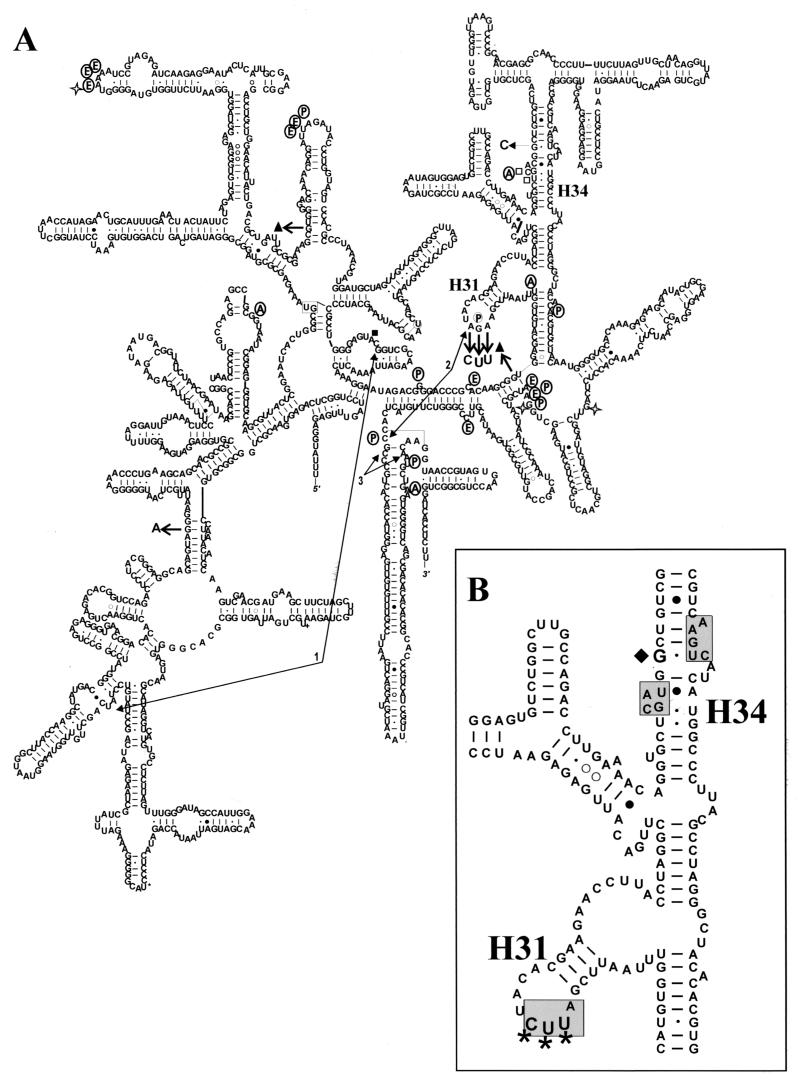
The 16S rRNA gene sequences of strains 108 and 109 were compared with published H. pylori 16S sequences in Fig. 1, which shows a sequence alignment of the 108 and 109 16S rRNA genes with a consensus sequence derived from aligning the other H. pylori 16S rRNA genes. None of the strains had the exact same sequence. For the tetracycline-resistant strains, there were four unique nucleotide changes in strain 108 and six in 109. The numbering of the 16S rRNA nucleotides is based on the E. coli 16S rRNA and is used throughout. Both strains had a mutation of G360 to A in domain I and a triplet mutation of AGA965-967TTC in domain III. An additional two nucleotides were deleted in strain 109, G771 in domain II and G942 in domain III. These changes are depicted by arrows on a secondary structure model of the H. pylori 16S rRNA (Fig. 2) together with information on tetracycline interactions and tRNA binding. The 16S rRNA genes from strains 108 and 109 were identical along their entire length, with the exception of the two extra deletions in 109, and differed from the rest of the H. pylori 16S sequences, a strong indication that they are both derivatives of the same strain.
FIG. 1.
Sequence alignment of the tetracycline-resistant H. pylori and the consensus H. pylori 16S rRNA gene sequences. The consensus H. pylori 16S rRNA gene sequence was derived from a sequence alignment of all of the H. pylori sequences over 1,200 nucleotides long in the GenBank database (accession no. U01332, U01331, U01328, U01329, and U01330 [7]; U00697 [6]; NC_00921 [nucleotides 1189529 to 1188022] [1]; NC_00915 [nucleotides 1512634 to 1511130] [34]; Z25742 and Z25841 [M. Khan, B. Drasar, and N. Stoker, unpublished data]; U08906 [9]; AF361935 [D. Queiroz, A. Oliveira, G. Rocha, S. Moura, E. Camargos, P. Valle, L. Bicalho, and R. Dani, unpublished data]). Small partial sequences or those from strains not specified as H. pylori were excluded, as were duplicate sequences of the same strain. Alignments were generated using the LASERGENE software package. The 108 and 109 16S rRNA gene sequences were aligned with the consensus H. pylori sequence. Sequence variations are boxed, and the mutation from the tetracycline-resistant P. acnes isolates is marked with an arrow.
FIG. 2.
Secondary structure model of the 16S rRNA from H. pylori 109. (A) This model of the 16S rRNA from 109 is based on the H. pylori 16S rRNA model found on the Comparative RNA website (http://www.rna.icmb.utexas.edu/) and modified with permission. The sequence deviations unique to strain 109 are marked by short, thick arrows with the altered residue indicated (a solid triangle denotes deletion of a residue). Tetracycline cross-links to the 16S rRNA are denoted by open stars (22). The joined arrows 1, 2, and 3 indicate rRNA cross-links that are disrupted by tetracycline binding (21). Nucleotides displaying altered reactivity to chemical modification in the presence of tetracycline are indicated by squares (solid squares, protected; open squares, enhanced cleavage) (19). Circled letters indicate nucleotides that contact tRNA molecules in the 70S crystal structure (40) at the A site (A), the P site (P), or the E site (E). (B) Close-up of the region involved in tetracycline resistance. The boxed residues of h34 and h31 make up the primary or Tet-1 binding site (4, 23). The triple mutation AGA965-967UUC is indicated by the triple asterisk. The G1058C mutation is marked by a solid diamond.
Transfer of tetracycline resistance by natural transformation.
H. pylori strains are naturally competent and take up exogenous DNA, incorporating it into the chromosome by homologous recombination (8). Natural transformation was used to determine the ability of the 16S rRNA gene from strain 109 to mediate tetracycline resistance. The gene was amplified by PCR using oligonucleotides Hp1 and Hp9 to give a 1.5-kb product that did not include the promoter region. This was then transferred to the tetracycline-susceptible strain 26695, and transformants were selected by plating strains on 2 μg of tetracycline/ml. A higher level of tetracycline was used originally for selection (8 μg/ml), but no transformants were isolated, suggesting that perhaps recombination into both copies of the 16S rRNA genes must take place to give high-level resistance. Time may also be required for the mutation to establish itself, i.e., for resistant populations of ribosomes to accumulate. A level of 2 μg of tetracycline/ml represents a fourfold increase over the MIC of tetracycline for strain 26695. Transformants were isolated for which MICs of tetracycline were 4 and 16 μg/ml. Sequencing of the 16S rRNA genes on the chromosome was attempted, but more than one sequence was present, likely due to the deleted residues present in the strain 109 gene compared to the native strain 26695 16S rRNA genes. With four regions of sequence substitutions, it is possible that not all changes were incorporated into both of the chromosomal genes, as the entire DNA fragment may not have been involved in the recombination. A mixture of susceptible wild-type and resistant mutant ribosomes would account for intermediate levels of resistance to tetracycline. Negative controls in which no DNA was added to the cells did not give rise to any colonies.
Mutagenesis of the H. pylori 26695 16S rRNA genes.
Rather than deconvoluting the mixed results from the above transformants, we decided to determine which of the nucleotide changes in the 16S rRNA genes were involved in tetracycline resistance by mutagenesis of a single residue at a time. Each mutation was introduced individually into the 16S rRNA gene. The 16S rRNA gene (including approximately 600 bp upstream and 300 bp downstream) was amplified by PCR using a high-fidelity polymerase, Pfx. This gene was cloned into the vector pOK12, sequenced, and used as the template for mutagenesis and as a negative control. The mutations G360A, AGA965-967TTC, A965T, G966T, and A967C and the deletions of G771 and G942 were constructed by recombinant PCR, again using Pfx to minimize errors. The P. acnes 16S rRNA mutation, G1058C, was also recreated in the H. pylori 16S rRNA gene. Mutant 16S rRNA genes were cloned and then sequenced to ensure that only the desired mutation was present. The plasmids containing the 16S rRNA genes did not mediate tetracycline resistance in the host E. coli strain.
These plasmids then served as templates to amplify the 16S rRNA genes, either the complete 16S rRNA gene minus the promoter (Hp1/Hp9) or a small fragment of about 300 bp containing the mutation (Hp1/Hp7 for G360A or Hp3/Hp8 for the remainder). These DNA fragments were used to transform 26695. Tetracycline-resistant mutants were selected by plating on 2 or 4 μg of tetracycline/ml. At 4 μg of tetracycline/ml only DNA encoding the triple mutation AGA965-967TTC was able to confer resistance on 26695. Both the long and short 16S rRNA gene fragments were taken up and recombined into 26695. Transformants were passaged on tetracycline to ensure their stability, and then the incorporation of the triple mutation into the chromosomal 16S rRNA genes was confirmed by DNA sequencing. In all cases there was no sequence heterogeneity, indicating that the mutant DNA had recombined and replaced both 16S rRNA genes. At a concentration of 2 μg of tetracycline/ml it was possible to isolate colonies from the other transformation reactions but these were false positives, as they did not grow when restreaked on the same concentration of tetracycline. The MICs of tetracycline for the 16S AGA965-967TTC transformants were determined to be 32 to 64 μg/ml; some clones were twofold-more resistant than others were. The triple mutant grew as well as did the wild-type 26695, and the mutation and tetracycline resistance were retained over several months, several passages, and storage at −80°C. These results were also duplicated using strain NCTC 11639 and selection on 4 μg of tetracycline/ml, as the MIC of tetracycline for this strain is 1 μg/ml. Only DNA containing the AGA965-967TTC mutation was able to confer tetracycline resistance on NCTC 11639 (Table 2). No spontaneous tetracycline resistance mutations of either NCTC 11639 or strain 26695 were ever isolated (mutation frequency of >10−8 mutants/number of cells plated).
Metronidazole resistance in tetracycline-resistant H. pylori.
Previous studies have linked tetracycline and metronidazole resistance and their transfer to susceptible organisms (12, 14, 18). We determined the metronidazole susceptibilities of the tetracycline-resistant H. pylori and the transformants. As metronidazole becomes mutagenic when reduced, spontaneous mutations are observed and complicate MIC determinations (11, 28). We used two different methods, but both methods use a lower inoculum than that for the standard NCCLS procedure, and this seems to give more reproducible results. Both our standard method and the one proposed by Jeong et al. (11) gave similar results, with the second method typically giving a value twofold less than that of our standard method (Table 2). When heavier inocula were tested, the MIC of metronidazole increased two- to fourfold (data not shown). Both strain 108 and strain 109 were highly resistant to metronidazole, with MICs being 64 to 128 μg/ml. H. pylori 26695 was metronidazole susceptible, with the MIC being 1 to 2 μg/ml. All of the tetracycline-resistant transformants of strains 26695 and NCTC 11639 were susceptible to metronidazole, indicating that the tetracycline resistance phenotype is not genetically linked to metronidazole resistance. The rdxA gene from these strains was sequenced and found to be identical in the two strains. There were two unique mutations in the amino acid sequence of RdxA that were not found in other RdxA sequences as revealed by a Blast search of the protein databases, A118S and a deletion of Y141. An A118T mutation was seen previously for a metronidazole-susceptible strain (30), and so it appears that the Y141 deletion may be responsible for resistance. Transfer of the rdxA genes from strains 108 and 109 to H. pylori 26695 did confer metronidazole resistance on transformants but did not confer tetracycline resistance.
Selection of tetracycline-resistant H. pylori.
The presence of only two rRNA operons in H. pylori increases the likelihood of isolating strains with tetracycline resistance mutations in the 16S rRNA by serial passage on increasing concentrations of tetracycline. To increase the likelihood of being able to isolate mutants, a strain was constructed from 26695 in which one 16S rRNA gene was replaced by a chloramphenicol resistance gene. This strain, UA1374, grew very slowly, taking an average of 10 days to grow versus the 3 to 4 days for 26695. The deletion strain was also twofold-more susceptible to tetracycline; the MIC of tetracycline for UA1374 was 0.25 μg/ml, reduced from 0.5 μg/ml for strain 26695. Two additional strains of H. pylori were tested, NCTC 11639 and A12 (a metronidazole-resistant derivative of 11639 for which the MIC was 64 μg/ml), to determine if the prior metronidazole resistance increased the likelihood of isolating tetracycline-resistant mutants.
Strains were serially passaged on increasing concentrations of tetracycline from a level well below the MIC (0.063 μg/ml) until they would no longer grow. The MICs of tetracycline increased fourfold for all the strains (eightfold for 11639) and would go no further. Metronidazole was added at 0.5 μg/ml, a concentration just below the MIC (1 to 2 μg/ml) for the strains 26695, 11639, and UA1374, but no tetracycline-resistant mutants were isolated. Mutants displaying high-level resistance to tetracycline were not isolated even with strain A12 in the presence of 4 μg of metronidazole/ml. When the 16S rRNA genes from these passaged strains were sequenced, they were all found to have a deletion at G942.
DISCUSSION
We have shown that mutations found in the 16S rRNA genes of two tetracycline-resistant clinical isolates of H. pylori can confer tetracycline resistance on susceptible strains. Two strains of H. pylori, 26695 and 11639, were made resistant to tetracycline by transformation with the 16S rRNA gene from strain 109. We identified several unique (among H. pylori strains) sequence variations in the 16S rRNA genes of this strain, and these have been placed on a secondary structure model of the H. pylori 16S rRNA in Fig. 2 (indicated by thick arrows). The mutations AGA965-967UUC (h31 loop) and the G942 deletion are located in domain III. Several nucleotides in this region interact with tRNA molecules. In particular, the base G966 directly contacts the anticodon loop of the tRNA in the P site (40). Tetracycline has been shown previously to enhance the chemical reactivity of bases at positions 1052 and 1054 (19) and to photolabel bases 1300 and 1338 (22). Like G966, nucleotides 1338 and 1339 are in close proximity to the P-site tRNA (40) and are near the G942 deletion. Nucleotide 1054 contacts the A-site tRNA (40) and is very near G1058, the P. acnes tetracycline resistance mutation. Tetracycline binding also disrupts a 16S rRNA cross-link between C967 and C1400 in the E. coli 30S ribosomal subunit (21). The crystal structures of the 30S ribosome-tetracycline complexes show tetracycline interacting primarily with atoms of the phosphodiester backbone of nucleotides in h34 and the loop of h31, the boxed residues in Fig. 2B (4, 23). In contrast, the G360A mutation (domain I) and the G771 deletion (domain II) are in regions of the 16S rRNA not closely associated with either tetracycline or tRNA binding.
Of the several sequence variations present in the 16S rRNA genes of strain 109 only DNA encoding the triple AGA965-967TTC mutation was found to confer tetracycline resistance on a susceptible strain. These mutations overlap the primary tetracycline binding site (Fig. 2B) and part of the P site (G966). The A site is in close proximity as well, with the base C1054 contacting the A-site tRNA and the backbone forming part of the tetracycline binding site (4, 23, 40). A phylogenic analysis of the 16S rRNA from eubacteria showing the conservation of each base at every position can be found at www.rna.icmb.utexas.edu/CSI. Among the nucleotides of the h31 loop, nucleotide 965 was the least conserved of the three, with a U present in 53% of sequences, the remainder being A (37%), C (9%), and G (2%). Nucleotide 966 was the most conserved, with G being present in 97.5% of sequences. Nucleotide 967 was a C in 85% of sequences and an A in only 13%. The triple mutation changed the rRNA from AGA to UUC; the first and last bases were changed to the eubacterial preferred nucleotides, but the middle substitution (U966) is found in only 0.15% of eubacterial 16S rRNA genes (www.rna.icmb.utexas.edu/CSI). Organisms with the 16S rRNA sequence of UGC at 965 to 967 (such as E. coli) are tetracycline susceptible, so it is likely that the substitution of the middle G residue is primarily responsible for resistance. However, transformation with a mutant 16S rRNA gene containing only the G966T substitution did not give rise to resistant colonies when colonies were selected on 2 to 4 μg of tetracycline/ml. The presence of the additional mutations may add some aspect of stability or a functional advantage to the rRNA that is lacking with a sequence of AUA. The tetracycline-resistant H. pylori strains 108 and 109 grow much faster than did H. pylori 26695; thus, the mutations in the 16S rRNA do not seem to be inhibitory. We predict that bacteria with a residue other than G at position 966 may be intrinsically resistant to tetracycline.
The exact mechanism by which the h31 loop mutations mediate tetracycline resistance was not determined in this study. As mentioned, both 30S ribosome crystal structures include four residues of the h31 loop, 964 to 967, in the primary or Tet-1 binding site (4, 23). In these structures, however, it is primarily the phosphodiester backbones of h34 and the h31 loop that interact with tetracycline. There is a hydrogen bond between tetracycline and the O2P of G966 (4) that may be lost by a nucleotide substitution at this position. The 30S ribosomal subunits in the crystal structures were purified from Thermus thermophilus and have the sequence AGC at positions 965 to 967 of the 16S rRNA. Both structures identified more than one tetracycline binding site, two in one structure (4) and six in the other (23). Only the primary or Tet-1 site was fully occupied, and it is the most probable site at which tetracycline inhibits protein synthesis. The most likely explanation for the tetracycline resistance conferred by this 16S rRNA mutation is the loss or weakening of tetracycline binding due to alteration of the functional binding site. A decrease in the ribosome's affinity for tetracycline may increase the ability of the tRNA to compete for binding. The residue identified in the tetracycline-resistant P. acnes mutants is directly adjacent to the primary binding site (4, 23, 25), but this mutation did not mediate tetracycline resistance in H. pylori. The effects of this mutation were quite varied in P. acnes, with MICs of tetracycline ranging from 2 to 64 μg/ml. This residue may be too far from the tetracycline binding site to significantly interfere with binding.
Tetracycline-resistant H. pylori isolates are also resistant to metronidazole (14, 18). Metronidazole resistance in the overwhelming majority of H. pylori strains is due to mutations in the rdxA gene, which codes for an NADPH nitroreductase (11), and may involve mutations of an NADPH flavin oxidoreductase, FrxA (13). Strains 108 and 109 have a unique deletion of amino acid Y141 in the RdxA protein that is likely the cause of metronidazole resistance. In a previous study transformation of susceptible strains with genomic DNA generated only isolates with both metronidazole and tetracycline resistance (14). Analysis of the H. pylori genome sequences (1, 34) indicates that the smallest distances between rdxA and one of the 16S rRNA genes are 193 kb in strain 26695 and 205 kb in strain J99. The frxA gene and the 16S rRNA genes are even further apart, at minimum distances of 519 and 537 kb in 26695 and J99, respectively. The metronidazole resistance determinants were not identified in the study of tetracycline and metronidazole resistance by Kwon et al. (14), but it is possible that the distances between rdxA and the 16S rRNA genes may not be as great in these isolates, as H. pylori is noted for its genetic diversity (2). In this study we have separated these two phenotypes. Transfer of the tetracycline resistance phenotype by transformation with the isolated 16S rRNA gene from a resistant organism demonstrates that this trait is not physically linked to metronidazole resistance and that it is due to mutations in the 16S rRNA gene.
Mutation of the residues in h31 did not appear to affect the growth rate of H. pylori, but replacement of one of the 16S rRNA genes with the cat gene did. Serial passage on increasing concentrations of tetracycline did not select any high-level-tetracycline-resistant organisms. After several passages, strains for which MICs of tetracycline displayed a fourfold increase were isolated. The 16S rRNA gene deletion strain did not demonstrate any increased frequency of mutation to tetracycline resistance. The finding that all characterized tetracycline-resistant H. pylori strains are also metronidazole resistant (14, 18) suggests that there may be a progressive acquisition of resistance where metronidazole resistance may be required before tetracycline resistance can develop. Metronidazole in H. pylori is converted to an active, mutagenic form (28) that could potentially increase mutations in the 16S rRNA genes. However, although we could increase the MIC of tetracycline for a metronidazole-resistant mutant to 8 μg/ml, we did not isolate any resistant mutants comparable to strains 108 and 109. The strains with increased resistance to tetracycline all had a deletion of G942. This residue is located at the Tet-4 binding site identified in one of the 30S structures (23) and is deleted in strain 109. Additionally, G942 is located near nucleotides that make contacts with P- and E-site tRNAs (40), as seen in Fig. 2A. The occupancy of the Tet-4 site by tetracycline in the 30S subunit-tetracycline crystal structure is only half of that of the Tet-1 site (23). This site may contribute to tetracycline inhibition, but it is not the major, high-affinity site of tetracycline action. No mutations in h31 or h34 were identified by this method. No spontaneous tetracycline resistance mutations were ever isolated in these studies. Their rare emergence in a clinical setting may be due to the circumstances of treatment and the nature of the gastric environment.
Tetracycline together with metronidazole and bismuth is often used as a low-cost treatment or a rescue therapy (with a proton pump inhibitor) to resolve H. pylori infections (17, 27), and thus tetracycline resistance in this organism is increasingly a concern (12, 39). In this study, we have identified mutations in h31 of the 16S rRNA of clinical isolates of H. pylori that when transferred to susceptible strains mediated tetracycline resistance. These mutations are located at the primary tetracycline binding site and are likely to alter the affinity of the ribosome for this antibiotic. The H. pylori strains in this study did not acquire the h31 mutations from serial passage on tetracycline, perhaps reflecting the rarity of the emergence of tetracycline-resistant H. pylori in clinical settings. However, once exposed to DNA containing the mutations they easily took it up and recombined it into the chromosome, thus acquiring resistance. The 16S rRNA triple mutation was not deleterious to the organism's growth and was stably maintained. Tetracycline resistance in H. pylori is due to ribosomal mutations and can be spread through natural transformation events.
Acknowledgments
We thank Peter Midolo for providing the H. pylori strains. We also thank Trevor Wilson and Ping He for carrying out the manual sequencing and Monika Keelan for assistance with the metronidazole MIC determinations.
This work was supported by a Fellowship to C.A.T. from the Canadian Association of Gastroenterology, Abbott Laboratories, and the Canadian Institutes of Health Research. D.E.T. is an Alberta Heritage Foundation for Medical Research Scientist. We also thank the Natural Sciences and Engineering Research Council for support.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Blaser, M. J., and D. E. Berg. 2001. Helicobacter pylori genetic diversity and risk of human disease. J. Clin. Investig. 107:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser, M. J., and J. Parsonnet. 1994. Parasitism by the “slow” bacterium Helicobacter pylori leads to altered gastric homeostasis and neoplasia. J. Clin. Investig. 94:4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 5.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drazek, E. S., A. Dubois, and R. K. Holmes. 1994. Characterization and presumptive identification of Helicobacter pylori isolates from rhesus monkeys. J. Clin. Microbiol. 32:1799-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckloff, B. W., R. P. Podzorski, B. C. Kline, and F. R. Cockerill. 1994. A comparison of 16S ribosomal DNA sequences from five isolates of Helicobacter pylori. Int. J. Syst. Bacteriol. 44:320-323. [DOI] [PubMed] [Google Scholar]
- 8.Ge, Z., and D. E. Taylor. 1997. H. pylori DNA transformation by natural transformation and electroporation, p. 145-152. In C. L. Clayton and H. L. T. Mobley (ed.), Methods in molecular medicine: Helicobacter pylori protocols. Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 9.Handt, L. K., J. G. Fox, F. E. Dewhirst, G. J. Fraser, B. J. Paster, L. L. Yan, H. Rozmiarek, R. Rufo, and I. H. Stalis. 1994. Helicobacter pylori isolated from the domestic cat: public health implications. Infect. Immun. 62:2367-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 11.Jeong, J. Y., A. K. Mukhopadhyay, J. K. Akada, D. Dailidiene, P. Hoffman, and D. E. Berg. 2001. Roles of FrxA and RdxA nitroreductases of Helicobacter pylori in susceptibility and resistance to metronidazole. J. Bacteriol. 183:5155-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, J. J., R. Reddy, M. Lee, J. G. Kim, F. A. El-Zaatari, M. S. Osato, D. Y. Graham, and D. H. Kwon. 2001. Analysis of metronidazole, clarithromycin and tetracycline resistance of Helicobacter pylori isolates from Korea. J. Antimicrob. Chemother. 47:459-461. [DOI] [PubMed] [Google Scholar]
- 13.Kwon, D. H., K. Hulten, M. Kato, J. J. Kim, M. Lee, F. A. El-Zaatari, M. S. Osato, and D. Y. Graham. 2001. DNA sequence analysis of rdxA and frxA from 12 pairs of metronidazole-sensitive and -resistant clinical Helicobacter pylori isolates. Antimicrob. Agents Chemother. 45:2609-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon, D. H., J. J. Kim, M. Lee, Y. Yamaoka, M. Kato, M. S. Osato, F. A. K. El-Zaatari, and D. Y. Graham. 2000. Isolation and characterization of tetracycline-resistant clinical isolates of Helicobacter pylori. Antimicrob. Agents Chemother. 44:3203-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, X.-Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1:1311-1315. [DOI] [PubMed] [Google Scholar]
- 17.Mégraud, F., and B. J. Marshall. 2000. How to treat Helicobacter pylori. First-line, second-line, and future therapies. Gastroenterol. Clin. N. Am. 29:759-773. [DOI] [PubMed] [Google Scholar]
- 18.Midolo, P. D., M. G. Korman, J. D. Turnidge, and J. R. Lambert. 1996. Helicobacter pylori resistance to tetracycline. Lancet 347:1194-1195. [PubMed] [Google Scholar]
- 19.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 20.Nikaido, H. 1996. Multidrug efflux pumps in gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noah, J. W., M. A. Dolan, P. Babin, and P. Wollenzien. 1999. Effects of tetracycline and spectinomycin on the tertiary structure of ribosomal RNA in the Escherichia coli 30S ribosomal subunit. J. Biol. Chem. 274:16576-16581. [DOI] [PubMed] [Google Scholar]
- 22.Oehler, R., N. Polacek, G. Steiner, and A. Barta. 1997. Interaction of tetracycline with RNA: photoincorporation into ribosomal RNA of Escherichia coli. Nucleic Acids Res. 25:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pioletti, M., F. Schlünzen, J. Harms, R. Zarivach, M. Glühmann, H. Avila, A. Bashan, H. Bartels, T. Auerbach, C. Jacobi, T. Hartsch, A. Yonath, and F. Franceschi. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20:1829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reese, R. E., and R. F. Betts. 1993. Handbook of antibiotics. Little, Brown and Company, Boston, Mass.
- 25.Ross, J. I., E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob. Agents Chemother. 42:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Shiotani, A., Z. Z. Nurgalieva, Y. Yamaoka, and D. Y. Graham. 2000. Helicobacter pylori. Med. Clin. N. Am. 84:1125-1136. [DOI] [PubMed] [Google Scholar]
- 28.Sisson, G., J.-Y. Jeong, A. Goodwin, L. Bryden, N. Rossler, S. Lim-Morrison, A. Raudonikiene, D. E. Berg, and P. S. Hoffman. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori rdxA+ (nitroreductase) gene. J. Bacteriol. 182:5091-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloan, J., L. M. McMurry, D. Lyras, S. B. Levy, and J. I. Rood. 1994. The Clostridium perfringens Tet P determinant comprises two overlapping genes: tetA(P), which mediates active tetracycline efflux, and tetB(P), which is related to the ribosomal protection family of tetracycline-resistance determinants. Mol. Microbiol. 11:403-415. [DOI] [PubMed] [Google Scholar]
- 30.Solca, N. M., M. V. Bernasconi, and J. C. Piffaretti. 2000. Mechanism of metronidazole resistance in Helicobacter pylori: comparison of the rdxA gene sequences in 30 strains. Antimicrob. Agents Chemother. 44:2207-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speer, B. S., L. Bedzyk, and A. A. Salyers. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J. Bacteriol. 173:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, D. E., Z. Ge, D. Purych, T. Lo, and K. Hiratsuka. 1997. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob. Agents Chemother. 41:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, D. E., Q. Jiang, and R. N. Fedorak. 1998. Antibiotic susceptibilities of Helicobacter pylori strains isolated in the Province of Alberta. Can. J. Gastroenterol. 12:295-298. [DOI] [PubMed] [Google Scholar]
- 34.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 35.Trieber, C. A., N. Burkhardt, K. H. Nierhaus, and D. E. Taylor. 1998. Ribosomal protection from tetracycline mediated by Tet(O): Tet(O) interaction with ribosomes is GTP-dependent. Biol. Chem. 379:847-855. [DOI] [PubMed] [Google Scholar]
- 36.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 37.Wang, G., T. J. Wilson, Q. Jiang, and D. E. Taylor. 2001. Spontaneous mutations that confer antibiotic resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 45:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 39.Wu, H., X. D. Shi, H. T. Wang, and J. X. Liu. 2000. Resistance of Helicobacter pylori to metronidazole, tetracycline and amoxycillin. J. Antimicrob. Chemother. 46:121-123. [DOI] [PubMed] [Google Scholar]
- 40.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. D. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]