Abstract
Flavobacterium johnsoniae moves rapidly over surfaces by a process known as gliding motility. The mechanism of this form of motility is not known. Four genes that are required for F. johnsoniae gliding motility, gldA, gldB, gldD, and ftsX, have recently been described. GldA is similar to the ATP-hydrolyzing components of ATP binding cassette (ABC) transporters. Tn4351 mutagenesis was used to identify two additional genes, gldF and gldG, that are required for cell movement. gldF and gldG appear to constitute an operon, and a Tn4351 insertion in gldF was polar on gldG. pMK314, which carries the entire gldFG region, restored motility to each of the gldF and gldG mutants. pMK321, which expresses GldG but not GldF, restored motility to each of the gldG mutants but did not complement the gldF mutant. GldF has six putative membrane-spanning segments and is similar in sequence to channel-forming components of ABC transporters. GldG is similar to putative accessory proteins of ABC transporters. It has two apparent membrane-spanning helices, one near the amino terminus and one near the carboxy terminus, and a large intervening loop that is predicted to reside in the periplasm. GldF and GldG are involved in membrane localization of GldA, suggesting that GldA, GldF, and GldG may interact to form a transporter. F. johnsoniae gldA is not closely linked to gldFG, but the gldA, gldF, and gldG homologs of the distantly related gliding bacterium Cytophaga hutchinsonii are arranged in what appears to be an operon. The exact roles of F. johnsoniae GldA, GldF, and GldG in gliding are not known. Sequence similarities of GldA to components of other ABC transporters suggest that the Gld transporter may be involved in export of some material to the periplasm, outer membrane, or beyond.
Gliding motility, the movement of cells over surfaces without the aid of flagella, is a common form of movement exhibited by diverse bacteria. Bacterial gliding motility was first observed nearly 200 years ago (54). Since then a number of mechanisms have been proposed to explain this form of cell propulsion (6, 28, 35, 50). Recently it has become clear that different types of “motors” are used by different groups of gliding bacteria (28). Mycoplasma gliding is thought to rely on the cell cytoskeleton (53), whereas gliding of filamentous cyanobacteria may be powered by polysaccharide secretion (18). Myxococcus xanthus has two independent motility systems. “S” or social gliding motility requires type IV pili and is probably related to the twitching motility of Pseudomonas aeruginosa and other bacteria (23, 45, 55, 58). Extension and retraction of pili appear to be responsible for this type of cell movement (30, 52). The other form of motility employed by M. xanthus is “A” or adventurous gliding motility (16, 50). The mechanism of M. xanthus A motility is not known, but it does not appear to involve pili (50).
Numerous bacteria which belong to the Cytophaga-Flavobacterium-Bacteroides (CFB) branch of the eubacterial phylogenetic tree exhibit rapid gliding motility (4, 42). This form of gliding does not appear to involve pili (15, 42). Lapidus and Berg proposed that cells of Cytophaga U67 have a system of tracks anchored to the peptidoglycan. In their model, outer membrane components are driven along these tracks by periplasmic and cell membrane proteins that obtain energy from the proton motive force (PMF) (27). Other models to explain gliding of members of the CFB group include contraction or expansion of fibrils in the periplasm or cytoplasm (5), the functioning of rotary motors (36), the generation of waves in the outer membrane (8), or the movement of “conveyor belts” made of polysaccharide or protein along the cell surface (28).
Flavobacterium johnsoniae (formerly Cytophaga johnsonae) (4) is a common gliding bacterium that belongs to the CFB group. Cells of F. johnsoniae glide at rates of 2 to 10 μm/s (35). Cells also occasionally attach to a surface by one pole and rotate in place at frequencies of about 2 revolutions per s. Techniques to genetically manipulate F. johnsoniae were recently developed (29) and have been used to identify four genes, gldA, gldB, gldD, and ftsX, that are required for gliding motility (2, 21, 22, 24).
GldA exhibits sequence similarity to ATP binding cassette (ABC) transport proteins (2). ABC transporters are common in bacteria, archaea, and eukarya. They utilize ATP hydrolysis as a source of energy to transport substances across one or more membranes into or out of cells (9, 19, 60). Molecules that are transported by different ABC transporters include amino acids, peptides, proteins, polysaccharides, lipids, inorganic ions, and various drugs. Bacterial ABC transporters often consist of several subunits. GldA is similar to the ATP-hydrolyzing subunits. It does not have any obvious hydrophobic regions and probably requires the presence of integral membrane proteins to form a functional transporter. The genes that code for these membrane proteins do not appear to be closely linked to gldA (2). In this report, we describe the identification and characterization of two genes, gldF and gldG, that are required for F. johnsoniae gliding motility. GldF and GldG are membrane proteins that appear to interact with GldA to form an ABC transporter that is required for gliding.
MATERIALS AND METHODS
Bacterial and bacteriophage strains, plasmids, and growth conditions.
F. johnsoniae UW101 (ATCC 17061) was the wild-type strain used in these studies, and all mutants were derived from this strain. The 50 nonmotile mutants of F. johnsoniae (obtained from J. Pate) were previously described (7, 22, 57). The bacteriophages active against F. johnsoniae that were used in this study were φCj1, φCj7, φCj13, φCj23, φCj29, φCj42, φCj48, and φCj54 (7, 38, 57). The Escherichia coli strains used were DH5αMCR (Gibco BRL Life Technologies), S17-1 (46), and BW19851 (31). E. coli strains were grown in Luria-Bertani medium at 37°C, and F. johnsoniae strains were grown in Casitone-yeast extract medium at 30°C, as previously described (29). To observe colony spreading, F. johnsoniae was grown on PY2 agar medium (2) at 25°C. Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml; cefoxitin, 100 μg/ml; chloramphenicol, 30 μg/ml; erythromycin, 100 μg/ml; and tetracycline, 20 μg/ml. Plasmids and primers used in this study are listed in Table 1.
TABLE 1.
Plasmids and primers used in this studya
| Plasmid or primer | Description and primer sequence | Source or reference |
|---|---|---|
| Plasmids | ||
| pBC SK(+) | ColE1; Cmr | Stratagene |
| pCR-Script Cam SK(+) | ColE1 ori; Cmr | Stratagene |
| pSTBlue-1 | ColE1 ori; Apr | Novagen |
| pT7Blue | ColE1 ori; Apr | Novagen |
| pSPORT1 | ColE1 ori; Apr | Gibco BRL |
| pMAL-c2 | MalE fusion protein expression vector; Apr | New England Biolabs |
| pCP29 | E. coli-F. johnsoniae shuttle plasmid; Apr(Cfr Emr) | 24 |
| pSA21 | gldA in E. coli-F. johnsoniae shuttle plasmid pCP23; Apr(Tcr) | 2 |
| pMK312 | 3.1-kb gldFG PCR product cloned into the SrfI site of pCR-Script Cam SK(+); Cmr | This study |
| pMK313 | 3.1-kb gldFG fragment of pMK312 cloned as a NotI-SalI fragment into pSPORT1; Apr | This study |
| pMK314 | 3.1-kb gldFG fragment of pMK313 cloned as a KpnI-SphI fragment into pCP29; Apr(Cfr Emr) | This study |
| pMK315 | 1.3-kb PstI fragment of pMK312 containing gldF cloned into pCP29; Apr(Cfr Emr) | This study |
| pMK316 | 1.3-kb PstI fragment of pMK312 containing gldF cloned into pCP29 (opposite orientation of pMK315); Apr(Cfr Emr) | This study |
| pMK317 | 2.5-kb EagI fragment of pMK312 containing gldG, cloned into pBC SK(+); Cmr | This study |
| pMK321 | 2.6-kb fragment spanning gldFG, with an in-frame deletion of 540 bp in gldF, cloned into pCP29; Apr(Cfr Emr) | This study |
| pMM274 | 2.5-kb fragment of pMK317 containing gldG cloned into pCP29; Apr(Cfr Emr) | This study |
| pDH254 | 1.3-kb fragment containing 3′ end of gldG in EcoRV site of pSTBlue-1; Apr | This study |
| pDH289 | gldG expression plasmid, 1.3-kb HindIII-PstI fragment of pDH254 in pMAL-c2; Apr | This study |
| pDH264 | 923-bp fragment containing gldA in EcoRV site of pT7Blue; Apr | This study |
| pDH265 | gldA expression plasmid, 905-bp EcoRI-XbaI fragment of pDH264 in pMAL-c2; Apr | This study |
| Primers | ||
| 340 | 5′ GACTTGGATACCTCACGCC 3′; primer in IS 4351 | |
| 341 | 5′ TTGGAAATTTTCTGGGAGG 3′; primer in ermF of Tn 4351 | |
| 396 | 5′ AAAATACGAAATGCAGACGG 3′; primer in fjo15 | |
| 405 | 5′ AGTTTCAAGGTCTGAAGCCG 3′; primer in dnaN | |
| 410 | 5′ AGACGTTTACTGCAGGAAACCAAACAA 3′; primer for amplification of 3′ end of gldG | |
| 411 | 5′ TATTAACATAAGCTTACTGTACTTTCT 3′; primer for amplification of 3′ end of gldG | |
| 430 | 5′ ACCTGTATTCAATATATTGTAGTCTCC 3′; primer for construction of in-frame deletion of gldF | |
| 431 | 5′ GGCGTTATTGATACCCGC 3′; primer for construction of in-frame deletion of gldF | |
| 444 | 5′ AAATAAAAAGAATTCTCGATAGAAGTA 3′; primer for amplification of gldA | |
| 445 | 5′ AACTGAGGCTCTAGATTATTTAGTAAT 3′; primer for amplification of gldA | |
| 491 | 5′ CCGCGTTGTGCAATCAACC 3′; primer in C. hutchinsonii gldG | |
| 493 | 5′ TTGTGTCTATACATCTGCCGG 3′; primer in C. hutchinsonii gldF |
Antibiotic resistance phenotypes: Apr, ampicillin; Cfr, cefoxitin; Cmr, chloramphenicol; Emr, erythromycin; and Tcr, tetracycline. Unless indicated otherwise, antibiotic resistance phenotypes are those expressed in E. coli. Antibiotic resistance phenotypes listed in parentheses are those expressed in F. johnsoniae but not in E. coli.
Tn4351 mutagenesis of F. johnsoniae and identification of sites of insertion in gldF and gldG.
Tn4351 was introduced into wild-type F. johnsoniae, and nonmotile mutants were isolated as described previously (22). The sites of transposon insertion in the nonmotile mutants CJ776, CJ785, CJ787, and CJ788 were determined by cloning the Tn4351-disrupted genes essentially as previously described (24) and by inverse PCR (33). For inverse PCR, chromosomal DNA was digested with HindIII and ligated, resulting in the formation of closed circles. Tn4351-specific primers 340 and 341 were used to amplify the DNA adjacent to the site of the insertion using Taq polymerase.
Nucleic acid sequencing.
Nucleic acid sequencing was performed by the dideoxy nucleotide procedure using an automated (Applied Biosystems) sequencing system. Sequences were analyzed with the MacVector and AssemblyLign software (Oxford Molecular Group Inc., Campbell, Calif.), and comparisons to database sequences were made using the BLAST (3) and FASTA (40) algorithms. Predictions regarding cellular localization and topology of membrane proteins were made using PSORT (http://PSORT.NIBB.AC.JP [32]) and TMpredict (17).
Amplification and analysis of the Cytophaga hutchinsonii gldF and gldG genes.
Preliminary sequence data for C. hutchinsonii were obtained from The DOE Joint Genome Institute at http://jgi.doe.gov/. The region spanning the end of gldF and the beginning of gldG was amplified with primers 491 and 493 to allow completion of the sequence.
Construction of plasmids for complementation of gldF and gldG mutants.
A 3.1-kb region of F. johnsoniae DNA which spans gldFG was amplified by PCR using Elongase (Gibco BRL Life Technologies) and primers 396 and 405. This PCR product was cloned into the SrfI site of pCR-Script Cam SK(+) to generate pMK312 and was transferred to pSPORT1 as a NotI-SalI fragment to form pMK313. pMK313 was digested with KpnI and SphI, and the fragment containing gldFG was introduced into pCP29 to generate pMK314 (Fig. 1). pMK315, which carries just gldF, was constructed by cloning the 1.3-kb PstI fragment from pMK312 into pCP29 (Fig. 1). pMK316 is identical to pMK315, except that the gldF fragment is inserted in pCP29 in the opposite orientation. pMK317, which contains gldG but not gldF, was constructed by transferring the 2.5-kb EagI fragment from pMK312 into pBC SK(+). The 2.5-kb fragment of pMK317 was cloned as a NotI-BamHI fragment into pSPORT1 and was then transferred as a KpnI-SphI fragment into pCP29 to generate pMM274 (Fig. 1).
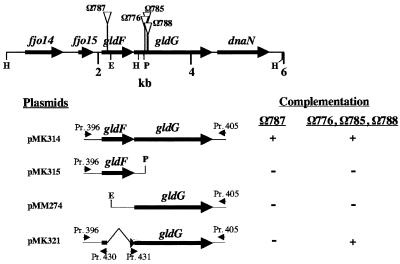
FIG. 1.
Map of the gldFG region of F. johnsoniae. The sites of the Tn4351 insertions in CJ776, CJ785, CJ787, and CJ788 are indicated by triangles. Complementation of gldF and gldG mutants of F. johnsoniae by fragments cloned into pCP29 is indicated beneath the map. Primers (Pr.) used to amplify fragments for plasmid construction are shown. Restriction sites: E, EagI; H, HindIII; and P, PstI.
pMK321, which contains a 540-bp in-frame deletion within the 723-bp coding region of gldF (Fig. 1), was constructed to allow production of GldG in the absence of functional GldF. pMK313 was digested with PvuII to release a 3.1-kb F. johnsoniae fragment containing gldF and gldG flanked by vector sequences. The gldFG fragment was treated with T4 DNA ligase to form closed circles, and the ends of gldF and the entire gldG gene were amplified using primers 430 and 431 and Vent polymerase. The resulting PCR product was incubated with T4 DNA ligase to form closed circles, was digested with KpnI and SphI, which cut in the polycloning sites flanking gldFG, and was introduced into pCP29 to generate pMK321.
For complementation analyses, plasmids were introduced into the F. johnsoniae mutants by conjugation or electroporation as previously described (22, 29).
Microscopic observations of cell movement.
Wild-type and mutant cells of F. johnsoniae were examined for movement over glass and agar surfaces and for their ability to propel polystyrene latex spheres by phase-contrast microscopy as previously described (21, 22).
Measurements of phage sensitivity.
Sensitivity to F. johnsoniae phage was determined essentially as previously described by spotting 2.5 to 10 μl of phage lysates (2 × 107 PFU/ml) onto lawns of cells in overlay agar (22). The plate contents were incubated for 24 h at 25°C to observe lysis.
Protein expression and antibody production.
A 923-bp fragment coding for GldA was amplified by PCR using Vent polymerase (New England Biolabs) with primers 444 and 445. The amplified fragment was cloned into pT7Blue (Novagen) to produce pDH264. The gldA fragment was isolated as an EcoRI-XbaI fragment and ligated into pMAL-c2 to produce pDH265. A fragment which codes for the C-terminal 439 amino acids of GldG was amplified as described above, using primers 410 and 411, and cloned into pSTBlue-1 (Novagen) to generate pDH254. The gldG fragment was isolated from pDH254 as a HindIII-PstI fragment and was ligated into pMAL-c2 to produce pDH289. Both fusion constructs were verified by DNA sequencing. Expression of fusion proteins was induced in E. coli DH5αMCR by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside. Fusion proteins were purified by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and antibodies were generated as previously described (22). Anti-GldA antiserum was affinity purified essentially as previously described (22). Anti-GldG antiserum was purified by selective removal of antibodies that reacted with antigens in extracts of the gldG mutant CJ776. Cells of CJ776 were disrupted with a French press, and the cell extract was boiled for 5 min. The extract was applied to a polyvinyl difluoride membrane and was incubated for 16 h at 4°C. The membrane was blocked with 5% skim milk in Tris-buffered saline (20 mM Tris hydrochloride [pH 7.5] and 0.15 M NaCl) for 4 h and washed with Tris-buffered saline, and crude antiserum was added and incubated for 16 h at 4°C.
Cell fractionation and Western blot analyses.
F. johnsoniae cells were disrupted with a French press and fractionated into soluble and membrane fractions as described (22) except that 0.6 mM phenylmethylsulfonyl fluoride was added to cell extracts. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blot analyses were performed as previously described (22), except that GldG was separated on gels containing 10% acrylamide, and antigens were detected using goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate and Opti-4CN (Bio-Rad, Hercules, Calif.).
Genetic nomenclature.
Genes required for gliding motility were given the name gld followed by a letter. Open reading frames (ORFs) that exhibited strong sequence similarity to genes of known function were named after the corresponding genes. F. johnsoniae ORFs of unknown function that did not exhibit strong similarity to previously described genes were given the provisional name fjo (for F. johnsoniae ORF) followed by a number. C. hutchinsonii ORFs of unknown function that did not exhibit similarity to previously described genes were named cho followed by a number.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the GenBank database under accession no. AY036615.
RESULTS
Tn4351 mutagenesis of F. johnsoniae.
F. johnsoniae was mutagenized with Tn4351, and 29 nonmotile mutants were isolated. Eleven of these had defects in cell division and were not considered further in this study. Complementation analysis revealed that five of the remaining mutants had Tn4351 insertions in previously identified gliding motility genes (one in gldA and four in gldB). Southern blot analyses revealed that five other mutants had multiple Tn4351 insertions. The remaining eight mutants were analyzed further.
Identification of gldF and gldG.
To identify the regions disrupted by Tn4351 in each of the mutants, the DNA flanking the sites of insertion was cloned or amplified as described in Materials and Methods. Sequence analysis revealed that four of the eight nonmotile mutants contained insertions in a 1-kb region of the genome. A 5.9-kb fragment of DNA spanning this region was sequenced. Analysis of this sequence identified five ORFs (Fig. 1). CJ787 contained a Tn4351 insertion in gldF, and CJ776, CJ785, and CJ788 contained insertions in gldG. The stop codon of gldF and the start codon of gldG overlap by 1 nucleotide, which suggests that gldF and gldG are organized as an operon.
The predicted protein product of gldF is 241 amino acids long and has a molecular weight of 27 kDa. Hydrophilicity analyses suggest that GldF has six transmembrane domains. GldF exhibits sequence similarity to a hypothetical protein that we refer to as C. hutchinsonii GldF (46% identity over 241 amino acids). F. johnsoniae GldF also exhibits sequence similarity (27 to 36% amino acid identity) to several transmembrane channel- forming proteins of ABC transporters, including Bradyrhizobium japonicum NosY (27% identity over 208 amino acids) (GenBank accession no. AJ002531) and Sinorhizobium meliloti NosY (30% identity over 153 amino acids) (20).
GldG is 561 amino acids long and has a predicted molecular weight of 63.7 kDa. Hydrophilicity analyses suggest that GldG has two transmembrane regions, one at the N terminus and one at the C terminus. The large hydrophilic region between these transmembrane domains is predicted to reside in the periplasm based on sequence analysis. GldG exhibits similarity to a protein that we refer to as C. hutchinsonii GldG (32% amino acid identity over 549 residues) and to several putative membrane-anchored accessory proteins of ABC transporters, such as Borrelia burgdorferi BB0752 (23% identity over 561 amino acids) (10) and P. aeruginosa PA3670 (23% identity over 259 amino acids) (51).
fjo15 lies upstream of gldF. The stop codon of fjo15 and the start codon of gldF are separated by 179 nucleotides. The predicted protein product of fjo15 does not exhibit amino acid sequence similarity to any proteins of known function in the databases. An 18-nucleotide inverted repeat starts 3 nucleotides after the predicted fjo15 stop codon (5′ GTTTCAAGTTTCAGGTTT…//107 nucleotides//…AAACCTGAAACTTGAAAC 3′). In the 107 nucleotides between the inverted repeats, there are two copies of a 9-nucleotide direct repeat (5′ CGCAAAGAA 3′). The functions of the direct and inverted repeats are not known. The transcriptional start site for gldF is likely to reside in the region between fjo15 and gldF. The region of DNA between the inverted repeat and the gldF start codon is AT rich (83% AT over 33 nucleotides), as is often observed in promoter regions of other bacteria (44). fjo14 lies upstream of fjo15. The predicted product of fjo14 is not similar to any proteins of known function in the databases. F. johnsoniae dnaN is predicted to start 107 nucleotides downstream of the gldG stop codon. F. johnsoniae DnaN is 25% identical to E. coli DnaN (34) and 27% identical to Vibrio cholerae DnaN (14). DnaN is the β subunit of bacterial DNA polymerase III. There is no evidence indicating that fjo14, fjo15, or dnaN is involved in gliding motility.
Complementation of Tn4351-induced gldF and gldG mutants of F. johnsoniae.
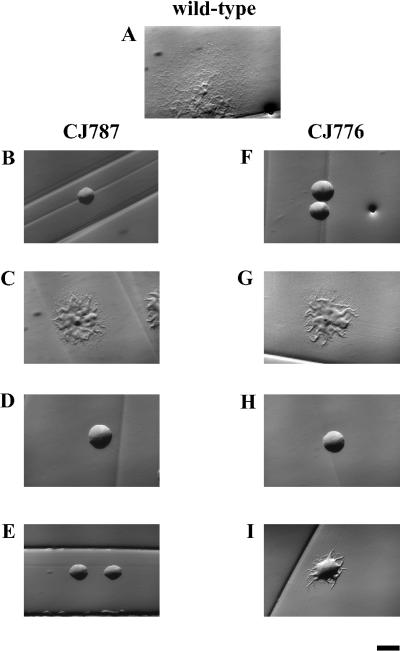
Introduction of pMK314, which carries gldF and gldG, into the gldF mutant CJ787 and into the gldG mutants CJ776, CJ785, and CJ788 resulted in complementation of each (Fig. 1 and 2C and G). The resulting colonies spread over agar, and cells exhibited rapid gliding motility. Plasmids containing just gldF (pMK315 and pMK316) failed to complement any of the mutants (Fig. 1 and 2D). Similarly, pMM274, which contains gldG and 527 bp of upstream DNA but lacks the promoter and translational start site of gldF, failed to complement any of the mutants (Fig. 1 and 2H). pMK321, which contains the putative gldF promoter region and gldG but has an in-frame deletion within gldF, was constructed to allow production of GldG in the absence of functional GldF. Introduction of pMK321 restored motility to each of the Tn4351-induced gldG mutants but failed to complement the gldF mutant CJ787 (Fig. 1 and 2E and I). pMK321 is a derivative of pCP1 and has a copy number of approximately 10 in F. johnsoniae. gldG mutants complemented with pMK321 did not spread as well as wild-type colonies. This may be due to an imbalance in the ratio of GldF to GldG in these strains. It is also possible that GldF and GldG have difficulty assembling to form a functional complex unless they are transcribed and translated together. The results of the complementation analyses described above indicate that GldF and GldG are both needed for gliding motility. They also support the idea that gldF and gldG are transcriptionally coupled, since the transposon insertion in gldF was polar on gldG.
FIG. 2.
Photomicrographs of F. johnsoniae colonies. Colonies were grown for 2 days at 25°C on PY2 agar media. Photomicrographs were taken with a Diagnostic Instruments RT color digital camera mounted on a Nikon Diaphot inverted phase-contrast microscope. Bar indicates 1 mm. (A) Wild-type F. johnsoniae UW101. (B) gldF mutant CJ787. (C) CJ787 complemented with pMK314, which carries gldF and gldG. (D) CJ787 with pMK315, which carries gldF. (E) CJ787 carrying pMK321, which expresses gldG but not gldF. (F) gldG mutant CJ776. (G) CJ776 complemented with pMK314, which carries gldF and gldG. (H) CJ776 with pMM274, which carries gldG but lacks the transcriptional start site of gldF. (I) CJ776 carrying pMK321, which expresses gldG but not gldF.
Identification of gldF and gldG point mutants.
Pate and colleagues isolated a large number of spontaneous and chemically induced nonmotile mutants (7, 57). We introduced pMK314 into 50 of these mutants to determine if any has defects in gldF or gldG. pMK314 restored motility to 5 of the 50 mutants (UW102-25, UW102-34, UW102-39, UW102-75, and UW102-77). Colonies of the complemented mutants spread over agar, and cells exhibited wild-type motility in wet mounts. pMK321 (which expresses gldG but not gldF) restored motility to UW102-25, UW102-34, and UW102-75 but failed to complement the other two mutants. pMK315 (which express gldF but not gldG) failed to complement any of the mutants. These results suggest that UW102-25, UW102-34, and UW102-75 have mutations in gldG and that UW102-39 and UW102-77 have mutations in gldF that are polar on gldG. The exact sites of the mutations in each mutant were determined by amplification and sequencing of the gldFG region. Each mutation was the result of insertion or deletion of 1 or 2 bases. As expected, UW102-39 and UW102-77 had mutations in gldF, and UW102-25, UW102-34, and UW102-75 had mutations in gldG. UW102-39 had a T inserted at position 91 (numbered from the A of the gldF start codon), while UW102-77 had a deletion of nucleotides 17 and 18 (TA). UW102-25 had a G deleted at position 951 (numbered from the A of the gldG start codon), and UW102-34 and UW102-75 each had a single base deletion within a run of A's starting at position 162.
Expression and localization of GldG.
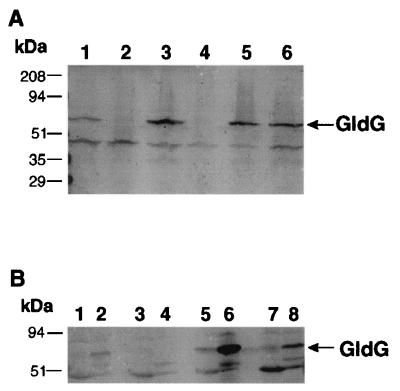
Antisera were used to detect GldG in extracts of wild-type and mutant cells by Western blot analyses (Fig. 3A). GldG was detected in wild-type cells and migrated with an apparent molecular weight of approximately 62 kDa (Fig. 3A, lane 1) but was absent from the gldG mutant CJ776 (Fig. 3A, lane 2). Introduction of pMK314, which carries gldF and gldG, into CJ776 restored production of GldG (lane 3). Expression of GldG in this strain was higher than seen in wild-type cells, as expected, since pMK314 has a copy number of approximately 10 in F. johnsoniae. The gldF mutant CJ787 failed to produce GldG protein (Fig. 3A, lane 4), indicating that the transposon insertion in gldF was polar on gldG. Strains with frameshift mutations in gldF (UW102-39 and UW102-77) also failed to produce GldG (data not shown). Introduction of pMK314 into CJ787 restored production of GldG protein (Fig. 3A, lane 5). Introduction of pMK321, which carries gldFG with an in-frame deletion within gldF, resulted in production of GldG in the absence of GldF (Fig. 3A, lane 6). GldG was found primarily in membrane fractions of wild-type cells (Fig. 3B, lanes 1 and 2) or of cells which expressed GldG in the absence of GldF (Fig. 3B, lanes 5 and 6) or GldA (Fig. 3B lanes 7 and 8).
FIG. 3.
Expression and localization of GldG. (A) Cell extracts were examined for GldG by Western blot analysis. Lane 1, wild-type cells; lane 2, CJ776 (Tn4351 insertion in gldG); lane 3, CJ776 with pMK314, which carries gldFG; lane 4, CJ787 (Tn4351 insertion in gldF); lane 5, CJ787 with pMK314, which carries gldFG; and lane 6, CJ787 with pMK321, which carries the gldFG region with an in-frame deletion in gldF. (B) Cell fractions were examined for GldG by Western blot analysis. Odd-numbered lanes contain soluble fractions, while even-numbered lanes contain membrane fractions. Lanes 1 and 2, wild-type cells; lanes 3 and 4, CJ787 (Tn4351 insertion in gldF); lanes 5 and 6, CJ787 with pMK321, which carries the gldFG region with an in-frame deletion in gldF; and lanes 7 and 8, CJ101-288 (insertion mutation in gldA). kDa, kilodaltons.
GldF and GldG are involved in membrane localization of GldA.
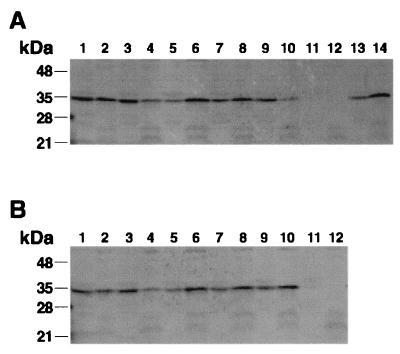
Sequence analyses of GldA did not identify any putative transmembrane domains, but it was suggested that GldA is probably associated with membrane proteins to form an ABC transporter (2). Western blot analyses demonstrated that GldA is found in both the membrane and soluble fractions of wild-type cells (Fig. 4A and B, lanes 1 and 2). In contrast, GldA was found primarily in the soluble fraction of extracts of the gldF mutant CJ787 (Fig. 4A, lanes 3 and 4) or the gldG mutant CJ776 (Fig. 4B, lanes 3 and 4), suggesting that GldA may interact with GldF or GldG. Introduction of pMK314, which expresses gldF and gldG, into cells of the wild type, gldF mutant CJ787, or gldG mutant CJ776 resulted in most of the GldA being sequestered to the membrane fraction (Fig. 4A, lanes 5, 6, 13, and 14, and B, lanes 5 and 6). Introduction of pMK315 (which expresses gldF) to CJ787 or CJ776 resulted in partial localization of GldA to the membrane fraction (Fig. 4A and B, lanes 7 and 8). Introduction of pMK321 (which expresses GldG) restored membrane localization of GldA to the gldG mutant CJ776, (Fig. 4B, lanes 9 and 10) but not to the gldF mutant CJ787 (Fig. 4A, lanes 9 and 10). Apparently GldF and GldG participate either directly or indirectly in localization of GldA to the membrane.
FIG. 4.
Localization of GldA in wild-type cells and in gldF and gldG mutants. Cell fractions were examined for GldA by Western blot analysis. Odd-numbered lanes contain soluble fractions, while even-numbered lanes contain membrane fractions. An equal amount, based on total cell protein measurements prior to fractionation, was loaded in each lane. (A) Lanes 1 and 2, wild-type cells; lanes 3 and 4, CJ787 (Tn4351 insertion in gldF); lanes 5 and 6, CJ787 with pMK314, which carries gldFG; lanes 7 and 8, CJ787 with pMK315, which carries gldF; lanes 9 and 10, CJ787 with pMK321, which carries the gldFG region with an in-frame deletion in gldF; lanes 11 and 12, CJ288 (gldA mutant); and lanes 13 and 14, wild-type cells with pMK314. (B) Lanes 1 and 2, wild-type cells; lanes 3 and 4, CJ776 (Tn4351 insertion in gldG); lanes 5 and 6, CJ776 with pMK314; lanes 7 and 8, CJ776 with pMK315; lanes 9 and 10, CJ776 with pMK321; and lanes 11 and 12, CJ288 (gldA mutant).
Phage resistance of mutants.
It has previously been reported that many nonmotile mutants of F. johnsoniae are resistant to infection by a number of F. johnsoniae bacteriophages (56). The reason for this pleiotropy is not known. We tested the sensitivity of F. johnsoniae strains UW101, CJ787, CJ776, CJ880 (CJ787 carrying pMK314), and CJ878 (CJ776 carrying pMK314) to the F. johnsoniae bacteriophages φCj1, φCj7, φCj13, φCj23, φCj29, φCj42, φCj48, and φCj54. F. johnsoniae UW101 was readily lysed by these phages, whereas CJ787 and CJ776 were resistant to infection by each of them. Introduction of pMK314 into CJ787 and CJ776 restored sensitivity to each of these phages in addition to restoring gliding motility.
gldF and gldG mutants fail to move latex spheres.
Wild-type cells of F. johnsoniae and related bacteria bind latex spheres and propel them along their surfaces (27, 36). We examined the ability of cells of the F. johnsoniae strains UW101, CJ787, CJ776, CJ880 (CJ787 carrying pMK314), and CJ878 (CJ776 carrying pMK314) to bind and propel latex spheres. Wild-type cells propelled the spheres, whereas cells of the gldF or gldG mutants CJ787 and CJ776 did not. Introduction of pMK314 into CJ787 and CJ776 restored the ability to propel spheres, in addition to restoring gliding motility.
C. hutchinsonii gldF and gldG are adjacent to gldA.
As noted above, F. johnsoniae gldF and gldG have homologs in the C. hutchinsonii genome. We used PCR amplification and DNA sequencing to determine the arrangement of the C. hutchinsonii genes. C. hutchinsonii gldF and gldG appear to be arranged in an operon with gldA and one additional gene. The order of the C. hutchinsonii genes is gldA, gldF, gldG, and cho1. The protein product of C. hutchinsonii gldA exhibits 49% identity with F. johnsoniae GldA over 297 amino acids. Cho1, which is predicted to be a periplasmic or outer membrane protein, exhibits no significant similarity to protein sequences in the databases.
DISCUSSION
The mechanism responsible for F. johnsoniae gliding motility is not known. Previously, we identified four genes, gldA, gldB, gldD, and ftsX, that are required for gliding (2, 21, 22, 24). gldA codes for a protein that is similar in sequence to the ATP-hydrolyzing components of ABC transporter complexes. GldB and GldD are anchored in the cytoplasmic membrane. The exact functions of these proteins in gliding motility are not known. FtsX is needed for both cell division and gliding motility. The inability of ftsX mutants to glide may be an indirect effect of the defects in cell division. The results presented in this paper identify two additional genes, gldF and gldG, that are required for gliding. Transposon insertions in gldF or gldG eliminated gliding motility, and introduction of a wild-type copy of gldFG on a plasmid restored motility to the mutants.
gldF and gldG appear to be organized in an operon, and both genes are required for motility. Insertional inactivation of gldF by Tn4351 was polar on gldG. The gldF mutant failed to produce detectable levels of GldG protein, and complementation of gliding required the introduction of both gldF and gldG. Expression of either gene alone did not restore motility. The polarity of the Tn4351 insertion in gldF on gldG was unexpected. Tn4351 has outward-reading promoters at each end (47). These promoters appear to function in F. johnsoniae, since insertion of Tn4351 in either orientation in the first gene of the gldBC operon did not eliminate expression of the downstream gene (22). The polarity of the Tn4351 insertion in gldF may be the result of disruption of translational coupling. The overlapping stop and start codons of gldF and gldG, and lack of an obvious ribosome binding site upstream of gldG are consistent with this explanation.
F. johnsoniae GldF is predicted to be a cytoplasmic membrane protein with six membrane-spanning domains. It exhibits amino acid sequence similarity to proteins that are thought to be cytoplasmic membrane channel-forming components of ABC transporters. F. johnsoniae GldF is most similar to the putative C. hutchinsonii GldF. It also exhibits similarity to S. meliloti NosY, which is involved in maturation of periplasmic nitrous oxide reductase NosZ (20). NosY is thought to form an ABC transporter with NosF (the ATP binding protein of the transporter) and NosD (a periplasmic protein which is thought to interact with NosZ) (20, 61). Mutations in nosD, nosF, or nosY prevent assembly of the Cu center of the reductase. It is not known whether these proteins function in Cu import or export or in some other process required for NosZ maturation.
F. johnsoniae GldG is a cytoplasmic membrane protein that exhibits similarity to proteins that are thought to be accessory components of ABC transporters. The functions of these transporters are not known. GldG has two predicted transmembrane domains, one near its amino terminus and the other near its carboxy terminus, and a large hydrophilic loop which is predicted to reside in the periplasm. Some polysaccharide exporters of the ABC-2 family employ membrane proteins, such as E. coli KpsE, that have similar topology and may perform similar roles (39, 60).
ABC transporters typically have two ATP binding domains located in the cytoplasm. Neither GldF or GldG has an obvious ATP binding domain. GldA, another protein that is required for gliding, is similar to ATP binding components of ABC transporters. GldA was predicted to be a component of an ABC transporter, but the membrane-spanning components of this transporter were not identified. The results reported in this paper suggest that GldF and GldG are the membrane proteins that interact with GldA to form an ABC transporter. F. johnsoniae GldA was found in the membrane and soluble fractions of wild-type cells. In contrast, GldA was found primarily in the soluble fraction in extracts of cells with mutations in gldF or gldG. Introduction of gldFG on a plasmid into the mutants restored membrane localization to GldA. Increased expression of gldF and gldG (approximately 10-fold) resulted in increased localization of GldA to the membrane fraction. Both GldF and GldG were needed to obtain maximal sequestration of GldA in the membrane fraction. This suggests that GldA interacts with GldF and GldG to form an ABC transporter that is required for gliding motility.
The genes encoding components of ABC transporters are usually arranged in operons, but F. johnsoniae gldA is not closely linked to gldFG. However, the homologous genes from C. hutchinsonii appear to be organized as an operon. F. johnsoniae GldA, GldF, and GldG exhibit 49, 46, and 32% amino acid identity, respectively, with their C. hutchinsonii homologs. C. hutchinsonii exhibits rapid gliding motility and is distantly related to F. johnsoniae. Although it has not been demonstrated, it is likely that the C. hutchinsonii genes are involved in gliding motility. C. hutchinsonii gldA, gldF, and gldG may be cotranscribed with one additional gene, cho1. cho1, which lies downstream of gldG, is predicted to encode a periplasmic or outer membrane protein. Cho1 may be an additional component of the ABC transporter and may be required for gliding motility. (Cho1 is not likely to be the cargo of the transporter, since it appears to have a cleavable signal peptide and is probably transported by the general secretory machinery.) We do not know whether F. johnsoniae has a homolog to Cho1 or if additional proteins are part of the F. johnsoniae Gld transporter.
GldA exhibits sequence similarity to a large number of ATP-hydrolyzing components of ABC transporters (2). Homologs of known function include Bacillus subtilis SpaF (40% identity over 220 amino acids), Bacillus licheniformis BcrA (33% identity over 295 amino acids), S. meliloti NodI (33% identity over 299 amino acids), S. meliloti NosF (38% identity over 216 amino acids), and M. xanthus PilH (38% identity over 213 amino acids), among others. SpaF (25) and BcrA (41) are involved in resistance to the peptide antibiotics subtilin and bacitracin, respectively, presumably by exporting them. NodI is thought to be involved in lipochitin oligosaccharide export (48, 49). NosF is thought to interact with NosD and NosY (GldF homolog) to form a transporter that is involved in maturation of the periplasmic nitrous oxide reductase NosZ (20, 61). M. xanthus PilH is the ATP binding component of a transporter involved in pilus biogenesis and social gliding motility (59). Pili have not been observed in F. johnsoniae and related gliding bacteria (15, 42; M. J. McBride, unpublished data), and genes similar to those required for pilus assembly and function (other than pilH) have not been identified. While it appears unlikely that pili are responsible for F. johnsoniae motility, GldA and M. xanthus PilH may perform analogous roles in the assembly, modification, or functioning of their respective motility machineries.
It is not known whether GldA, GldF, and GldG function in import or export, but the similarities listed above suggest that GldA may be involved in export of some material to the periplasm, cell surface, or beyond. Several cell surface molecules have been implicated in F. johnsoniae gliding. Sulfonolipids are found in the outer membrane of F. johnsoniae, and several mutants that fail to make sulfonolipids fail to glide (1, 11, 12). Sulfonolipids may be directly involved in gliding or may provide an environment that allows proper assembly and functioning of the motility apparatus. Cell surface polysaccharides and glycoproteins have also been implicated in gliding (11, 13, 35, 37). GldA, GldF, and GldG may be involved in export of sulfonolipids or polysaccharides or in formation of cell surface glycoproteins. Alternatively they might be involved in export of protein components of the gliding machinery to the periplasm or outer membrane. The gld ABC transporter may be needed for assembly or modification of the motility apparatus, as suggested above, or may have a direct role in gliding by importing or exporting conveyor belts that run along the cell surface. It is unlikely that ATP hydrolysis by GldA is the sole driving force for cell movement, however, because others have demonstrated that PMF is required for gliding of F. johnsoniae and related bacteria (36, 43). Gliding could require the coordinated activity of two transporters with different energy requirements or a single transporter which requires both ATP and PMF. The HlyA hemolysin exporter of E. coli is an example of an ABC transporter that may require both ATP hydrolysis and PMF for transport (26, 60).
Gliding motility is thought to be the result of movement of cell surface components. The movement of these components can be observed by adding latex spheres which bind to cells and are propelled along their surfaces. Spheres move at approximately the same speed as gliding cells, and metabolic poisons and uncouplers that reduce PMF inhibit motility and block sphere movement (36). Nonmotile mutants fail to propel spheres (2, 7, 21, 22, 24). Our results confirm and extend this finding. Cells of gldF or gldG mutants failed to propel latex spheres, but complementation of the motility defect by introduction of gldFG on a plasmid restored this ability. These results support the suggestion that the machinery that propels latex spheres along the cell surface is also responsible for cell movement.
Many nonmotile mutants of F. johnsoniae are resistant to bacteriophage that infect wild-type cells (56). In addition, chemicals that dissipate PMF block gliding motility of wild-type cells (36) and also block adsorption of phage (56). Cells with mutations in gldF or gldG were resistant to bacteriophage infection. Restoration of motility by introduction of gldFG on a plasmid restored sensitivity to bacteriophage. These results are similar to those obtained for strains with mutations in gldA, gldB, gldD, or ftsX (2, 21, 22, 24) and indicate that mutations that disrupt gliding motility generally result in resistance to diverse F. johnsoniae bacteriophages. Bacteriophage usually attach to specific receptors on the cell surface, such as outer membrane proteins, polysaccharides, lipopolysaccharides, pili, or flagella. The receptors for F. johnsoniae bacteriophages have not been identified. If the Gld ABC transporter has an outer membrane component, it could provide a phage attachment site and the transporter could function as a conduit for entry of phage nucleic acid into the cytoplasm. Alternatively, the transporter may be needed to assemble or modify cell surface components which function as phage receptors and as components of the motility apparatus. Finally, phage resistance may be a nonspecific result of the defect in motility. In the absence of cell surface movement, phage receptors may be inaccessible.
Pate and colleagues isolated a bank of 50 independent spontaneous or chemically induced mutants that are completely deficient in gliding motility (7, 22, 57). Complementation and sequence analyses indicate that four of these mutants are nonmotile because of mutations in gldA (2), four fail to glide because of mutations in gldB (22), three others have mutations in gldD (21), and five are nonmotile because of mutations in gldF or gldG (this study). Complementation of 16 of 50 nonmotile mutants by just five genes suggests that a relatively small number of genes, perhaps less than 20, are required specifically for gliding motility.
The mechanism of F. johnsoniae gliding motility remains a mystery. Genetic analyses have uncovered six genes (gldA, gldB, gldD, gldF, gldG, and ftsX) that are required for F. johnsoniae gliding motility. The results presented in this paper suggest that the products of gldA, gldF, and gldG form a complex that functions as an ATP- dependent transporter that is required for cell movement. Further analysis of GldA, GldF, and GldG should elucidate the role of this transporter in F. johnsoniae gliding motility.
Acknowledgments
This research was supported by a grant from the National Science Foundation (MCB-9727825) and by a Shaw Scientist Award to M.J.M. from The Milwaukee Foundation.
DNA sequencing was performed by the Automated DNA Sequencing Facility of the University of Wisconsin-Milwaukee Department of Biological Sciences. Preliminary sequence data for C. hutchinsonii were obtained from The DOE Joint Genome Institute at http://jgi.doe.gov. We thank D. Saffarini for careful reading of the manuscript.
REFERENCES
- 1.Abbanat, D. R., E. R. Leadbetter, W. Godchaux III, and A. Escher. 1986. Sulphonolipids are molecular determinants of gliding motility. Nature 324:367-369. [Google Scholar]
- 2.Agarwal, S., D. W. Hunnicutt, and M. J. McBride. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. USA 94:12139-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bernardet, J.-F., P. Segers, M. Vancanneyt, F. Berthe, K. Kersters, and P. Vandamme. 1996. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga aquatilis Strohl and Tait 1978). Int. J. Syst. Bacteriol. 46:128-148. [Google Scholar]
- 5.Burchard, R. P. 1984. Gliding motility and taxes, p. 301. In E. Rosenberg (ed.), Myxobacteria, development and cell interactions. Springer-Verlag, New York, N.Y.
- 6.Burchard, R. P. 1981. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu. Rev. Microbiol. 35:497-529. [DOI] [PubMed] [Google Scholar]
- 7.Chang, L. Y. E., J. L. Pate, and R. J. Betzig. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 159:26-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duxbury, T., B. A. Humphrey, and K. C. Marshall. 1980. Continuous observations of bacterial gliding motility in a dialysis microchamber: the effects of inhibitors. Arch. Microbiol. 124:169-175. [Google Scholar]
- 9.Fath, M. J., and R. Kolter. 1993. ABC transporters: bacterial exporters. Microbiol. Rev. 57:995-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 11.Godchaux, W., III, L. Gorski, and E. R. Leadbetter. 1990. Outer membrane polysaccharide deficiency in two nongliding mutants of Cytophaga johnsonae. J. Bacteriol. 172:1250-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godchaux, W., III, and E. R. Leadbetter. 1988. Sulfonolipids are localized in the outer membrane of the gliding bacterium Cytophaga johnsonae. Arch. Microbiol. 150:42-47. [Google Scholar]
- 13.Godchaux, W., III, M. A. Lynes, and E. R. Leadbetter. 1991. Defects in gliding motility in mutants of Cytophaga johnsonae lacking a high-molecular-weight cell surface polysaccharide. J. Bacteriol. 173:7607-7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. A. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henrichsen, J., and J. Blom. 1975. Examination of fimbriation of some Gram-negative rods with and without twitching and gliding motility. Acta Pathol. Microbiol. Scand. Sect. B 83:161-170. [DOI] [PubMed] [Google Scholar]
- 16.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacteriales): two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 17.Hofmann, K., and W. Stoffel. 1993. TMbase--a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 18.Hoiczyk, E., and W. Baumeister. 1998. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 8:1161-1168. [DOI] [PubMed] [Google Scholar]
- 19.Holland, I. B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293:381-399. [DOI] [PubMed] [Google Scholar]
- 20.Holloway, P., W. McCormick, R. J. Watson, and Y. K. Chan. 1996. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J. Bacteriol. 178:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunnicutt, D. W., and M. J. McBride. 2001. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes gldD and gldE. J. Bacteriol. 183:4167-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunnicutt, D. W., and M. J. McBride. 2000. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes, gldB and gldC. J. Bacteriol. 182:911-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kempf, M. J., and M. J. McBride. 2000. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J. Bacteriol. 182:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, C., and K. D. Entian. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koronakis, V., C. Hughes, and E. Koronakis. 1991. Energetically distinct early and late stages of HlyB/HlyD-dependent secretion across both Escherichia coli membranes. EMBO J. 10:3263-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapidus, I. R., and H. C. Berg. 1982. Gliding motility of Cytophaga sp. strain U67. J. Bacteriol. 151:384-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 29.McBride, M. J., and M. J. Kempf. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 178:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 31.Metcalf, W. W., W. Jiang, L. L. Daniels, S.-K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 32.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 33.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohmori, H., M. Kimura, T. Nagata, and Y. Sakakibara. 1984. Structural analysis of the dnaA and dnaN genes of Escherichia coli. Gene 28:159-170. [DOI] [PubMed] [Google Scholar]
- 35.Pate, J. L. 1988. Gliding motility. Can. J. Microbiol. 34:459-465. [Google Scholar]
- 36.Pate, J. L., and L.-Y. E. Chang. 1979. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelopes. Curr. Microbiol. 2:59-64. [Google Scholar]
- 37.Pate, J. L., and D. M. De Jong. 1990. Use of nonmotile mutants to identify a set of membrane proteins related to gliding motility in Cytophaga johnsonae. J. Bacteriol. 172:3117-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pate, J. L., S. J. Petzold, and L.-Y. E. Chang. 1979. Phages for the gliding bacterium Cytophaga johnsonae that infect only motile cells. Curr. Microbiol. 2:257-262. [Google Scholar]
- 39.Pazzani, C., C. Rosenow, G. J. Boulnois, D. Bronner, K. Jann, and I. S. Roberts. 1993. Molecular analysis of region 1 of the Escherichia coli K5 antigen gene cluster: a region encoding proteins involved in cell surface expression of capsular polysaccharide. J. Bacteriol. 175:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 41.Podlesek, Z., A. Comino, B. Herzog-Velikonja, D. Zgur-Bertok, R. Komel, and M. Grabnar. 1995. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16:969-976. [DOI] [PubMed] [Google Scholar]
- 42.Reichenbach, H. 1992. The order Cytophagales, p. 3631-3675. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. M. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 43.Ridgway, H. F. 1977. Source of energy for gliding motility in Flexibacter polymorphus: effects of metabolic and respiratory inhibitors on gliding movement. J. Bacteriol. 131:544-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishahama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 45.Semmler, A. B. T., C. B. Whitchurch, and J. S. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145:2863-2873. [DOI] [PubMed] [Google Scholar]
- 46.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 2:784-791. [Google Scholar]
- 47.Smith, C. J., M. B. Rogers, and M. L. McKee. 1992. Heterologous gene expression in Bacteroides fragilis. Plasmid 27:141-154. [DOI] [PubMed] [Google Scholar]
- 48.Spaink, H. P. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54:257-288. [DOI] [PubMed] [Google Scholar]
- 49.Spaink, H. P., A. H. M. Wijfjes, and B. J. J. Lugtenberg. 1995. Rhizobium NodI and NodJ proteins play a role in the efficiency of secretion of lipochitin oligosaccharides. J. Bacteriol. 177:6276-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spormann, A. M. 1999. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol. Mol. Biol. Rev. 63:621-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 52.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 53.Trachtenberg, S. 1998. Mollicutes: wall-less bacteria with internal cytoskeletons. J. Struct. Biol. 124:244-256. [DOI] [PubMed] [Google Scholar]
- 54.Vaucher, J. P. 1803. Histoire des conferves d'eau douce, contenant leurs differens modes de reproduction, et la description de leurs principales especes, suivie de l'histoire des tremelles et des ulves d'eau douce. J. J. Paschoud, Geneva, Switzerland.
- 55.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 56.Wolkin, R. H., and J. L. Pate. 1986. Phage adsorption and cell adherence are motility-dependent characteristics of the gliding bacterium Cytophaga johnsonae. J. Gen. Microbiol. 132:355-367. [Google Scholar]
- 57.Wolkin, R. H., and J. L. Pate. 1985. Selection for nonadherent or nonhydrophobic mutants co-selects for nonspreading mutants of Cytophaga johnsonae and other gliding bacteria. J. Gen. Microbiol. 131:737-750. [Google Scholar]
- 58.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 59.Wu, S. S., J. Wu, Y. L. Cheng, and D. Kaiser. 1998. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol. Microbiol. 29:1249-1261. [DOI] [PubMed] [Google Scholar]
- 60.Young, J., and I. B. Holland. 1999. ABC transporters: bacterial exporters—revisited five years on. Biochim. Biophys. Acta 1461:177-200. [DOI] [PubMed] [Google Scholar]
- 61.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]