Abstract
Rhodospirillum centenum is a purple photosynthetic bacterium that is capable of differentiating from vibrioid swimming cells that contain a single polar flagellum into rod-shaped swarming cells that have a polar flagellum plus numerous lateral flagella. Microscopic studies have demonstrated that the polar flagellum is constitutively present and that the lateral flagella are found only when the cells are grown on solidified or viscous medium. In this study, we demonstrated that R. centenum contains two sets of motor and switch genes, one set for the lateral flagella and the other for the polar flagellum. Electron microscopic analysis indicated that polar and lateral flagellum-specific FliG, FliM, and FliN switch proteins are necessary for assembly of the respective flagella. In contrast, separate polar and lateral MotA and MotB motor subunits are shown to be required for motility but are not needed for the synthesis of polar and lateral flagella. Phylogenetic analysis indicates that the polar and lateral FliG, FliM, and FliN switch proteins are closely related and most likely arose as a gene duplication event. However, phylogenetic analysis of the MotA and MotB motor subunits suggests that the polar flagellum may have obtained a set of motor genes through a lateral transfer event.
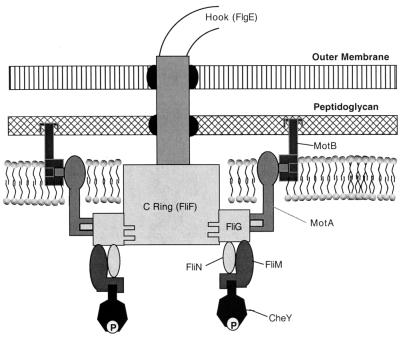
While a great deal is known about the motor and switch components of the bacterial flagellum (for recent reviews, see references 9 and 10), the mechanism for control of its rotation is a complex problem. Figure 1 is a diagram showing what is known about the structural features of the Escherichia coli flagellum as it passes through the inner membrane, the peptidoglycan layer, and the outer membrane. FliG, FliM, and FliN are parts of the rotor and components of the switch complex or C ring. These proteins are necessary for flagellar assembly, rotation, and control of the direction of rotation (9, 10). Switching from counterclockwise (CCW) to clockwise (CW) rotation is controlled by interactions between FliM and phosphorylated CheY (4, 38). Binding of CheY∼P to FliM causes CW flagellar rotation, leading to cell tumbling and a subsequent random change in orientation. By interrupting runs of smooth swimming (CCW rotation) with periods of tumbling (CW rotation), the cells are able to perform biased walks up or down gradients of attractants or repellents, respectively. The motor components that are responsible for flagellar rotation are MotA and MotB. Unlike the switch components, which rotate with the flagellum, MotA and MotB are part of the stator, which does not rotate. MotA and MotB are thought to form a transmembrane proton channel with the translocation of protons promoting movement of the switch component. The interaction of the motor with the switch complex leads to conversion of a proton potential into mechanical rotation of the flagellum (2, 37).
FIG. 1.
Structural features of the bacterial flagellum. See introduction for details.
Rhodospirillum centenum is a recently described α-purple photosynthetic bacterium that undergoes a dimorphic differentiation from swimming cells to swarming cells (28, 30, 31). In liquid growth medium, R. centenum cells exhibit a vibrioid shape with a single polar flagellum. However, when grown on an agar-solidified medium, the cells differentiate into a rod shape and produce lateral flagella, as well as a polar flagellum. In many respects, R. centenum differentiation is similar to that observed in Vibrio parahaemolyticus, an aquatic and enteric bacterium (25). In both species, a sheathed polar flagellum is constitutively synthesized by both cell types. Synthesis of separate unsheathed lateral flagella occurs upon contact with a solid surface or by the addition of viscosifying agents to a liquid medium (25, 30). For V. parahaemolyticus, it has been demonstrated that the lateral and polar flagella have separate motor genes. The lateral flagellar motor proteins are LafT and LafU, which are homologs of MotA and MotB, respectively (26). The sodium-driven polar flagellum motor is more complex, involving four proteins, MotA, MotB, and two additional proteins termed MotX and MotY (1, 23, 24). V. parahaemolyticus also produces separate sets of lateral and polar FliG, FliM, and FliN switch proteins that are necessary for flagellar assembly and rotation (5). One notable difference between R. centenum and. V. parahaemolyticus is that a functional polar flagellum is not necessary for swarming motility of V. parahaemolyticus whereas it is required for R. centenum swarming motility (5, 18). Microscopic analysis indicates that lateral flagellum rotation occurs in the absence of a functional polar flagellum but that surface movement is impeded (unpublished observation). This suggests that coordination of lateral flagellum rotation, such as proper formation of a flagellar bundle, is defective in defective swarming cells that contain a nonfunctional polar flagellum.
The focus of this study was to determine whether R. centenum polar and lateral flagella share any motor or switch components. For this investigation, we cloned, sequenced, and mutationally disrupted lateral and polar flagellum motor and switch components. Phylogenetic analysis of individual R. centenum motor and switch proteins indicates that lateral and polar switch genes arose as a consequence of a gene duplication event but that polar motor subunits most likely were acquired by lateral gene transfer.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are listed in Table 1. R. centenum wild-type swarming cells (ATCC 51521) were grown in CENS liquid medium or 0.8% agar-solidified PYVS plates at 42°C as described previously (26). E. coli DH5α and JM109 (Promega, Madison, Wis.) were used for routine DNA manipulations. JM109 (λ pir) (27) was used to maintain plasmids with an incomplete R67K replicon, and S-17 (λ pir) (29, 33) was used conjugally to deliver suicide plasmids into R. centenum. E. coli strains were grown in Luria-Bertani medium at 37°C and with SOC medium used for recovery of E. coli cells after electroporation (33).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference(s) or source |
|---|---|---|
| Strains | ||
| R. centenum | ||
| ATCC 51521 | Wild-type swarming strain | 31 |
| Q3-13 | motAP::Tn5-Spr | This study |
| L9-1 | motBP::Tn5-Spr | This study |
| Q9-8 | fliMP::Tn5-Spr | This study |
| R8-24 | fliML::Tn5-Spr | This study |
| ZJP246 | fliNP::Tn5-Spr | 18 |
| Q5-34 | fliGL::Tn5-Spr | This study |
| B8-35 | fliGP::Tn5-Spr | This study |
| R7-13 | motAL::Tn5-Spr | This study |
| JM-LG | fliGL::Spr | This study |
| JM-PA | motAP::Spr | This study |
| JM-PN | fliNP::Spr | This study |
| JM-PG | filGP::Spr | This study |
| 243-1a | fliNL::Gmr insertion of pBGR243-1 | This study |
| E. coli | ||
| DH5α | Bethesda Research Laboratories | |
| JM109 (λ pir) | 27 | |
| S-17 (λ pir) | 29, 34 | |
| Plasmids | ||
| pBluescript SK+ | Cloning vector | Stratagene |
| pZJD3 | Suicide vector; Gmr | 17 |
| pZJD9 | Derivative of pZJD3 also called pGmLacZ in reference 17 | 17 |
| pZJD9-fliMP::Spr | XbaI-XhoI restriction fragment containing fliMP::Spr segment | This study |
| pZJD9-fliNP::Spr | XbaI-XhoI restriction fragment containing fliNP::Spr segment | This study |
| pZJD9-fliGP::Spr | XbaI-XhoI restriction fragment containing fliGP::Spr segment | This study |
| pZJD9-fliGL::Spr | XbaI-XhoI restriction fragment containing fliGL::Spr segment | This study |
| pBGR242 | Internal 269-bp ClaI-EcoRI fliNL fragment in pBluescript SK+; Apr | This study |
| pBGR243 | SalI-EcoRI fliNL fragment from pBGR242 in pZJD3; Gmr | This study |
| pBGR243-1 | pBGR243 derivative containing 201-bp fliNL fragment; Gmr | This study |
Ampicillin and spectinomycin were added to the E. coli medium at a final concentration of 100 μg/ml. For R. centenum cultures, spectinomycin and gentamicin were used at 10 μg/ml.
Cloning and DNA sequencing of mini-Tn5 spectinomycin resistance (Spr)-disrupted genes.
An initial screen for genes that were disrupted by the mini-Tn5-Spr transposon (18) involved inverse PCR amplification with primers GGGCTTTACTA AGCTGATCC and CGAGCAGGGGAATTGATCC, which hybridized to the ends of the transposon, coupled with sequence analysis of the PCR product. For this analysis, 5 ml of the mutagenized cells was inoculated and grown aerobically overnight in the presence of spectinomycin at 50 μg/ml. Genomic DNA was then isolated as described by Jiang et al. (17) with 5 μg of DNA digested with SacII and or PstI. After inactivation of the restriction enzyme, a dilute ligation reaction was performed with 8 ml of ligase buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg of bovine serum albumin per ml) in the presence of 2,000 U of T4 DNA ligase for 4 to 8 h at 16°C. An inverse PCR fragment was subsequently gel purified (Qiaex II; Qiagen, Inc.) and sequenced with an ABI automated DNA sequencer (Applied Biosystems model 373; Perkin-Elmer) with the same primers that were used in the inverse PCR amplification step. Sequence information was used to identify potential motor or switch genes on the basis of homology to previously sequenced genes.
Complete sequence analysis of both strands of the motor and switch genes motAP, motALBL, and fliNP was accomplished by cloning a genomic DNA fragment from the mini-Tn5-disrupted strains. To obtain the mutated genes, restriction enzyme-digested genomic DNA fragments from mutant strains were ligated into a pBluescript SK+ cloning vector (Stratagene) with the mini-Tn5-disrupted gene obtained by selecting for Spr. Clones were subsequently sequenced on both strands by primer walking. For all other gene sequences, an inverse PCR was performed to amplify DNA segments flanking the transposon (39). One strand of the inverse PCR fragment was directly sequenced until both ends of the genes were reached. PCR amplification was then used to amplify the gene from wild-type R. centenum genomic DNA. The opposite strand was then sequenced with the full-length PCR fragment as a template. Sequence data were analyzed and assembled with the Sequencher program (Genes Codes, Ann Arbor, Mich.). Codon preference analysis was performed to assist in the identification of open reading frames (ORFs) and to ensure that undetected frameshift mutations were not present. For this analysis, a codon preference table was created with previously sequenced R. centenum genes.
The evolutionary relationship of motor and switch genes were analyzed by performing PILEUP sequence comparisons and unbranched tree alignments on the protein sequences compared to their respective homologs. The sequence and tree alignments were completed with the ClustalX program with trees plotted with NJPLOT (32).
Construction of gene disruptions.
A defined Spr disruption of fliMP (fliMP::Spr) was constructed by amplifying an internal segment of fliMP with primers TGCTCTAGAGAGGCGGACGCGGACG GCATAGAC and CCGCTCGAGCTC CTGATGCGGGTCAATAC via PCR. These primers also encode sequences for the restriction enzymes XbaI and XhoI (underlined), respectively. After PCR amplification, the fragment was digested with XbaI and XhoI, gel purified, and cloned into the corresponding sites of the PCR script vector (Stratagene Corp.). The fliMP clone was digested with NarI (located at codon 60), and this site was used for ligation to an SmaI DNA fragment that codes for Spr (17). The XbaI-XhoI restriction fragment containing the fliMP::Spr segment was cloned into suicide vector pZJD9 to form the final construct pZJD9-fliMP::Spr (Table 1) (suicide vector pZJD9 is described in reference 17 as pGmLacZ).
A similar process was used to construct defined Spr disruptions of fliNP, fliGP, and fliGL. To construct an fliNP::Spr disruption, primers TGCTCTAGAGCCTCG GCCGCGGCGGG and CCGCTCGAGCCGGCGGGCTGCCGGCC were used to amplify an internal segment of fliNP. After cloning of the amplified segment into PCRscript, the Spr fragment was cloned into a unique RsrII site that is located at codon 17 of fliNP and the fliNP::Spr disruption was subcloned into suicide vector pZJD9. To construct an fliGP::Spr disruption, primers CTAGTCTAGAACACGGCCAGCGCCTTC and CCCGCTCGAGATCGGCTAC GACCTGGG were used to amplify an internal segment of fliGP, which was cloned as described above. The Spr fragment was then cloned into a unique SphI site located at codon 167 in fliGP, and the fliGP::Spr disruption was then subcloned into suicide vector pZJD9. For construction of an fliGL::SpR disruption, primers TGCTCTAGACTCCGCCATGCGACGGGTCC and CCGCTCGAGTGCTGGCCGCCATCGACCCG were used to amplify an internal segment of fliGL. Following cloning, the Spr fragment was inserted into a unique NcoI site located at codon 148 of fliGL. The fliGL::Spr disruption was then subcloned into suicide vector pZJD9.
The suicide plasmids containing individual Spr knockouts were transformed into S17-1 λ pir (34) and filter mated with wild-type R. centenum (ATCC 51521) on nonselective PYVS medium for 4 h. The cells were then streaked onto CENS medium containing spectinomycin and 5% (wt/vol) 5-bromo-4 chloro-3 indolyl-β-d-galactopyranoside. Since the suicide plasmid contains lacZ and a gentamicin resistance-encoding gene, double recombination events that recombined the Spr-encoding gene into the chromosome were screened as red colonies lacking lacZ that were Spr and gentamicin sensitive. Recombinants were checked for proper integration by PCR amplification of appropriate-size fragments with primers that hybridized to specific gene segments that flanked the Spr-encoding gene. Strain names are listed in Table 1.
Production of fliNL mutant strains.
To construct mutations in the putative fliNL gene, a 269-bp ClaI-EcoRI fragment from a clone isolated in a different study (unpublished results) was inserted into pBluescript SK+ (Stratagene Corp.). The resulting vector, pBGR242, contains an internal portion of fliN that is truncated by 18 nucleotides at the 5′ end and 15 nucleotides at the 3′ end. This fragment was subcloned into a suicide vector, pZJD3 (17), with the internal EcoRI site and the polylinker site SalI, creating pBGR243. Since single recombination events with this plasmid generate nearly full-length fliN copies, we created a derivative of pBGR243 by digestion with DrdI and EcoRI, filling in of the ends to blunt the fragments (33), and allowing for religation of the plasmids. This procedure results in pBGR243-1, which has a further deletion of the 3′ end of the gene, leaving an internal 201-bp ClaI-DrdI fliN fragment. Filter matings of suicide plasmid-containing strains with wild-type R. centenum were performed overnight on CENS medium and followed by selective plating on gentamicin and kanamycin. Recombinants were isolated and checked for proper recombination by Southern blot analysis.
Analysis of motility.
For analysis of liquid motility, 200 μl of saturated aerobically grown cells was added to 2 ml of CENS medium and incubated for 3 h at 37°C with moderate shaking. Samples were viewed for motility with a Nikon Optiphot-2 microscope at ×400 magnification.
For analysis of surface motility, liquid cultures were grown photosynthetically for 2 days at 42°C under anaerobic conditions in the presence of light. Samples were spotted onto a 0.8% agar CENS plate and incubated at 42°C for a minimum of 6 h. Cells were viewed microscopically on the plate with a 40× objective lens with a long working distance.
Immunoblot assays.
Immunoblot (Western blot) assays were performed as described by Jiang et al. (18) with polyclonal anti-lateral flagellum serum that cross-reacts with both the lateral and polar flagellar subunits (18).
Transmission electron microscopy.
To observe polar and lateral flagella in swarming cells, a 100-fold-concentrated CENS medium culture of liquid-grown swimming cells was spotted in a 10-μl volume onto PYVS-0.8% agar plates. The plates were placed in the dark in a 42°C incubator for a few minutes, until the spots were dry, and then illuminated with a single tungsten light source for a minimum of 6 h to ensure production of lateral flagella. Cells in the colony were then gently resuspended by addition of 10 μl of 0.1 M KPO4 (pH 6.8). After 1 min, the cell suspension was transferred to the Formvar-coated side of a copper grid with a sterile inoculating loop. The suspension was allowed to sit on the grid for 30 min, rinsed with a 50-μl drop of water, blotted with Whatman paper, placed into a 7.5% uranyl acetate solution for 1 min, and blotted again. The grids were then washed and blotted five more times in water and dried at room temperature.
Nucleotide sequence accession numbers.
The nucleotide sequences generated by analysis in this study have been submitted to the GenBank database and assigned accession numbers AF220007 (motAPBP), AF220006 (fliGL), AF220005 (motBL), AF220004 (fliGP), AF220003 (fliNP) AF332661 (fliNL and motAP), AF220001 (fliMP), and AF220002 (fliML).
RESULTS
Identification of motor and switch genes.
In a previous study, we described the isolation of a collection of mutants generated by mini-Tn5-Spr insertion mutagenesis that were defective in swimming cell and/or swarming cell motility (18). Approximately 200 strains that were defective in motility were subsequently screened for the ability to synthesize lateral and polar flagella by Western blot analysis of swimming and swarming cells with antibodies that cross-react with lateral and polar flagellin proteins (data not shown). This screen allowed the segregation of mutants into three classes: (i) cells that were defective in polar flagellum synthesis, (ii) cells that were defective in lateral flagellum synthesis, and (iii) cells that were immobile when grown in liquid and/or on agar-solidified plates but still capable of lateral and polar flagellum synthesis. The first two classes include mutations in flagellum structural genes and mutations in genes involved in flagellar subunit transport, genes involved in assembly or genes involved in the control of flagellar gene expression. The third class includes mutations in motor subunits.
Approximately 140 mutants from these three classes were additionally characterized by performing sequence analysis of DNA that flanked the transposon. The sequence information was then used to perform BLAST searches for homologous sequences in the GenBank database. As discussed below, this approach resulted in the identification of putative motor and switch genes for both the polar and lateral flagella.
Sequence and mutational analysis of motor genes.
DNA sequence analysis of mutants that were capable of synthesizing lateral and polar flagella but were also defective in either liquid or swarming cell motility resulted in the identification of four putative motor genes. Sequence analysis of DNA flanking the transposon in strain R7-13 indicated that the mini-Tn5-Spr transposon was inserted into codon 211 of an ORF that codes for a protein with extensive sequence similarity to the MotA motor subunit that has been characterized in other species (Fig. 2A). As indicated in Table 2, disruption of this ORF resulted in a defect in swarming cell motility but not swimming cell motility, indicating that this motor subunit is involved in lateral but not polar flagellum rotation. Consequently, we have designated this gene motAL. A codon preference plot of the DNA sequence from this region also indicated the existence of a second ORF located immediately (3 bp) downstream of motAL. A homology search of the polypeptide encoded by this second ORF exhibited a high degree of sequence similarity to the second motor subunit, MotB (Fig. 2B). This observation was not unexpected since, in many species, motA and motB are coexpressed in an operon with motA preceding motB (14, 15, 35). Consequently, we have designated the gene coding for this second ORF motBL.
FIG. 2.
Alignment of deduced polypeptide sequences of ORFs that were sequenced in this study with selected homologous sequences in the GenBank database. (A) Alignment of R. centenum (R. cen) MotAL and MotAP. (B) Alignment of R. centenum MotBL and MotBP. (C) Alignment of R. centenum FliML and FliMP. (D) Alignment of R. centenum FliGL and FliGP. (E) Alignment of R. centenum FliNL and FliNP. Inverse lettering (white on black) indicates identity to one or both of the R. centenum flagellar subunits. Filled circles and brackets highlight functionally significant regions as covered in the Discussion. V. para, V. parahaemolyticus.
TABLE 2.
Phenotypes of bacterial strains derived from this study
| Strain | Genotype | Swimminga | Swarmingb | Polar flagellumc | Lateral flagellad |
|---|---|---|---|---|---|
| JM-PM | fliMP::Spr | No | No | Reduced | Yes |
| R8-24 | fliML::Tn5-Spr | Yes | No | Yes | No |
| JM-PN | fliNP::Spr | No | No | Reduced | Yes |
| 243-1a | fliNL::Gmr | Yes | No | Yes | No |
| JM-PG | fliGP::Spr | No | No | Reduced | Yes |
| JM-LG | fliGL::Spr | Yes | No | Yes | No |
| R7-13 | motAL::Tn5-Spr | Yes | No | Yes | Yes |
| Q3-13 | motAP::Tn5-Spr | No | No | Yes | Yes |
| L9-1 | motBP::Tn5-Spr | No | No | Yes | Yes |
Swimming was assayed by microscopic observation of the motility of cells from a liquid-grown culture, with “No” indicating that <1% of the cells were motile.
Swarming motility was assayed by microscopic observation of the motility of cells in a colon.
The presence or absence of polar and lateral flagella was based on a combination of Western blot analysis using polar and lateral flagellum-specific polyclonal antiserum, visual microscopic observation using a flagellum stain, and electron microscopic analysis.
“No” indicates that most of the cells in the population synthesized no visible lateral flagella and a few cells were capable of synthesizing <1% of the lateral flagellum level that is observed with wild-type swarming cells.
Previous studies have indicated that a functional polar flagellum is required for both swimming and swarming cell motility whereas functional lateral flagella are required only for swarming cell motility (18). Thus, to identify polar flagellum motor genes, we sequenced mutants that were incapable of swimming cell (liquid) and swarming cell (plate) motility but were still capable of synthesizing both lateral and polar flagella, as shown by Western blot assays and microscopic analysis of cells with stained flagella (data not shown). Sequence analysis of strain Q3-13 indicated that the mini-Tn5-Spr transposon was inserted into codon 102 of an ORF that exhibits significant sequence similarity to MotA (Fig. 2A). On the basis of both the sequence similarity and the phenotype observed upon disruption of this ORF, we have designated this gene motAP. Unlike MotAL, sequence analysis upstream and downstream of motAP did not show the presence of an motB homolog. However, sequence analysis of the mini-Tn5-Spr-disrupted gene in a second mutant strain (L9-1), which had a phenotype similar to that of strain Q3-13, indicted that strain L9-1 contained a transposition insertion into codon 75 of an ORF that coded for a polypeptide with significant sequence similarity to MotB (Fig. 2B). This second motB-like gene has been designated motBP. The observed motility phenotypes (Table 2) indicate that disruption of the polar flagellar motor genes, motAP and motBP, gives rise to defects in both swimming and swarming cell motility.
Sequence and mutational analysis of switch genes.
Identification of switch genes involved an approach similar to that used for the identification of motor genes, with the exception that null mutations in the switch genes also produce a defect in flagellum assembly (5, 40, 41). Thus, to identify switch genes for the polar flagellum, we performed a Western blot analysis with antisera raised to the polar flagellar subunit to screen for mutants that showed no, or severely reduced, synthesis of the polar flagellum. Sequence analysis of DNA segments flanking more than 50 mini-Tn5 insertion mutations in this class gave rise to the identification of insertions in genes that exhibited significant sequence similarity to fliG, fliM, and fliN. As shown by the alignment in Fig. 2C, strain B8-35 contained a mini-Tn5 insertion into codon 235 of an ORF that has significant sequence similarity to fliG. This strain and strain JM-PG (Table 1), which was constructed with a defined spectinomycin resistance insertion mutation in the same ORF, exhibited similar phenotypes. Specifically, disruption of this ORF did not affect induction of lateral flagellum synthesis upon growth on agar-solidified medium, indicating that this ORF does not code for a component of the lateral flagellum (Fig. 3C). However, the majority of the liquid-grown swimming cells with fliG disrupted exhibited no motility (>95% were nonmotile) (Table 2). Electron microscopic analysis also showed an absence of polar flagellum synthesis in most (79%) of the observed mutant cells (Fig. 3D). This is in contrasted to the presence of a polar flagellum in most (>95%) of the wild-type cells (Fig. 3A and B). Disruption of this gene did exhibit a leaky phenotype since there were some observable motile swimming cells in liquid-grown fliG mutant strains. Furthermore, electron microscopic analysis also indicated that a portion of the cells with fliG disrupted (∼20%) were still capable of synthesizing a polar flagellum (data not shown). Since disruption of this ORF only affects polar flagellum synthesis, we have designated this gene fliGP to indicate its role as a switch component of the polar flagellum.
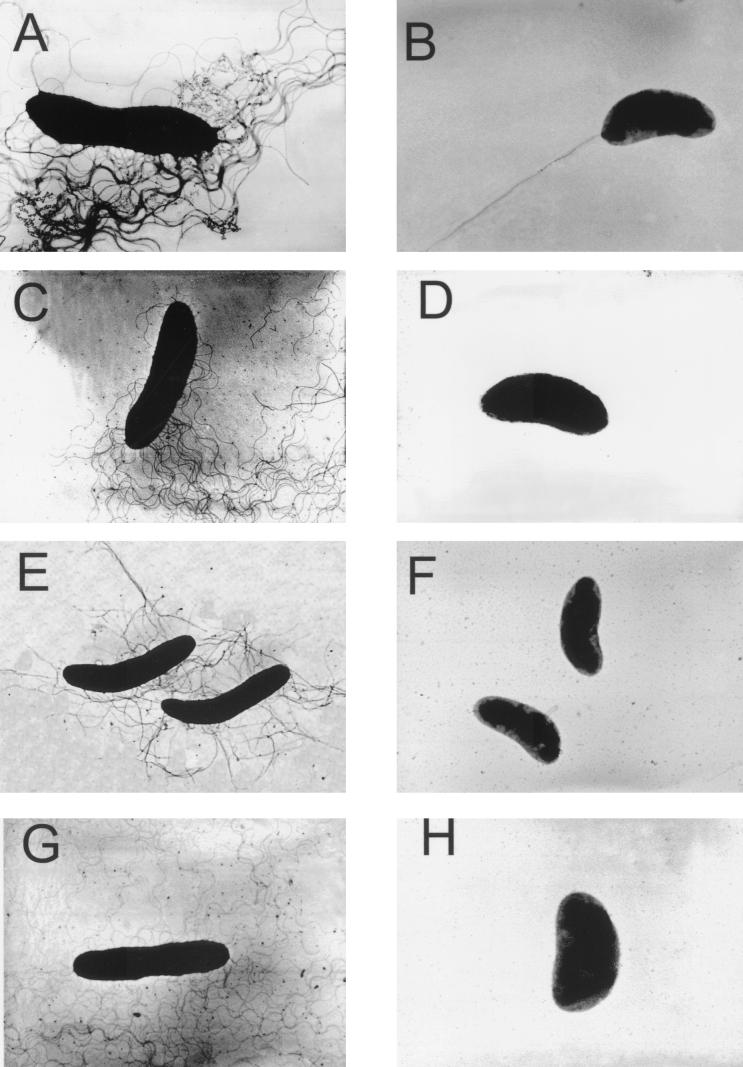
FIG. 3.
Transmission electron microscopic analysis of polar and lateral flagella synthesized by R. centenum polar switch mutants. (A and B) Agar- and liquid medium-grown wild-type swarming and swimming cells, respectively. (C and D) Agar- and liquid medium-grown fliGP::Spr strain JM-PG, respectively. (E and F) Agar- and liquid medium-grown fliNP::Spr strain JM-PN, respectively. (G and H) Agar- and liquid medium-grown fliMP::Spr strain JM-PM, respectively. The electron microscopic photographs represent the most predominant cell type that was observed.
Similar sequence analysis of strain ZJP-246 (18), which contains a mini-Tn5-Spr insertion (codon 106), resulted in the identification of an ORF exhibiting sequence similarity to fliN (designated fliNP) (Fig. 2D). This strain and strain JM-PN, which we constructed and which contains a defined spectinomycin resistance disruption in fliNP, exhibited the same motility (Table 2) and flagellar biosynthesis phenotypes as described for strain B8-35. Specifically, these cells exhibited normal lateral flagellum synthesis (Fig. 3E) and a loss of polar flagellum synthesis (Fig. 3F) in the vast majority (86%) of the cells with fliNP disrupted.
Despite sequencing numerous mutants that were defective in synthesis of the polar flagellum, we did not identify any strains that contained specific disruptions of a putative polar fliM gene. However, sequence analysis of this class of polar flagellum-defective mutants did indicate that strain R5-18 had a disruption in an ORF with considerable sequence similarity to fliL (data not shown) and that downstream of fliL was an ORF that codes for a protein with considerable sequence similarity to fliM (Fig. 2E). We subsequently constructed a defined Spr disruption of this fliM homolog (strain JM-PM). Analysis of JM-PM indicated that this strain has motility (Table 2) and flagellar biosynthesis phenotypes similar to those described for fliGP and fliNP mutants. Specifically, electron microscopic analysis indicated that disruption of this ORF produced no effect on lateral flagellum synthesis (Fig. 3G) and an absence of polar flagellum synthesis (Fig. 3H) in 76% of the cells (Table 2). Consequently, we designated this gene fliMP.
For identification of lateral switch genes, we screened for loss of the lateral flagellum by performing Western blot analysis on cells harvested from agar plates with antiserum raised to the lateral flagellar subunit (data not shown). By sequencing DNA segments flanking the transposon from more than 50 mutants, we were able to identify mutations that disrupted two of the three putative lateral switch genes. As shown by the alignment in Fig. 2E, strain R8-24 contained a mini-Tn5-Spr insertion (codon 110) in an ORF that has significant sequence similarity to FliM. This strain exhibited normal swimming cell motility but a defect in swarming cell motility (Table 2). Electron microscopic analysis indicated that R8-24 swimming cells contain a normal polar flagellum in liquid medium and agar-solidified medium and that the majority (>98%) of R8-24 swarming cells are defective in lateral flagellum synthesis (Fig. 4B). In a few cases (1 to 2%), there appeared to be a few agar-grown R8-24 swarming cells that contained one or two lateral flagella in addition to the polar flagellum (data not shown). This value is in contrast to that of wild-type cells, which exhibited numerous (>100) lateral flagella per swarming cell (Fig. 4A). Since disruption of this gene affects lateral but not polar flagellum synthesis, we have designated this ORF fliML. A similar analysis demonstrated that strain Q5-34 contained a mini-Tn5 insertion in codon 199 of an ORF that codes for a protein that has significant sequence similarity to FliG (Fig. 2C). Since disruption of this ORF resulted in a phenotype similar to that described for fliML (Table 2; Fig. 4C), we have designated this gene fliGL.
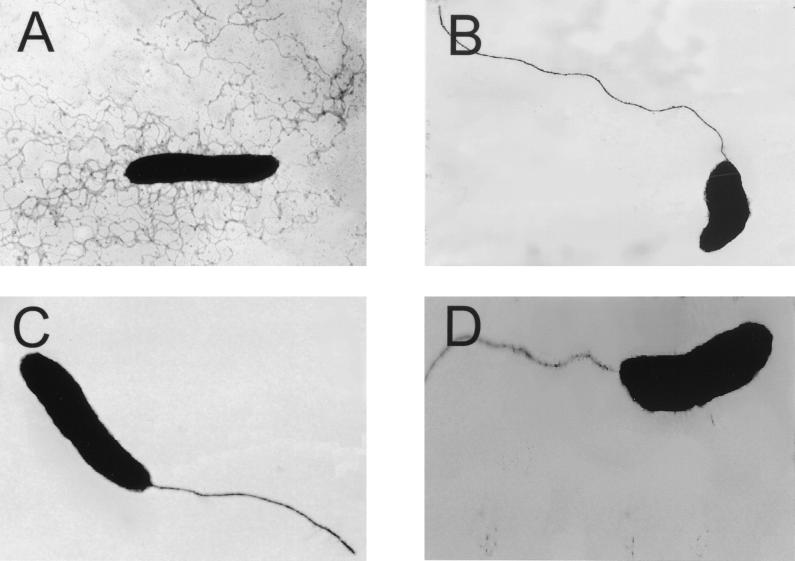
FIG. 4.
Transmission electron microscopic analysis of R. centenum lateral switch mutants grown on agar-solidified medium. Panels: A, wild-type cell; B, fliGL::Spr strain JM-LG; C, fliNL::Gmr strain 243-1a; D, fliML::Tn5-Spr strain R8-24. The electron microscopic photographs represent the most predominant cell type that was observed.
Sequence analysis of additional strains defective in lateral flagellum synthesis did not result in the identification of a disruption of a fliN homolog that is necessary for lateral flagellum synthesis. However, in a separate study involving sequence analysis of respiratory electron transfer components (B. Rushing and C. Bauer, unpublished results), an ORF was discovered that encodes a protein with significant sequence similarity to FliN from Caulobacter crescentus and other species, including R. centenum FliNP.
To determine if this second fliN-like gene is involved in lateral or polar flagellum synthesis, we constructed a gentamicin-resistant disruption of this gene by selecting for single recombination with a suicide plasmid containing an internal portion of the putative fliN gene. Upon homologous chromosomal recombination, this plasmid creates a tandem duplication of the fliN-like gene, of which one contains a deletion of the promoter and the amino-terminal region and the other contains a deletion of the carboxyl-terminal region. Immunoblot and electron microscopy analyses of recombinant strain 243-1a showed that this strain synthesizes a polar flagellum in liquid medium and agar-solidified medium. However, like that observed in the fliML and fliGL mutants, synthesis of the lateral flagellum is severely reduced in strain 243-1a (Fig. 4D). This gene has therefore been designated fliNL. An alignment of FliNL with FliNP and other FliN homologs is shown in Fig. 2D.
Phylogenetic analysis.
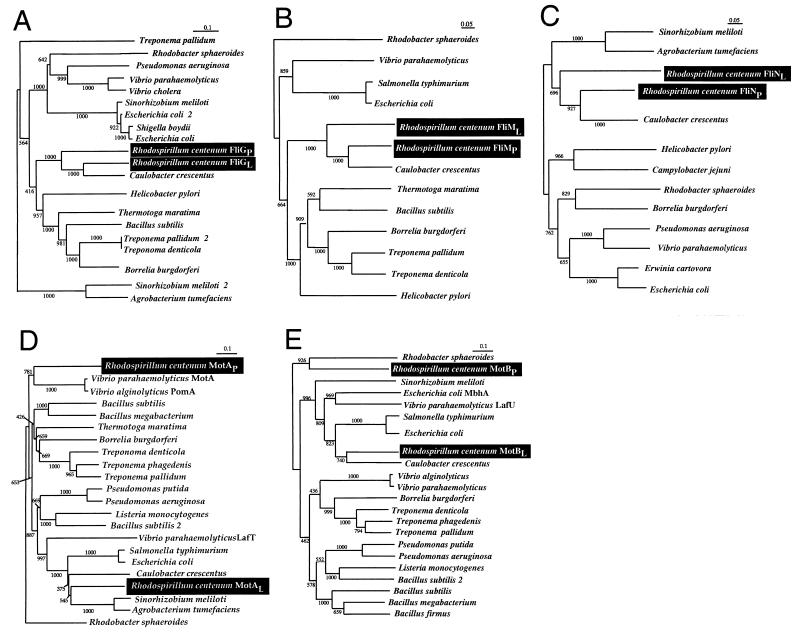
To determine the evolutionary relationship between the duplicated motor and switch genes, we constructed phylogenetic trees of the genes identified in this study with various motor and switch homologs that are present in the GenBank database. The first feature we noticed from our protein sequence alignments is that the R. centenum lateral and polar switch subunits exhibit high sequence similarity to each other, as well as to C. crescentus homologs (Fig. 2). The result of our phylogenetic analyses confirms this observation by demonstrating that the fliGL fliMP and fliNL pair of R. centenum switch genes form a separate clade with the respective Caulobacter homologs (Fig. 5A to C). This result suggests that these closely related species derived their genes from the same source. Moreover, the duplicated R. centenum switch genes probably arose through one or more gene duplication events rather than obtaining a duplicate set of switch genes via multiple lateral transfers. Overall, the clades formed in the switch gene phylogenetic analyses are similar to 16S rRNA analyses in which enteric bacteria such as Shigella boydii, E. coli, and Erwinia carotovora typically form a clade and the spirochetes Borrelia burgdorferi, Treponema pallidum, and T. denticola also group together (data not shown).
FIG. 5.
Phylogenetic analysis. Protein sequences were aligned with the Genetics Computer Group PILEUP software. Phylogenetic trees were made from these alignments with ClustalX with 1,000 bootstraps. Trees were plotted with NJPLOT. Panels: A, FliG; B, FliM; C, FliN; D, MotA; E, MotB.
Phylogenetic analysis of the motor trees gave strikingly different results. As shown in Fig. 5D and E, the lateral motor subunits MotAL and MotBL both show a close relationship with the C. crescentus motor subunits. This is consistent with the trees obtained with the switch genes. However, the polar motor subunits MotAP and MotBP no longer form a clade with Caulobacter (Fig. 5D and E). For both MotAP and MotBP, there is a close relationship with the Rhodobacter sphaeroides motor subunits. Interestingly, MotAP actually showed the strongest sequence similarity to the sodium-driven motor of V. parahaemolyticus. The fact that the R. centenum MotAP and MotBP proteins belong to different clades leads us to conclude that the polar motor genes may not be a result of a gene duplication event, as appears to be case of the switch genes. Instead, these results indicate that one or more lateral transfer events may have given rise to the R. centenum polar motor subunits.
DISCUSSION
By sequencing a large number of random mini-Tn5 insertion mutations, we identified two sets of flagellar motor and switch genes, one set that controls lateral flagellar rotation and another that controls the rotation of the polar flagellum. Microscopic studies indicate that the lateral and polar switch genes are necessary for lateral and polar flagellum assembly and that the motor genes are not needed for assembly. This observation is in agreement with similar results obtained with enteric bacteria (40, 41).
In many respects, the results we obtained with R. centenum are similar to those obtained with V. parahaemolyticus, which also undergoes similar differentiation from swimming cells to swarming cells. Like R. centenum, V. parahaemolyticus contains separate lateral and polar genes for the motor and switch proteins (5, 19). These two species are distinct from other swarming cell differentiating species, such as Proteus mirabilis and Serratia marcescens, which only contain a single set of flagellar genes. In these latter species, swarming cells simply express more of the same flagellar type that is observed in swimming cells. The results of our phylogenetic analyses indicate that the lateral and polar flagellar switch components most likely arose via a gene duplication event. This observation is in contrast to phylogenetic analyses of the motor subunits that indicate that the lateral and polar subunits are highly diverged. The MotAL and MotBL subunits actually show a close relationship to the C. crescentus motor subunits. A comparable close relationship to Caulobacter is seen in the switch components, indicating that the lateral motor genes have an ancestral lineage that is similar to that of the lateral motor genes. The significant divergence of the polar motor subunits suggests that these genes may have been obtained via lateral transfer.
We also observed that disruption of the polar switch components gave rise to a leaky phenotype in that a significant proportion (14 to 25%) of the cells were still capable of synthesizing a polar flagellum, as shown by electron microscopic evaluation of the mutant cells. A similar incomplete phenotype was also observed for disruptions of many different polar flagellar genes in V. parahaemolyticus (19). In contrast to the electron microscopic results, only a small fraction (<1%) of the cells with a polar gene disruption are motile when grown in liquid culture, indicating that a significant proportion of the polar flagellum that is synthesized by the flagellum appears to be nonfunctional. One possible explanation for this is that there is a significant basal level of lateral switch components in swimming cells and that the lateral components are capable of compensating for the loss of the polar subunits. Another possibility is that the R. centenum polar flagellum is capable of assembly, at a low frequency, in the absence of the switch components.
Features of the lateral and polar motor subunits.
Several studies have indicated regions of the MotA and MotB motor subunits that are involved in conversion of the influx of ions into the production of torque. Amino acid residue Asp32, which is located in the proton channel near the cytoplasmic face of MotB, is thought to play a critical role in torque generation (44). Alignment of 21 MotB homologs in the GenBank data base indicated that this residue is universally conserved (data not shown). Asp32 is followed by a large, conserved, hydrophobic patch in 20 of 21 MotB proteins as well. The one protein that does not have the conserved hydrophobic patch just downstream of Asp21 is R. centenum MotBP. A highly conserved patch of residues near the MotB carboxyl terminus (GHTD in E. coli residues 194 to 197; NWELS—RA in E. coli residues 210 to 218, and NRR in E. coli residues 257 to 259) are nearly identical across all 21 MotB proteins. This region is believed to be involved in the binding of MotB to the peptidoglycan, providing an anchor for the stator (8).
Analysis of critical residues in MotA has also received considerable attention (3, 12, 13, 36). In addition to forming a torque-generating ion channel with MotB, MotA also functions as a gear that transmits torque to the flagellum by interacting with the carboxyl terminus of FliG (16, 21). There are several conserved residues in the MotA protein that are essential for torque generation (6). E. coli residues Pro222 and Pro173 are conserved among all MotA proteins, including R. centenum MotAL and MotAP. These residues, which are located on the cytoplasmic side of the membrane (43), are thought to be located near Asp32 of MotB (6). E. coli Arg90 and Glu98 have also been shown to be important for motor rotation and are thought to interact with charged residues of FliG (20, 42, 44). Arg90 was present in both of the R. centenum MotAL and MotAP proteins, as well as in 24 of 26 MotA homologs. Glu98 is present in 22 of the 26 homologs, including R. centenum MotAL, but is not present in MotAP.
Features of the lateral and polar switch subunits.
Switch protein FliG is thought to interact with MotA, promoting the transfer of torque from the motor to the flagella. Mutational analysis of E. coli FliG indicates that Arg279, Asp286, and Asp287 are involved in torque transfer along with of Lys262 and Arg295, which have lesser roles (20). All of these residues are conserved in the R. centenum FliGL and FliGP proteins (Fig. 2C). The N-terminal portion of FliG, which is thought to bind to the MS ring, is also highly conserved among the R. centenum homologs (11). A conserved region of note is the first 45 amino acids of FliG, which are necessary for binding of FliG to FliF (22). The same 45 amino-terminal residues are not necessary for binding of FliG to FliM.
FliN is the smallest of the five motor switch components. Although no specific role for FliN is known, it is clear that it is necessary for proper assembly of the flagella and control of the direction of flagellar rotation. Sequence homology exists between FliN and components of the type III export system, suggesting one possible role for this protein. The alignment in Fig. 2D indicates that there are several conserved amino acids among FliN homologs. Some mutational analysis of FliN has been undertaken (16); however, the involvement of specific residues in the formation of an active switch complex is still uncertain.
One of the major functions of FliM is the interaction with phosphorylated CheY that affects the direction of flagellar rotation. Mutational analysis indicates that the first 16 amino acids of FliM are necessary for Che∼P binding (7). This amino-terminal region is highly conserved among all 13 FliM homologs in the GenBank database, with the exception of C. crescentus FliM and the two R. centenum FliM proteins (Fig. 2E). Although these closely related FliM proteins contain the conserved sequence, it starts at residue 52 rather than directly at the amino terminus. This result supports the phylogenetic analysis, which indicates that the R. centenum FliM proteins are closely related to C. crescentus FliM. Amino acid residues 283 to 384 of E. coli FliM have also been implicated in the interaction with FliN (22). A few conserved motifs are found in this area, with Arg311 being the only residue that is the same in all 13 FliM proteins.
Acknowledgments
We thank Lisa Ragatz for comments regarding the manuscript and Ze-Yu Jiang for technical help.
This study was supported by grant RO1 GM58050 from the National Institutes of Health. B.G.R was supported by a National Science Foundation postdoctoral fellowship.
REFERENCES
- 1.Atsumi, T., L. McCarter, and Y. Imae. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182-184. [DOI] [PubMed] [Google Scholar]
- 2.Berg, H. C. 1995. Torque generation by the flagellar rotary motor. Biophys. J. 68:163-166. [PMC free article] [PubMed] [Google Scholar]
- 3.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton conducting component of the flagellar motor. Cell 60:439-449. [DOI] [PubMed] [Google Scholar]
- 4.Blair, D. F. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49:489-522. [DOI] [PubMed] [Google Scholar]
- 5.Boles, B. R., and L. L. McCarter. 2000. Insertional inactivation of genes encoding components of the sodium-type flagellar motor and switch of Vibrio parahaemolyticus. J. Bacteriol. 182:1035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, T. F., S. Paulson, J. B. Gully, J. C. Empey, S. Van Way, A. Putnam, and D. F. Blair. 1999. Function of the proline residue of MotA in torque generation by the flagellar motor of Escherichia coli. J. Bacteriol. 181:3542-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bren, A., and M. Eisenbach. 1998. The N terminus of the flagellar switch protein, FliM is the binding domain of the chemotactic response regulator, CheY. J. Mol. Biol. 278:507-514. [DOI] [PubMed] [Google Scholar]
- 8.DeMot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation of between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 9.DeRosier, D. J. 1998. The turn of the screw: the bacterial flagellar motor. Cell 93:17-20. [DOI] [PubMed] [Google Scholar]
- 10.Eisenbach, M., and S. R. Caplan. 1998. Bacterial chemotaxis: unsolved mystery of the flagellar switch. Curr. Biol. 8:444-446. [DOI] [PubMed] [Google Scholar]
- 11.Francis, N. R., V. M. Irukura, S. Yamaguchi, D. J. DeRosier, and R. M. Macnab. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. USA 89:6304-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garza, A. G., P. A. Bronstein, P. A. Valdez, L. W. Harris-Haller, and M. D. Manson. 1996. Extragenic suppression of motB missense mutations of Escherichia coli J. Bacteriol. 178:6116-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garza, A. G., L. W. Harris-Haller, R. A. Stoebner, and, M. D. Manson. 1995. Motility protein interactions in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 92:1970-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge, Y., and N. W. Charon. 1997. Identification of a large motility operon in Borrelia burgdorferi by semi-random PCR chromosome walking. Gene 189:195-201. [DOI] [PubMed] [Google Scholar]
- 15.Hueck, C., A. Kraus, and W. Hillen. 1994. Sequences of ccpA and two downstream Bacillus megaterium genes with homology to the motAB operon from Bacillus subtilis. Gene 143:147-148. [DOI] [PubMed] [Google Scholar]
- 16.Irkura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, Z.-Y., H. Gest, and C. Bauer. 1997. Chemosensory and photosensory perception in purple photosynthetic bacteria utilize common signal transduction components. J. Bacteriol. 179:5720-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, Z.-Y., B. G. Rushing, Y. Bai, H. Gest, and C. Bauer. 1998. Isolation of Rhodospirillum centenum mutants defective in phototactic colony motility by transposon mutagenesis. J. Bacteriol. 180:1248-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, Y.-K., and L. L. McCarter. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd, S. A., and D. F. Blair. 1997. Charged residues of the rotor protein FliG are essential for torque generation on the flagellar motor of Escherichia coli. J. Mol. Biol. 266:733-744. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd, S. A., H. Tang, X. Wang, S. Billings, and D. F. Blair. 1996. Torque generation in the flagellar motor of E. coli: evidence of a direct role for FliG but not FliM or FliN. J. Bacteriol. 178:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marykwas, D. L., S. A. Schmidt, and H. C. Berg. 1996. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J. Mol. Biol. 256:564-576. [DOI] [PubMed] [Google Scholar]
- 23.McCarter, L. L. 1994. MotY, a component of the sodium-type flagellar motor. J. Bacteriol. 176:4219-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarter, L. L. 1994. MotX, the channel component of the sodium-type flagellar motor. J. Bacteriol. 176:5988-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarter, L., and M. Silverman. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4:1057-1062. [DOI] [PubMed] [Google Scholar]
- 26.McCarter, L. L., and M. E. Wright. 1993. Identification of genes encoding components of the swarmer cell flagellar motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J. Bacteriol. 175:3361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nickens, D., C. J. Fry, L. R. Ragatz, C. E. Bauer, and H. Gest. 1995. Biotype of the nonsulfur purple photosynthetic bacterium Rhodospirillum centenum. Arch. Microbiol. 165:91-96. [DOI] [PubMed] [Google Scholar]
- 29.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 176:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragatz, L., Z.-Y. Jiang, H. Gest, and C. Bauer. 1994. Macroscopic phototactic behavior of the purple photosynthetic bacterium Rhodospirillum centenum. Arch. Microbiol. 163:1-6. [DOI] [PubMed] [Google Scholar]
- 31.Ragatz, L., Z.-Y. Jiang, H. Gest, and C. Bauer. 1994. Phototactic purple bacteria. Nature 370:104. [Google Scholar]
- 32.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning a laboratory manual, 2nd ed. Cold Springs Harbor Laboratory Press, Cold Springs Harbor, N.Y.
- 34.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 35.Stamm, L. V., and H. L. Bergen. 1999. Molecular characterization of a flagellar (fla) operon in the oral spirochete Treponema denticola ATCC 35405. FEMS Microbiol. Lett. 179:31-36. [DOI] [PubMed] [Google Scholar]
- 36.Stolz, B., and H. C. Berg. 1991. Evidence of interactions between MotA and MotB, torque-generating elements of the of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas, D. R., D. G. Morgan, and D. J. DeRosier. 1999. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. USA 96:10134-10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welch, M., K. Oosawa, S.-I. Aizawa, and Eisenbach. 1993. Phosphorylation-dependent binding of a single molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA 90:8787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong, J., K. Inoue, and C. Bauer. 1998. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis Proc. Natl. Acad. Sci. USA 95:14851-14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi, S., S.-I. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. MacNab. 1986. Genetic evidence for switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium flagellar basal body. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi, S., H. Fujita, A. Ishihara, S. Aizawa, and R. M. Macnab. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, J., and D. F. Blair. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273:428-439. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, J., R. T. Fazzio, and D. F. Blair. 1995. Membrane topology of the MotA protein of Escherichia coli. J. Mol. Biol. 251:237-242. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, J., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between the rotor and the stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 95:6436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]