Abstract
SecA is an essential ATP-driven motor protein that binds to presecretory or membrane proteins and the translocon and promotes the translocation or membrane integration of these proteins. secA is subject to a protein secretion-specific form of regulation, whereby its translation is elevated during secretion-limiting conditions. A novel mechanism that promotes this regulation involves translational pausing within the gene upstream of secA, secM. The secM translational pause prevents formation of an RNA helix that normally blocks secA translational initiation. The duration of this pause is controlled by the rate of secretion of nascent SecM, which in turn depends on its signal peptide and a functional translocon. We characterized the atypical secM signal peptide and found that mutations within the amino-terminal region specifically affect the secM translational pause and secA regulation, while mutations in the hydrophobic core region affect SecM secretion as well as translational pausing and secA regulation. In addition, mutational analysis of the 3′ end of secM allowed us to identify a conserved region that is required to promote the translational pause that appears to be operative at the peptide level. Together, our results provide direct support for the secM translational pause model of secA regulation, and they pinpoint key sequences within secM that promote this important regulatory system.
In bacteria nascent or fully synthesized presecretory or membrane proteins are selectively targeted to the translocon by interactions with SecB and SecA or the signal recognition particle and its receptor (3, 24, 40, 47, 50). These pathways converge at the translocon, which consists of the integral membrane proteins SecYEG and SecDFyajC and the peripheral membrane protein SecA ATPase. SecYE forms the preprotein channel and SecA receptor (10, 20, 27, 30), while SecG and SecDFyajC greatly enhance the rate of protein translocation by regulating SecA membrane cycling (11, 28, 34). SecA is central to protein translocation since it binds to the signal peptides or transmembrane segments of presecretory and membrane proteins, interacts with the SecB chaperone to promote release of the bound preprotein, and acts as a motor protein to drive protein translocation at the translocon (for a review, see reference 26). Considerable evidence suggests that SecA undergoes ATP-driven cycles of insertion and retraction at SecYE, thereby promoting the stepwise translocation of proteins across the plasma membrane (12, 13, 52).
The selectivity of the translocon for its protein cargo is remarkable, since erroneous translocation of cytoplasmic proteins is essentially undetectable. Current evidence suggests that the translocon possesses a proofreading activity that is responsible for aborting the translocation of preproteins that lack a functional signal peptide (for a review, see reference 7). prl alleles of secA, secY, secE, and secG have been isolated that allow translocation of preproteins with a defective signal peptide (4, 15, 17, 23, 48). A recent study suggested that the control of the ATP-dependent, preprotein insertion reaction by the SecA-SecYE complex may be the critical biochemical step that controls this proofreading activity (51).
Because it catalyzes what appears to be the first committed step in protein translocation, ATP-dependent insertion of the preprotein into the translocon, SecA occupies a pivotal position in this pathway. secA appears to be the only sec gene that is under protein secretion-specific regulation; inhibition of protein secretion by either genetic or biochemical means leads to 10-fold induction of secA translation (36, 38, 44). Analysis of this system has revealed that secA is the second gene in the secM secA operon and that translation of secA is coupled to translation of secM, since ribosomes translating the distal portion of secM are needed to disrupt an RNA repressor helix (helix II) that normally blocks secA translational initiation (29, 42, 45). Based on the recent findings that (i) secM encodes a presecretory protein, (ii) secM signal sequence defects render secA expression constitutive even during rapid secretion of other proteins, (iii) such secM signal sequence defects are suppressible by prlA (secY) mutations, and (iv) the secM signal sequence alleles are cis acting, the secM translational pause-arrest model for secA regulation was recently proposed (37, 41). This model postulates that there is coupling and feedback between SecM (secretion monitor) translation and secretion, whereby the frequency of secA translational initiation depends on a translational pause within the distal portion of secM and the length of the secM translational pause is governed by the rate of secretion of nascent SecM protein, which in turn depends on its signal peptide and interaction with SecA and the translocon. Recent biochemical analysis of this system has confirmed many of the basic features of this model (33). In particular, the presence of a natural translational pause within the distal portion of secM was demonstrated, and the length of the secM translational pause was shown to depend on the secretion of SecM protein; defects in the secM signal peptide or translocon promoted a prolonged arrest of secM translation and resulted in secA derepression.
Despite these recent advances in our understanding of secA regulation, a number of features of this system remain poorly defined. For example, a recently revised translational start site for secM indicated the presence of a signal peptide consisting of 37 amino acid residues with an unusually long 19-amino-acid amino-terminal region that contains a number of atypical amino acids (43). The importance of this unique signal peptide in controlling the secA regulatory system remains poorly explored. In particular, the effect that the existing secM signal sequence mutations have on the rate of secretion of SecM protein was not investigated, and only one secM signal sequence mutant was studied with the translational pause assay (33, 37). In addition, the precise location of the secM translational pause site has not been defined (33). Thus, the proximity of this site to the RNA helix that normally blocks secA translational initiation remains unclear, as does the peptide or RNA sequence that is required to promote the translational pause itself. Understanding these features is critical for confirming and elucidating this important protein secretion-specific regulatory system.
In the present study we utilized a combined genetic and biochemical approach to further characterize the atypical secM signal peptide and the 3′ end of the gene where the translational pause site is localized. Our results indicate that the amino-terminal region (N-region) and the hydrophobic core region (H-region) of the secM signal peptide have different functions with respect to promoting SecM protein secretion, the secM translational pause, and secA regulation. In addition, analysis of mutations at the end of secM that affect secA regulation allowed us to identify a conserved region that is required to promote the translational pause and to demonstrate that pausing at this site allows the stalled ribosome to block formation of the secA repressor helix.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and chemicals.
The bacterial strains and plasmids used in this study are listed in Table 1. M63 minimal medium and Luria-Bertani (LB) broth used for growth of bacteria have been described previously (31). 5-Bromo-4-chloro-3-indoylphosphate and isopropyl-1-thio-β-d-galactoside (IPTG) were obtained from Fisher Scientific, and cyclic AMP, o-nitrophenyl-β-d-galactoside, sodium azide, and protein A Sepharose were obtained from Sigma Chemical Co. DNA restriction enzymes were obtained from New England Biolabs, Inc., and were used as recommended by the supplier. Tran 35S label (∼1,100 Ci/mmol) was obtained from ICN Radiochemicals, and IgSorb was obtained from The Enzyme Center, Inc. The fluorographic reagent Amplify was obtained from Amersham Corp. (Piscataway, N.J.). 5-Bromo-4-chloro-3-indoyl-β-d-galactopyranoside and XAR film were purchased from Eastman Kodak Co. Oligonucleotides were purchased from Integrated DNA Technologies.
TABLE 1.
E. coli strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| AF118 | MC4100 lamBΔ111 | 18 |
| AF128 | MC4100 lamBΔ111 prlG1 zja::Tn10 | 18 |
| AF130 | MC4100 lamBΔ111 prlG3 zja::Tn10 | 18 |
| CG155 | MC1000 recA | Jon Beckwith |
| CG29 | MC1000 secD1(Cs) phoR recA1 srl::Tn10 | Jon Beckwith |
| CC118 | MC1000 phoAΔ20 rpsE rpoB argE(Am) recA1 | Jon Beckwith |
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(argF-lac)U169 deoR [φ80dlac Δ(lacZ)M15] | Laboratory stock |
| GN40 | MC4100 ompT::kan leu::Tn10 | 33 |
| JH101 | MC4100 lamB14D prlD22 leu::Tn10 | 23 |
| KB311 | MC4100 lamB14D prlD5 leu::Tn10 | 23 |
| MC4100 | F− Δ(argF-lac)U169 araD136 relA1 rpsL150 flbB5301 ptsF25 deoC1 thi | Jon Beckwith |
| MC4100.2 | MC4100 recA1 srl::Tn10 | Laboratory stock |
| SE6004 | MC4100 prlA4 lamBS60 | 15 |
| SE4014 | MC4100 prlA3 lamBS60 rpsE | 15 |
| Plasmids | ||
| pIF-A | Apr pBR322 derivative carrying secM φ(secA-lacZ)Hyb | 29 |
| pH+ | pACYC184 derivative, CmrprlH+ | 4 |
| pH5 | pH+prlH5 | 4 |
| pH6 | pH+prlH6 | 4 |
| pNH22 | Apr pUC118 derivative carrying secM-Met6 | 33 |
| pPhIF | pIF-A derivative carrying bacteriophage M13 replication origin | 29 |
| pSS1 | pPhIF φ(secM-phoA)Hyb | 43 |
| pSS6 | pPhIF secM3 | This study |
| pSS7 | pPhIF secM4 | This study |
| pSS8 | pPhIF secM6 | This study |
| pSS9 | pPhIF secM7 | This study |
| pSS10 | pPhIF secM8 | This study |
| pSS11 | pSS1 secM3 | This study |
| pSS12 | pSS1 secM4 | This study |
| pSS13 | pSS1 secM6 | This study |
| pSS14 | pSS1 secM7 | This study |
| pSS15 | pSS1 secM8 | This study |
| pSTD343 | Cmr pACYC184 derivative carrying lac1 | 33 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Cs, cold sensitive.
DNA manipulation and oligonucleotide-directed mutagenesis.
Mutations were made by using the QuikChange procedure as described by the manufacturer (Stratagene) and were verified at the University of Pennsylvania DNA Sequencing Facility. In order to generate secM-phoA fusions containing the secM3, secM4, secM6, secM7, and secM8 alleles, pSS6, pSS7, pSS8, pSS9, and pSS10 DNA were cut with HindIII and BstBI, and the 0.57-kb HindIII-BstBI DNA fragment containing the relevant secM allele was isolated and ligated to the 4.7-kb HindIII-BstBI DNA fragment from pSS1 (43) to generate pSS11, pSS12, pSS13, pSS14, and pSS15, respectively. After transformation of CC118, blue colonies on LB plates containing 100 μg of ampicillin per ml and 20 μg of 5-bromo-4-chloro-3-indoylphosphate per ml were isolated and purified, and the fusions were verified by restriction enzyme mapping and DNA sequence analysis of the relevant plasmid DNA.
RESULTS
Effects of secM signal sequence mutations on secA regulation and SecM secretion.
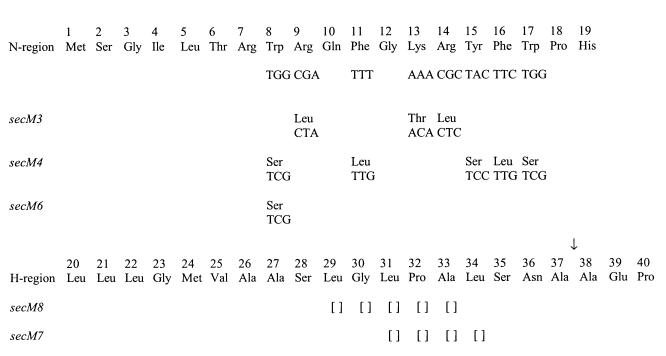
We have shown previously that secM possesses a signal peptide consisting of 37 amino acid residues with an unusually long N-region rich in basic and aromatic amino acids (Fig. 1) (43). In order to examine the importance of the N- and H-regions of the secM signal sequence in secA regulation, a number of mutations in these regions were constructed previously, and their effects on secA regulation were analyzed (43). The data indicated that both the N- and H-regions of the secM signal sequence are necessary for proper secA regulation. Truncation of the H-region by four residues (secM7) or five residues (secM8) resulted in constitutive secA expression during secretion-proficient conditions (Table 2). A mutation that reduced the positive charge within the N-region (secM3) while maintaining a single positively charged residue (Arg-7) that is often important for rapid protein secretion kinetics resulted in normal secA repression during secretion-proficient conditions but reduced induction during secretion-defective conditions. Mutations that reduced (secM6) or eliminated (secM4) the atypical aromatic amino acid residues within the N-region resulted in elevated secA expression during secretion-proficient conditions or an inability to fully induce secA expression during secretion-defective conditions (particularly for secM4).
FIG. 1.
secM signal sequence alleles used in this study. The N- and H-regions of the secM signal peptide are shown. The codon and amino acid substitutions for each allele are also shown. Empty brackets indicate deletions. An arrow indicates the presumed signal peptide processing site.
TABLE 2.
Effects of secM signal sequence alleles on secA regulation
| Mutation | Basal expression (%)a | Inductionb |
|---|---|---|
| Wild type | 100 | 6.42 |
| secM3 | 119 | 3.21 |
| secM4 | 445 | 0.77 |
| secM6 | 306 | 2.79 |
| secM7 | 513 | 1.21 |
| secM8 | 635 | 1.10 |
To determine basal expression, MC4100.2(pPhIF) (wild type) or an allelic derivative was grown in LB medium containing 100 μg of ampicillin per ml at 37°C to the mid-logarithmic phase, and then β-galactosidase assays were performed at 30°C in duplicate for each of two duplicate cultures as described previously (31). β-Galactosidase activities are expressed as percentages of the wild-type activity, which was defined as 100%.
Induction was the ratio of the β-galactosidase activities present in secretion-defective CG29 [secD1(Cs)] and secretion-proficient CG155 (sec+) strains carrying pPhIF (wild type) or on allelic derivative. Strains were grown in LB medium containing 100 μg of ampicillin per ml at 39°C to the mid-logarithmic phase, and then each culture was shifted to 23°C for 4 h. β-Galactosidase assays were performed as described above. Some data were taken from reference (43).
In order to investigate the effects that these alleles had on the protein secretion function of the secM signal peptide, isogenic strains carrying a secM-phoA fusion with a relevant allele were constructed by utilizing a previous fusion in which the first 157 codons of secM were fused to Tn phoA (41). The joint in this fusion should have been prior to the translational pause site in secM (see below), thereby allowing us to study SecM-PhoA secretion unimpeded by effects on translation. This approach was necessary also because SecM is an unstable periplasmic protein that is rapidly degraded by the C-terminus-specific Tsp protease soon after its synthesis and secretion (33). As shown recently, addition of a hexamethionine tag to the C terminus of SecM (SecM-Met6) stabilized it appreciably to proteolysis (33).
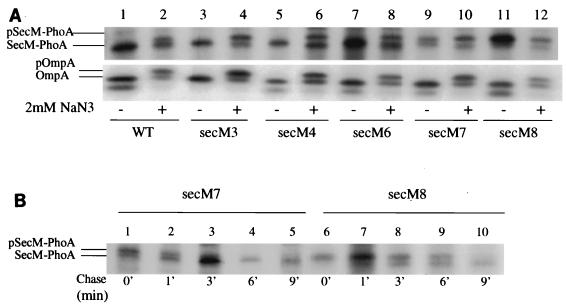
In order to assess the rate of secretion of the SecM-PhoA chimeras, the rate of processing of these chimeras was measured by utilizing pulse-chase radiolabeling methods. Since the catalytic domain of the leader peptidase is periplasmically disposed, signal peptide processing is a good method for measuring the initial rate of secretion of a secretory protein (53). As controls, portions of the cultures were also treated with sodium azide prior to labeling in order to inhibit protein secretion (36), and the synthesis and processing of OmpA protein were assessed as well. The latter control allowed us to compare the rates of synthesis of the various SecM-PhoA chimeras to the rate of synthesis of OmpA as an internal standard. Given the proximity of the secM signal sequence mutations to the translational initiation region, these alleles could have effects on the rate of secM translational initiation (19). The secM3, secM4, and secM6 mutants displayed rapid processing kinetics that were indistinguishable from those of the wild type (Fig. 2A). In contrast, the secM7 and secM8 mutants showed significant accumulation of the precursor form of SecM-PhoA during the 1-min pulse-labeling period. Additional pulse-chase analysis indicated that the secM7 and secM8 mutants had processing half-lives of the SecM-PhoA chimera of approximately 1 and 6 min, respectively (Fig. 2B). Of note were the relatively modest effects that the H-region truncations had on the secretion function of the secM signal peptide compared to the effects of similar mutations in other systems (2, 16). By contrast, the effects that these mutations had on secA regulation and secM translational pausing were more marked (Table 2) (see below). We also measured the levels of alkaline phosphatase activity of these strains, but our analysis was inconclusive due to the variable degrees of proteolysis of the SecM-PhoA chimeras (data not shown).
FIG. 2.
Effects of secM signal sequence mutations on processing. (A) MC4100.2 containing pSS1 (WT) or an allelic derivative was grown in M63 minimal medium containing 0.4% glucose, 2 μg of thiamine per ml, 20 μg of each of 18 amino acids (not including methionine and cysteine) per ml, and 20 μg of ampicillin per ml at 37°C until the mid-logarithmic phase. Sodium azide (NaN3) was not added (−) or was added to a final concentration of 2 mM (+), and labeling was initiated after 5 min. A 0.5-ml aliquot of each culture was pulse-labeled with 10 μCi of Tran 35S label (>1,000 Ci mmol−1) for 1 min, and then an equal volume of ice-cold 10% trichloroacetic acid was added to terminate labeling. Samples were processed for immunoprecipitation with antisera to alkaline phosphatase and OmpA and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and fluorography as described previously (36). The positions of the precursor and mature forms of the SecM-PhoA fusion proteins (pSecM-PhoA and SecM-PhoA, respectively) and OmpA (pOmpA and OmpA, respectively) are indicated on the left. Both the precursor and mature forms of OmpA migrated as two bands, which were the heat-modifiable and non-heat-modifiable forms (22). (B) Similar to panel A, except that a mixture of methionine and cysteine (final concentration of each, 200 μg/ml) was added after 1 min of labeling to initiate the chase (0 min) and aliquots were removed at different times and mixed with an equal volume of ice-cold 10% trichloroacetic acid to terminate the chase.
Effects of secM signal sequence mutations on translational pausing.
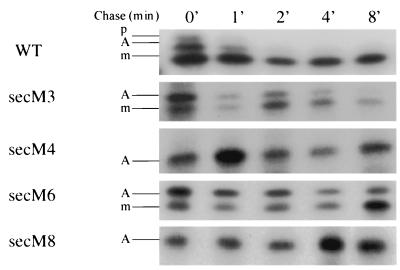
The existence of a translational pause within the distal portion of secM was demonstrated recently, and the duration of this pause was shown to be dependent on the activity of the secM signal sequence and secretion machinery (33). We studied the effects that the secM signal sequence mutations had on translational pausing utilizing the system developed by Nakatogawa and Ito (33). The wild-type strain synthesized three species of SecM protein after a 1-min pulse-labeling period, corresponding to preSecM-Met6, translationally paused SecM, and mature SecM-Met6 (Fig. 3), as observed previously (33). The paused species of SecM presumably still contained the SecM signal peptide, since it was located in the cytoplasm in the cell (33). Relatively small amounts of the first two species were present initially, and these species disappeared rapidly during the chase period along with a modest amount of mature SecM-Met6. In the case of the secM8 mutant essentially all of SecM protein was in the translationally paused form even during the 8-min chase period, as noted previously (33). A similar result was obtained for the secM4 mutant. The secM4 and secM8 mutants also gave similar results during an extended 20-min chase period in which only the translationally paused form of SecM was observed (data not shown). This result indicates that the secM4 and secM8 mutations have different effects on the secretion and translational pausing functions of the secM signal peptide and that the effects on the latter function are far more drastic than the effects on the former (compare Fig. 2 with Fig. 3). In contrast, although the secM3 and secM6 mutants displayed elevated levels of the translationally paused species compared to the wild type, this species did chase primarily into mature SecM-Met6. Taken together, our results are consistent with the proposal that the duration of the secM translational pause controls the frequency of secA translational initiation (33, 37). In particular, the two secM alleles that arrested translation (secM4 and secM8) exhibited strongly elevated levels of secA expression during secretion-proficient conditions (compare Fig. 3 with Table 2). By contrast, the two secM alleles that only delayed the release of the translational pause (secM3 and secM6) exhibited lower levels of secA expression, with the longer delay (secM6) corresponding to a higher level of secA expression. The latter results also suggest that the release of the secM translational pause must be significantly delayed in order to result in appreciable secA derepression.
FIG. 3.
Effects of secM signal sequence mutations on translational pausing. GN40(pSTD343) containing pNH22 (WT) or an allelic derivative was grown in M63 minimal medium supplemented with 0.4% glucose, 2 μg of thiamine per ml, 20 μg of each of 18 amino acids (not including methionine and cysteine) per ml, 20 μg of ampicillin per ml, and 10 μg of chloramphenicol per ml at 37°C until the mid-logarithmic phase, when IPTG and cyclic AMP were added at concentrations of 1 and 5 mM, respectively. Thirty minutes later an aliquot of each culture was pulse-labeled with 100 μCi of Tran 35S label (>1,000 Ci mmol−1) per ml for 1 min. Then a mixture of methionine and cysteine (final concentration of each, 200 μg/ml) was added to initiate the chase (0 min), and aliquots were removed at different times and mixed with an equal volume of ice-cold 10% trichloroacetic acid to terminate the chase. Samples were immunoprecipitated with a mixture of antisera against N- and C-terminal synthetic peptides of SecM (33) and analyzed on sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis gels by fluorography as described previously (35). p, preSecM-Met6; A, translationally paused SecM; m, mature SecM-Met6.
Effects of prl suppressors on the phenotype of secM signal sequence mutations.
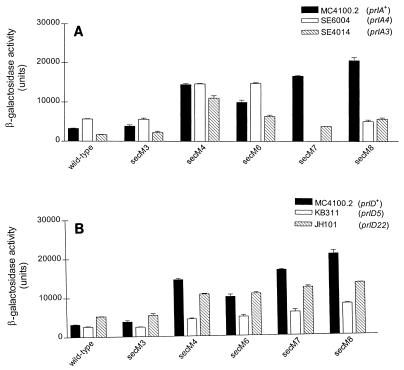
prl alleles of secA (prlD), secY (prlA), secE (prlG), and secG (prlH) that allow translocation of proteins with a defective signal peptide have been isolated (4, 15, 17, 23, 48). In order to genetically characterize the interaction of the secM signal peptide with the secretion machinery and its effect on secA regulation, we examined the effects that the secM signal sequence alleles had on secA regulation utilizing different prl suppressor strains. The most striking effects were observed for the prlA and prlD mutants. The H-region mutations were generally suppressed by the prlA alleles, as indicated by restoration of secA repression (Fig. 4A) (43). Interestingly, although prlA4 has been found to be a stronger suppressor of signal sequence defects in the H-region than prlA3 (14, 15), comparable suppression activities were observed for these two alleles with secM8. In addition, the secM7 allele was found to be synthetically lethal with prlA4, further confirming the importance of this interaction. This result may have been due to an unproductive interaction between the SecM7 protein and the PrlA4-containing translocon that led to translocon jamming, although further studies are required to explore this hypothesis. The N-region mutations showed little or no suppression with the prlA alleles. The most notable effects were with secM6; the two prlA alleles had modest but opposite effects on secA regulation in this case. In contrast to the results described above, both the N- and H-region mutations were suppressed by prlD alleles, particularly prlD5, although the degree of suppression of the H-region mutations was less than that observed for prlA (compare Fig. 4A and B). It has been noted previously that prlD alleles are efficient suppressors of defects within the N-region of signal sequences (39). The prlG and prlH suppressors had little or no effect on this system despite the fact that the strongest prlG and prlH alleles available (4, 18) were used (data not shown). Although these genetic studies were indirect, they did support the notion that proper interaction of the secM signal peptide with SecA and SecY proteins is important for control of secA regulation.
FIG. 4.
Effects of prl suppressors on the phenotypes of secM signal sequence mutations. A strain containing pPhIF (wild type) or its allelic derivative was grown in LB broth containing 100 μg of ampicillin per ml and 5 μg of chloramphenicol per ml, when necessary, at 37°C to the mid-logarithmic phase. β-Galactosidase assays were performed as described in Table 2, footnote a. (A) prlA suppressor; (B) prlD suppressor.
TPE mutations lie within the secM translational pause site.
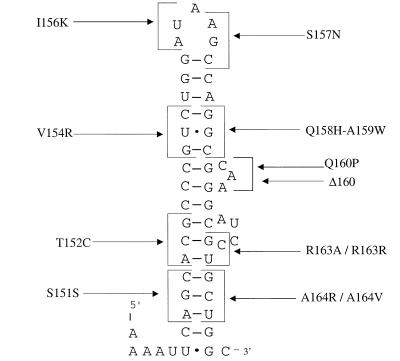
In previous analyses of secA regulation, two different classes of mutations within the distal portion of secM and the secM-secA intergenic region were studied (29, 42). Class II mutations lie within the repressor helix (helix II) that normally cloisters the secA Shine-Dalgarno sequence. It was predicted that these mutations would disrupt this helix, and they were found to render secA expression constitutive. Class I mutations were constructed within a second predicted helix (helix I) immediately upstream of helix II in order to test its importance in secA regulation. Certain class I mutations rendered secA expression noninducible, although the mutational pattern suggested that the RNA secondary structure may not be important for secA regulation (29). In particular, mutations on the 3′ side of helix I elicited the most defective phenotype. Below, we refer to the latter portion of helix I sequences as the TPE (three prime element) region.
Given the existence of a translational pause site in the distal portion of secM, we decided to reinvestigate the importance of helix I sequences, particularly the TPE region, in translational pausing and secA regulation. In particular, both the location and the phenotype of the TPE mutations were consistent with their lying within the secM translational pause site. Accordingly, utilizing Watson-Crick base pairing rules (including allowance for G-U base pairs) and the ambiguity of the genetic code, we designed additional class I mutations that either would disrupt the predicted RNA secondary structure while conserving a particular amino acid residue or, alternatively, would minimally perturb the predicted RNA secondary structure while altering a given amino acid residue. We reasoned that this approach might allow us to locate the secM translational pause site and determine whether RNA or peptide sequences (or both) are operative in the translational pausing mechanism.
A summary of class I mutations and their effects on secA regulation is shown in Table 3. Mutations in the 5′ portions of helix I sequences (at codons 151, 152, and 154) (Fig. 5) that either disrupted the predicted RNA secondary structure or altered a particular amino acid residue or both had little effect on secA regulation (29). The minor differences between mutants with these mutations and the wild type may have been due to reduced stability of secM-secA mRNA or another nonspecific effect. Similar results were obtained for mutants with mutations in the loop region of helix I (at codons 156 and 157) or the upper portion of the 3′ side of the helix (at codons 158 and 159). The rare codon AUA at position 156 did not appear to be important for secA regulation, since synonymous or nonsynonymous substitutions that utilized more abundant tRNAs had little effect on secA regulation. Deletion or an amino acid substitution in a predicted bulge (at codon 160) on the 3′ side of helix I resulted in a modest decline in secA basal expression and induction, suggesting that this region may help enhance secA expression and regulation by some means. Most strikingly, however, mutations in the distal portion of helix I sequences within the TPE region (at codons 163 and 164) resulted in a decline in secA basal expression and elimination of secA induction. In such cases it appeared that the amino acid sequence of SecM rather than the predicted RNA secondary structure or sequence was the important factor. For example, a mutation that maintained the former but perturbed the latter resulted in correct secA regulation (compare R163R with R163A), whereas the converse resulted in a loss of secA regulation (A164V).
TABLE 3.
secA regulation in class I mutants
| Mutant | Codon | Typea | Basal expression (%)b | Inductionc |
|---|---|---|---|---|
| Wild type | 100 | 6.26 | ||
| S151Sd | AGC→UCG | HD | 133 | 5.21 |
| T152Cd | ACG→UGC | AA + HD | 141 | 4.06 |
| T152M | ACG→AUG | AA | 83 | 5.89 |
| V154Rd | GUC→CGG | AA + HD | 75 | 6.26 |
| I156K | AUA→AAA | AA | 80 | 4.12 |
| I156R | AUA→AGA | AA | 94 | 3.57 |
| I156T | AUA→ACA | AA | 124 | 4.33 |
| I156T | AUA→ACC | AA | 86 | 3.36 |
| I156I | AUA→AUC | AA | 101 | 5.57 |
| S157N | AGC→AAC | AA | 87 | 4.74 |
| S157I | AGC→AUC | AA | 84 | 4.73 |
| S157T | AGC→ACC | AA | 106 | 4.42 |
| Q158H-A159Wd | GCG→CUG | AA + HD | 90 | 5.52 |
| Δ160d | CAA→Δ | AA | 49 | 3.26 |
| Q160P | CAA→CCA | AA | 31 | 3.50 |
| R163Ad | CGU→AGC | AA + HD | 30 | 1.26 |
| R163R | CGU→AGA | HD | 106 | 5.08 |
| A164Rd | GCU→CGA | AA + HD | 90 | 1.00 |
| A164V | GCU→GUU | AA | 64 | 0.68 |
FIG. 5.
Proposed RNA secondary structure for helix I. secM mutants are indicated by codons and single-letter codes for amino acids. The structure was taken from reference 29.
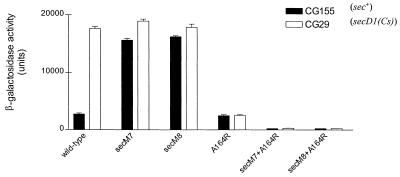
We next turned our attention to genetic and biochemical experiments to prove that the TPE region is the secM translational pause site and that TPE mutants are defective in translational pausing. If TPE mutations are defective in the secM translational pause, then they should be epistatic to secM signal sequence mutations. This is logical because the secM signal sequence is not required for initiation of the translational pause itself but rather is required for release of the pause (33). Accordingly, we constructed secM double mutants that contained both H-region (secM7 or secM8) and TPE (secM-A164R) mutations and analyzed their effects on secA regulation. While the H-region mutants were constitutive for secA expression and the TPE mutant was noninducible, the double mutants were also noninducible (Fig. 6). However, they displayed a lower level of secA expression than the TPE mutant, indicating that the secM signal sequence may have some effect on reducing TPE function.
FIG. 6.
Genetic interaction of secM signal sequence and TPE alleles. CG155 or CG29 carrying pPhIF (wild type) or an allelic derivative was grown and assayed for β-galactosidase activity as described in Table 2, footnote a.
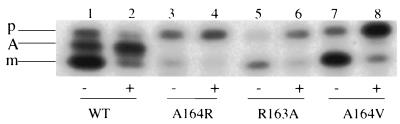
The class I mutants were also directly examined to determine their effects on the secM translational pause. While the wild-type strain synthesized the three SecM species, including the translationally paused species, the strains carrying the TPE mutations that eliminated secA induction synthesized only preSecM-Met6 and mature SecM-Met6 and therefore were defective in translational pausing (Fig. 7). This pausing defect may also account for the lower level of SecM-Met6 observed in this case, since it would have accelerated the kinetics of SecM-Met6 secretion to the periplasm, where it would have been subjected to limited proteolysis. Even pretreatment of the latter cultures with sodium azide to induce a protein secretion block did not result in appearance of the translationally paused species, although it did result in a greater accumulation of preSecM-Met6. Selected class I mutations in the 5′ region of helix I (T152C), the loop region (I156K), or the 3′ bulge (Q160P) gave patterns of translational pausing that were similar to that of the wild type (data not shown). These results demonstrate that codons 163 and 164 of secM are part of its translational pause site, and they agree with the observations of Nakatogawa and Ito that the secM translational pause site is located quite close to the 3′ end of secM (33). They also demonstrate that most of the helix I sequences are unimportant in the secM translational pause.
FIG. 7.
Effects of secM TPE mutations on translational pausing. GN40(pSTD343) containing pNH22 (WT) or an allelic derivative was grown and pulse-labeled with 100 μCi of Tran 35S label per ml for 1 min, and samples were processed and visualized as described in the legend to Fig. 3. Where indicated (+), sodium azide was added 5 min prior to labeling at a final concentration of 2 mM. p, preSecM-Met6; A, translationally paused SecM; m, mature SecM-Met6. The data are representative of the data obtained in three separate experiments.
DISCUSSION
In this work we investigated the role that the secM signal sequence plays in promoting SecM protein secretion, secM translational pausing, and secA regulation. A number of interesting and important conclusions were reached. It is clear that the N- and H-regions of the secM signal peptide have different functions with respect to these properties. Mutations in the H-region affected the rate of SecM protein secretion, the duration of the secM translational pause, and the fidelity of secA regulation, while mutations in the N-region affected only the latter two functions. This indicates that the N-region, which is unusually long and rich in basic and aromatic amino acids, plays a more exclusive role in promoting secA regulation by modulation of the secM pause-release cycle. While it is uncertain what this role is, analysis of the available SecM sequences showed that both the N-region and the early H-region of the secM signal peptide are highly conserved (Fig. 8). Recently, Nakatogawa and Ito showed that the secM signal peptide was not required for the secM translational pause but it was needed to promote the release through proper interaction with the secretion machinery (33). We speculate that the N-region makes important contact with one or more components of the Sec machinery in order to facilitate this event. While H-regions of signal peptides have been shown to interact with SecA and the translocon as well (32), it appears that in this specialized case H-region interaction is insufficient to promote proper signaling of the translation and secretion machinery and that an additional module (i.e., the atypical N-region) is required as well. Clearly, it will be of interest to investigate the detailed biochemical mechanisms that accomplish such coordination in order to facilitate the secretion-responsive regulation of secA.
FIG. 8.
Alignment of SecM proteins. SecM proteins from Escherichia coli (secMec) (43), Salmonella enterica serovar Typhi (secMst), Klebsiella pneumoniae (secMkp), and Yersinia pestis (secMyp) were multiply aligned by the Clustal X method (49). The S. enterica and Y. pestis sequences are available at ftp://ftp.sanger.ac.uk/pub/pathogens, and the K. pneumoniae sequence was produced by the Genome Sequencing Center at Washington University, St. Louis, Mo. (personal communication). Asterisks indicate completely conserved amino acid residues, while the arrow indicates the predicted signal peptide processing site.
Of note in our study were the relatively modest effects that the H-region truncations had on the secretion function of the secM signal sequence compared to the more dramatic effects observed for secM translational pausing and secA regulation. By comparison, similar mutations in other systems had more severe effects on secretion of the cognate protein. For example, truncation of the H-regions of the signal peptides of maltose-binding protein and lambda receptor led to strong secretion defects for these two proteins (2, 16). Genetic reversion analysis in the latter case, however, suggested that the proximity of α-helix-disruptive proline and glycine residues was responsible for the observed defect (16). It is important to note that the secM H-region is relatively rich in leucine residues and that four of six leucine residues remain in the secM7 and secM8 signal peptides. It also appears that conservation of the early portion of the secM H-region is more important than conservation of the later part, where the secM7 and secM8 alleles reside (Fig. 8). It has been shown that H-region function is highly dependent on net hydrophobicity, as well as mean hydrophobicity per residue, and that polyleucine-containing H-regions can therefore function efficiently when the regions are comparatively short (6, 8). This property should give the secM signal peptide a competitive advantage over other signal peptides for interacting with the Sec machinery. In this regard it has been found that relatively small differences in H-region hydrophobicity can have relatively large effects on preferential secretion of one protein over another (5). Preferential interaction of nascent preSecM with the Sec machinery could be important for the establishment of a low basal level of secA expression, since this association would promote release of the secM translational pause and resumption of secA translational repression.
In part of our genetic analysis we used prl suppressors in order to investigate the interaction of the secM signal peptide with the Sec machinery and its effect on secA regulation. While this type of study needs to be ultimately linked to biochemical investigations, some of our data are highly suggestive of specific interactions. In particular, the strong suppression and synthetic lethality of the H-region mutations with prlA, as well as the allele specificity noted for prlD and the N- and H-region mutations, suggest that SecY and SecA interact with these regions of the secM signal peptide. These inferences are consistent with the results of previous genetic and biochemical studies which indicated that SecYE and SecA interact with the H-region of signal peptides and with the N- and H-regions of signal peptides, respectively (1, 14, 23, 32, 39). By contrast, our data for the prlG and prlH suppressors were less striking, both from the standpoint of the strength of suppression and from the standpoint of allele specificity. The data suggest that the interaction of the secM signal peptide with SecA and SecY proteins is a key factor in controlling the steps that lead to correct secA regulation.
In part of our study we focused on the effects that mutations in the secM signal sequence and 3′ region had on the duration of the secM translational pause. This allowed us to directly correlate this property with the observed pattern of secA regulation and to obtain support for the secM translational pause-arrest model of secA regulation. We obtained evidence through examination of the different secM alleles that the duration of the secM translational pause was consistent with the observed secA expression levels. Our results may have been affected somewhat by the possibility that certain secM alleles could have other effects on secA expression, such as altering secM-secA mRNA folding or half-life. However, the consistency of our data, along with the observed prl-dependent suppression of these mutations, indicated that such considerations were minimal.
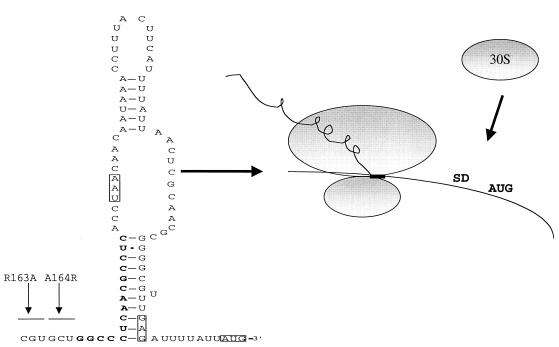
Our analysis of helix I sequences allowed us to locate residues at the 3′ end of secM within the TPE region that promote translational pausing, as well as to conclude that the pausing mechanism appears to be operative at the peptide level rather than the RNA level. The latter inference is consistent with previous results obtained in our laboratory, in which double frameshift mutations revealed the importance of the translational reading frame in the later portion of secM, as well as the results of Nakatogawa and Ito, who found that the proline analog azetidine inhibited the translational pause (29, 33). While the pausing mechanism itself remains to be elucidated, we note that the region identified (codons 163 and 164) falls within a hexapeptide sequence (GIRAGP) that is completely conserved in the available secM homologues (Fig. 8). There are no rare codons in this six-codon region or the last four codons of secM, which argues against the importance of limiting charged tRNAs in the pausing mechanism. Furthermore, the sequence in this region is more highly conserved at the amino acid level than at the RNA level, indicating the importance of a peptide in the translational pausing mechanism. Nascent peptide sequences that promote translational pausing have been found previously (for example, in control of chloramphenicol resistance in bacteria, which is referred to as translational attenuation [for a review, see reference 25). The precise location of the sequence is completely consistent with its ability to induce secA expression (Fig. 9). In particular, the translating ribosome, which should sequester at least 15 nucleotides 3′ of codon 164 of secM (21), should stall over mRNA sequences that normally comprise the 5′ portion of helix II, thereby activating the secA translational initiation region by exposure to the translational apparatus (29, 42).
FIG. 9.
Model for secA regulation. The proposed structure of the secA repressor helix (helix II) and its disruption by the secM translational pause are shown. The positions of the TPE mutations in codons 163 and 164 are indicated, along with the positions of 3′ nucleotides that would be sequestered by the translating ribosome (boldface type and box). The termination codon of secM (UAA) and the secA Shine-Dalgarno (SD) and the initiation codon (AUG) are enclosed in boxes. 30S, 30S ribosomal subunit. The structure of the secA repressor helix was taken from reference 29.
It is too early to say what specific elements control the duration of the secM translational pause beyond the need for appropriate N- and H-regions of the secM signal sequence and a functional interaction with the Sec machinery (33, 37, 43). One attractive model is that the pause is released simply by the mechanical action of SecA as it threads nascent preSecM into the translocon and dislodges the stall peptide from the translational apparatus. In this scenario secA induction occurs when the stalled nascent preSecM translational complex is prevented from docking with SecA and the translocon due to blockage of the latter components by other presecretory and membrane proteins. While our kinetic analysis of certain secM signal sequence mutants may appear to be at odds with this model (given the temporal disparity between rapid signal peptide processing and slower translational pause release), there is no reason that the translational pause release cannot occur later in the translocation of SecM protein, particularly given the loop model in which the N-region of the SecM signal peptide remains cytoplasmically exposed during SecM translocation (for a review, see reference 9). Furthermore, our results with the secM-phoA fusions probably do not depict the correct sequence of events since these fusions lack the secM translational pause site. Indeed, in the wild-type system there was no evidence of two translationally paused species that differed in the presence and absence of the secM signal peptide, indicating that translocation and processing are probably coordinated with the translational pause event (33; this study). More complex regulatory models can be envisioned as well; for example, the buildup of translocation intermediates of other presecretory and membrane proteins could titrate away a factor that is needed to promote the translational pause release. A role for SecA RNA helicase activity in promoting secA autoregulation has been ruled out recently, since secA helicase-defective mutants showed normal secA regulation (46). This observation precludes models in which the translational pausing agent is an RNA secondary or tertiary structure that is unwound by SecA helicase activity in order to release the secM translational pause. Clearly, additional genetic and biochemical analyses that are under way will be required to reveal many of the subtleties of this complex and fascinating system.
ADDENDUM IN PROOF
Nakatogawa and Ito have recently identified a similar SecM translational arrest peptide, FXXXXWIXXXXGIRAGP, that includes the specific arrest point (Pro), and they have also identified mutations in 23S rRNA and L22 protein near the ribosomal exit tunnel that bypass the translational arrest (H. Nakatogawa and K. Ito, Cell, in press).
Acknowledgments
We thank Hitoshi Nakatogawa and Koreaki Ito for their generous provision of plasmids and antisera to SecM and Kenneth Rudd for help with the SecM sequence alignment.
This work was supported by grant GM42033 from the National Institutes of Health to D.O.
REFERENCES
- 1.Akita, M., S. Sasaki, S. Matsuyama, and S. Mizushima. 1990. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J. Biol. Chem. 265:8164-8169. [PubMed] [Google Scholar]
- 2.Bankatis, V., B. Rasmussen, and P. J. Bassford. 1984. Intragenic suppressor mutations that restore export of maltose binding protein with a truncated signal peptide. Cell 37:243-252. [DOI] [PubMed] [Google Scholar]
- 3.Beck, K., L.-F. Wu, J. Brunner, and M. Muller. 2000. Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 19:134-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost, S., and D. Belin. 1997. prl mutations in the Escherichia coli secG gene. J. Biol. Chem. 272:4087-4093. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H., J. Kim, and D. A. Kendall. 1996. Competition between functional signal peptides demonstrates variation in affinity for the secretion pathway. J. Bacteriol. 178:6658-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou, M. M., and D. A. Kendall. 1990. Polymeric sequences reveal a functional interrelationship between hydrophobicity and length of signal peptides. J. Biol. Chem. 265:2873-2880. [PubMed] [Google Scholar]
- 7.Danese, P. N., and T. J. Silhavy. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 32:59-94. [DOI] [PubMed] [Google Scholar]
- 8.Doud, S. K., M. M. Chou, and D. A. Kendall. 1993. Titration of protein transport activity by incremental changes in signal peptide hydrophobicity. Biochemistry 32:1251-1256. [DOI] [PubMed] [Google Scholar]
- 9.Duffaud, G. D., S. K. Lehnhardt, P. E. March, and M. Inouye. 1985. Structure and function of the signal peptide. Curr. Top. Membr. Transp. 24:65-104. [Google Scholar]
- 10.Duong, F., and W. Wickner. 1997. Distinct catalytic roles of the SecYE, SecG, and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 16:2756-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duong, F., and W. Wickner. 1997. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 16:4871-4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Economou, A., J. A. Pogliano, J. Beckwith, D. B. Oliver, and W. Wickner. 1995. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83:1171-1181. [DOI] [PubMed] [Google Scholar]
- 13.Economou, A., and W. Wickner. 1994. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78:835-843. [DOI] [PubMed] [Google Scholar]
- 14.Emr, S. D., and P. J. Bassford. 1982. Localization and processing of outer membrane and periplasmic proteins in Escherichia coli strains harboring export-specific suppressor mutations. J. Biol. Chem. 257:5852-5860. [PubMed] [Google Scholar]
- 15.Emr, S. D., S. Hanley-Way, and T. J. Silhavy. 1981. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell 23:79-88. [DOI] [PubMed] [Google Scholar]
- 16.Emr, S. D., and T. J. Silhavy. 1983. Importance of secondary structure in the signal sequence for protein secretion. Proc. Natl. Acad. Sci. USA 80:4599-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fikes, J. D., and P. J. Bassford. 1989. Novel secA alleles improve export of maltose-binding protein synthesized with a defective signal peptide. J. Bacteriol. 171:402-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flower, A. M., R. C. Doebele, and T. J. Silhavy. 1994. PrlA and PrlG suppressors reduce the requirement for signal sequence recognition. J. Bacteriol. 176:5607-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gold, L. 1988. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem. 57:199-233. [DOI] [PubMed] [Google Scholar]
- 20.Hartl, F.-U., S. Lecker, E. Schiebel, J. P. Hendrick, and W. Wickner. 1990. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63:269-279. [DOI] [PubMed] [Google Scholar]
- 21.Hartz, D., D. S. McPheeters, R. Traut, and L. Gold. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164:419-425. [DOI] [PubMed] [Google Scholar]
- 22.Hindennach, I., and U. Henning. 1975. The major proteins of the Escherichia coli outer cell envelope membrane. Eur. J. Biochem. 59:207-213. [DOI] [PubMed] [Google Scholar]
- 23.Huie, J., and T. Silhavy. 1995. Suppression of signal sequence defects and azide resistance in Escherichia coli commonly result from the same mutations in secA. J. Bacteriol. 177:3518-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch, H.-G., T. Hengelage, C. Neumann-Haefelin, J. MacFarlane, H. Hoffschulte, K.-L. Schimz, B. Mechler, and M. Muller. 1999. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10:2163-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovett, P. S., and E. J. Rogers. 1996. Ribosome regulation by the nascent peptide. Microbiol. Rev. 60:366-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manting, E. H., and A. J. M. Driessen. 2000. Escherichia coli translocase: the unraveling of a molecular machine. Mol. Microbiol. 37:226-238. [DOI] [PubMed] [Google Scholar]
- 27.Manting, E. H., C. van der Does, H. Remigy, A. Engel, and A. J. M. Driessen. 2000. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 19:852-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto, G., H. Mori, and K. Ito. 1998. Roles of SecG and ATP- and SecA-dependent protein translocation. Proc. Natl. Acad. Sci. USA 95:13567-13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNicholas, P., R. Salavati, and D. Oliver. 1997. Dual regulation of Escherichia coli secA translation by distinct upstream elements. J. Mol. Biol. 265:128-141. [DOI] [PubMed] [Google Scholar]
- 30.Meyer, T., J.-F. Menetret, R. Breitling, K. Miller, C. Akey, and T. Rapoport. 1999. The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eucaryotic Sec61p complex. J. Mol. Biol. 285:1789-1800. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Mori, H., M. Araki, C. Hikita, M. Tagaya, and S. Mizushima. 1997. The hydrophobic region of signal peptides is involved in the interaction with membrane-bound SecA. Biochim. Biophys. Acta 1326:23-36. [DOI] [PubMed] [Google Scholar]
- 33.Nakatogawa, H., and K. Ito. 2001. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol. Cell 7:185-192. [DOI] [PubMed] [Google Scholar]
- 34.Nishiyama, K.-I., T. Suzuki, and H. Tokuda. 1996. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell 85:71-81. [DOI] [PubMed] [Google Scholar]
- 35.Oliver, D., and J. Beckwith. 1981. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 25:765-772. [DOI] [PubMed] [Google Scholar]
- 36.Oliver, D., R. Cabelli, K. Dolan, and G. Jarosik. 1990. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc. Natl. Acad. Sci. USA 87:8227-8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver, D., J. Norman, and S. Sarker. 1998. Regulation of Escherichia coli secA by cellular protein secretion proficiency requires an intact gene X signal sequence and an active translocon. J. Bacteriol. 180:5240-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver, D. B., and J. Beckwith. 1982. Regulation of a membrane component required for protein secretion in Escherichia coli. Cell 30:311-319. [DOI] [PubMed] [Google Scholar]
- 39.Puziss, J. W., J. D. Fikes, and P. J. J. Bassford. 1989. Analysis of mutational alterations in the hydrophilic segment of the maltose-binding protein signal peptide. J. Bacteriol. 171:2303-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi, H.-Y., and H. Bernstein. 1999. SecA is required for the insertion of inner membrane proteins targeted by the Escherichia coli signal recognition particle. J. Biol. Chem. 274:8993-8997. [DOI] [PubMed] [Google Scholar]
- 41.Rajapandi, T., K. M. Dolan, and D. Oliver. 1991. The first gene in the Escherichia coli secA operon, gene X, encodes a nonessential secretory protein. J. Bacteriol. 173:7092-7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salavati, R., and D. Oliver. 1997. Identification of elements on gene X-secA RNA of Escherichia coli required for SecA binding and secA auto-regulation. J. Mol. Biol. 265:142-152. [DOI] [PubMed] [Google Scholar]
- 43.Sarker, S., K. E. Rudd, and D. Oliver. 2000. Revised translation start site of secM defines an atypical signal peptide that regulates Escherichia coli secA expression. J. Bacteriol. 182:5592-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt, M., and D. B. Oliver. 1989. SecA protein autogenously represses its own translation during normal protein secretion in Escherichia coli. J. Bacteriol. 171:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt, M. G., E. E. Rollo, J. Grodberg, and D. B. Oliver. 1988. Nucleotide sequence of the secA gene and secA(Ts) mutations preventing protein export in Escherichia coli. J. Bacteriol. 170:3404-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt, M. O., R. M. Brosh, and D. B. Oliver. 2001. Escherichia coli SecA helicase activity is not required in vivo for efficient protein translocation or autogenous regulation. J. Biol. Chem. 276:37076-37085. [DOI] [PubMed] [Google Scholar]
- 47.Scotti, P., Q. Valent, E. Manting, M. Urbanus, A. Driessen, B. Oudega, and J. Luirink. 1999. SecA is not required for signal recognition particle-mediated targeting and initial membrane insertion of a nascent inner membrane protein. J. Biol. Chem. 274:29883-29888. [DOI] [PubMed] [Google Scholar]
- 48.Stader, J., L. J. Gansheroff, and T. J. Silhavy. 1989. New suppressors of signal-sequence mutations, prlG, are linked tightly to the secE gene of Escherichia coli. Genes Dev. 3:1045-1052. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian, H., D. Boyd, and J. Beckwith. 2000. A mutant hunt for defects in membrane protein assembly yields mutations affecting the bacterial signal recognition particle and Sec machinery. Proc. Natl. Acad. Sci. USA 97:4730-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Wolk, J., P. Fekkes, A. Boorsma, J. Huie, T. Silhavy, and A. Driessen. 1998. PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J. 17:3631-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Wolk, J. P. W., J. G. de Wit, and A. J. M. Driessen. 1997. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 16:7297-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfe, P. B., W. Wickner, and J. M. Goodman. 2080. 1983. Sequence of the leader peptidase gene of Escherichia coli and the orientation of leader peptidase in the bacterial envelope. J. Biol. Chem. 258:12073-12080. [PubMed] [Google Scholar]