Abstract
The stringent response in Bacillus subtilis was characterized by using proteome and transcriptome approaches. Comparison of protein synthesis patterns of wild-type and relA mutant cells cultivated under conditions which provoke the stringent response revealed significant differences. According to their altered synthesis patterns in response to dl-norvaline, proteins were assigned to four distinct classes: (i) negative stringent control, i.e., strongly decreased protein synthesis in the wild type but not in the relA mutant (e.g., r-proteins); (ii) positive stringent control, i.e., induction of protein synthesis in the wild type only (e.g., YvyD and LeuD); (iii) proteins that were induced independently of RelA (e.g., YjcI); and (iv) proteins downregulated independently of RelA (e.g., glycolytic enzymes). Transcriptome studies based on DNA macroarray techniques were used to complement the proteome data, resulting in comparable induction and repression patterns of almost all corresponding genes. However, a comparison of both approaches revealed that only a subset of RelA-dependent genes or proteins was detectable by proteomics, demonstrating that the transcriptome approach allows a more comprehensive global gene expression profile analysis. The present study presents the first comprehensive description of the stringent response of a bacterial species and an almost complete map of protein-encoding genes affected by (p)ppGpp. The negative stringent control concerns reactions typical of growth and reproduction (ribosome synthesis, DNA synthesis, cell wall synthesis, etc.). Negatively controlled unknown y-genes may also code for proteins with a specific function during growth and reproduction (e.g., YlaG). On the other hand, many genes are induced in a RelA-dependent manner, including genes coding for already-known and as-yet-unknown proteins. A passive model is preferred to explain this positive control relying on the redistribution of the RNA polymerase under the influence of (p)ppGpp.
Bacterial genes encoding products of similar adaptational functions are frequently coregulated. This organization ensures a balanced production of all proteins necessary for adaptation to a change in the environment. Two-dimensional (2D) protein gel electrophoresis is a powerful and highly sensitive tool for defining sets of coregulated proteins. Genes coding for such sets of proteins may form regulons, the basic modules of global gene expression. The sequence of the total Bacillus subtilis genome (49) is crucial for the rapid identification of protein spots on 2D gels by N-terminal sequencing (1, 7, 8) or by mass spectrometry (MS) techniques (e.g., matrix-assisted laser desorption ionization-time of flight [MALDI-TOF MS]) (3). These data were integrated into a B. subtilis proteome database called Sub-2D that is available via the World Wide Web (http://microbio2.biologie.uni-greifswald.de:8880/sub2d.htm).
In order to define the organization of a regulon, the protein synthesis pattern of the wild type has to be compared to that of a mutant strain carrying a null mutation in the corresponding global regulatory gene. Genes which are no longer induced or repressed in the mutant may belong to this regulon. In this way comprehensive 2D protein maps allow the allocation of proteins and/or genes to regulons. If the function of the regulon is still unknown, this approach can be used to predict its physiological role on the basis of the already-characterized proteins belonging to the regulon. This prediction, however, has to be proven by phenotypic analysis of the corresponding mutants. This approach was successfully used to dissect the heat stress stimulon of B. subtilis into distinct regulons and to predict the function of the σB-dependent general stress regulon (37-39).
In natural ecosystems, bacteria are subjected to a variety of stress and starvation conditions and have therefore developed a highly sophisticated network of adaptational responses to cope with these situations. One crucial component of this adaptational network is the (p)ppGpp-dependent stringent response (12, 14) that coordinates the global transcriptional pattern with the current growth conditions. In Escherichia coli, the relA gene encodes a ribosome-bound (p)ppGpp (guanosine tetra- and pentaphosphate) synthetase that catalyzes the transfer of a pyrophosphoryl group from ATP to the 3′-hydroxyl group of GTP. This protein acts as a sensor of amino acid starvation because it is activated by the arrival of an uncharged tRNA at the ribosome (12). In response to glucose starvation the product of the spoT gene, which is primarily responsible for (p)ppGpp degradation, catalyzes (p)ppGpp synthesis (12). The “alarmone” (p)ppGpp acts not only as an indicator of starvation but also as an organizer of the adaptive cellular response to starvation, i.e., the stringent response. Reactions appropriate for growing cells (e.g., ribosome synthesis) are switched off, and adaptive responses to nutrient starvation are induced. Recently, involvement of (p)ppGpp was also demonstrated for the induction of rpoS, which controls the general stress and starvation response in gram-negative bacteria (27, 51; for review on the regulation of rpoS, see reference 40). Therefore, the stringent control represents a crucial adaptive strategy essential for survival of E. coli cells in a nutrient-limited natural environment (12, 35).
In B. subtilis, the relA gene seems to represent the only (p)ppGpp synthetase, and it may be involved in amino acid, glucose, and oxygen starvation responses (36, 62, 89). In contrast to E. coli, the general stress response in B. subtilis, triggered by stress or energy depletion via the activation of the alternative sigma factor σB, is not induced by amino acid starvation or the stringent response (23, 56, 88). It is reasonable to assume that there is a considerable interplay between both responses in order to accomplish survival of extended periods of nutrient starvation. Whereas the stringent response is responsible for preventing the waste of nutrients during starvation, the general stress response provides the nongrowing cell with a preventive multiple stress resistance in “anticipation of future stress” (39, 70). Preliminary studies using the proteome approach to define the stringent response of B. subtilis were done by Hecker et al. (36) and Wendrich and Marahiel (89). For a better understanding of the stringent response, a more comprehensive analysis aiming for the detection of all stringently controlled genes is necessary. In this study, proteome and transcriptome analysis was combined in order to define the “RelA regulon.”
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The B. subtilis strains used in this study were BR16 (trpC2 lys), BR17 (trpC2 lys relA) (84), and BCE16 (trpC2 lys ΔrelA::mls). Strain BCE16 was constructed by transformation of strain BR16 with chromosomal DNA from strain TW30 (89). The genotype of strain BCE16 (ΔrelA) was confirmed by Southern hybridization and PCR. Cells were cultivated in minimal medium (82) supplemented with 0.2% (wt/vol) glucose, 50 μg of tryptophan/ml, 50 μg of lysine/ml, and for BCE16, also with 1 mM valine, isoleucine, and leucine. The medium contains 15 mM (NH4)2SO4 as nitrogen source. For induction of the stringent response, dl-norvaline (Sigma) was added at a final concentration of 500 μg/ml (wt/vol) to exponentially growing cells (optical density at 500 nm [OD500] = 0.4). The stringent (BR16) or relaxed (BR17) phenotypes were verified by measuring [3H]uridine incorporation into RNA as described earlier (34).
Pulse-labeling and 2D protein gel electrophoresis.
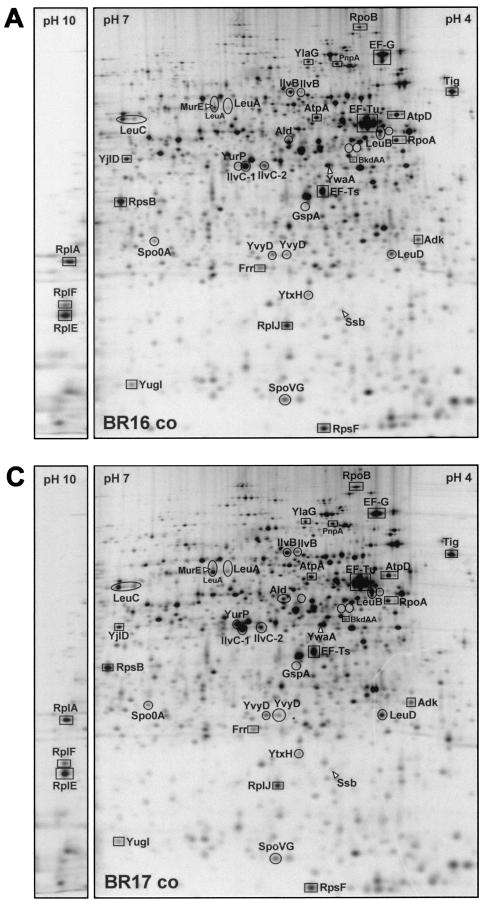
B. subtilis strains BR16, BR17, and BCE16 were grown in 50 ml of minimal medium. Then, 5-ml samples were harvested from exponentially growing cells (OD500 = 0.4) and from cells at several time points (5, 10, 20, 30, and 60 min) after treatment with norvaline and labeled for 5 min with 10 μCi of l-[35S]methionine (Amersham Pharmacia Biotech)/ml. Three such experiments were performed. Sample preparation and 2D protein gel electrophoresis were performed as described by Bernhardt et al. (8). Separation of 50 μg of radioactively labeled protein extracts was carried out on Immobiline strips (Amersham Pharmacia Biotech) in the pH ranges of 4 to 7, 4.5 to 5.5, and 3 to 10. Gels were silver stained, dried, exposed to storage Phosphor screens, and then scanned with a PhosphorImager SI (Molecular Dynamics) as described previously (8). Dual-channel images were created from the silver-stained gels and the corresponding autoradiograms by using the software DECODON Delta2D (DECODON GmbH Greifswald, Greifswald, Germany). The autoradiograms of the gels in the pH range from 4 to 7 were analyzed with the DECODON Delta2D software (DECODON GmbH Greifswald).
Identification of proteins by MALDI-TOF MS.
The dual-channel images of silver-stained gels and corresponding autoradiograms (described above) facilitate the identification of unknown protein spots by MALDI-TOF MS analysis because the content and synthesis rate of proteins of a bacterial culture are visualized in the same electropherogram (8). Protein extracts (500 μg) isolated from exponentially growing cells before and 30 min after norvaline addition were separated on Immobiline strips in pH ranges from 4.5 to 5.5 and from 4 to 7 and compared to dual-channel images. Protein spots of interest were cut out from the 2D gel after Coomassie blue staining and subsequently digested by Trypsine-Porcine (Promega). In-gel tryptic digestion was performed by using a peptide-collecting device (67). Sample template preparation for MALDI-TOF MS (Voyager DE-STR; PerSeptive Biosytems) was carried out by mixing 0.5 μl of the resulting peptide solution with an equal volume of saturated α-cyano-4-hydroxy cinnamic acid solution in 50% (vol/vol) acetonitrile-0.1% (wt/vol) trifluoroacetic acid. Peptide mass fingerprints were analyzed by using MS-Fit software (P. R. Baker and K. R. Clauser [http://prospector.ucsf.edu]).
Transcriptome analysis by DNA macroarray hybridization. (i) Preparation of RNA.
For the preparation of high-quality RNA, a modified protocol, originally developed for extraction of RNA from Saccharomyces cerevisiae (33), was used. B. subtilis BR16 and BR17 were grown aerobically in supplemented minimal medium (described above). Then, 30-ml samples were harvested by centrifugation (for 3 min at 7,155 × g at room temperature [RT]) from exponentially growing cultures (OD500 = 0.4 to 0.5) and from cultures treated for 10 min with norvaline at a final concentration of 0.05% (wt/vol). For mechanical disruption, the pellets were resuspended in 200 μl of supernatant, immediately dropped into the disruption Teflon vessel (filled and precooled with liquid N2), and then disrupted with a Mikro-Dismembrator S (B. Braun Biotech International, Melsungen, Germany) (2 min at 2,600 rpm). The resulting frozen powder was resuspended in 3 to 4 ml of prewarmed (50°C) lysis solution (4 M guanidine thiocyanate, 0.025 M sodium acetate [pH 5.2], 0.5% N-laurylsarcosine [wt/vol]) until the solution became clear at the tip of a 1-ml pipette tip. After complete lysis the solution was immediately transferred to Eppendorf tubes and placed on ice, and the total RNA was extracted as described previously (42).
(ii) Synthesis of radioactively labeled target cDNA.
For annealing of the specific oligonucleotide primers (complementary to the mRNAs specified by all B. subtilis genes), 2 μg of total RNA was hybridized to 4 μl of cDNA labeling mix (Sigma-Genosys) in hybridization buffer (10 mM Tris-HCl, pH 7.9; 1 mM EDTA; 250 mM KCl) in a total volume of 30 μl (1 h, 42°C). After annealing, 30 μl of reverse transcription premix (12 μl of 5× First Strand Buffer [Gibco-BRL], 6 μl of 0.1 mM dithiothreitol [Gibco-BRL], 2 μl of 10 mM dATP, 2 μl of 10 mM dGTP, 2 μl of 10 mM dTTP, 4.5 μl of [α-33P]dCTP [10 μCi/μl, NEN], 1.5 μl of reverse transcriptase [Superscript II; Gibco-BRL]) was added, and reverse transcription was carried out for 1.5 h at 42°C. Next, 2 μl of 0.5 M EDTA and 6 μl of 3.0 M NaOH were added, and the solution was incubated for 30 min at 65°C, followed by another 15 min at RT. The solution was neutralized with 20 μl of 1 M Tris-HCl (pH 8.0) and 6 μl of 2 N HCl, and cDNA was precipitated by the addition of 10 μl of 3 M sodium acetate (pH 5.2) and 400 μl of ethanol and freezing overnight at −20°C. cDNA was pelleted by centrifugation at 17,600 × g for 15 min at 4°C, washed with 70% (vol/vol) ethanol, dried, and resuspended in 100 μl of sterile water. Labeling efficiency was determined by liquid scintillation measurement.
(iii) Hybridization.
B. subtilis arrays (Sigma-Genosys; carrying PCR products which represent all B. subtilis protein-coding genes [n = 4,107]) were incubated for 10 min in 50 ml of SSPE buffer (0.18 M NaCl; 10 mM sodium phosphate, pH 7.7; 1 mM EDTA). Prehybridization was carried out in 10 ml of hybridization solution (5× Denhardt solution; 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]; 0.5% sodium dodecyl sulfate [SDS]; 100 μg of denaturated, salmon sperm DNA [Sigma]/ml) for 2 h at 65°C. Subsequently, hybridization was performed for 20 h at 65°C in 5 ml of hybridization solution containing the labeled cDNA probe which had been boiled for 5 min and rapidly cooled on ice before hybridization. Arrays were washed twice with 200 ml of 2× SSC and 0.1% (wt/vol) SDS (5 min at RT and 20 min at 65°C) and once with 200 ml of 0.2× SSC-0.1% (wt/vol) SDS (60 min at 65°C). Finally, arrays were air dried for 2 min, sealed in plastic bags, and exposed to PhosphorImager screens.
(iv) Data analysis.
Exposed PhosphorImager screens were scanned by using a Storm 860 PhosphorImager (Molecular Dynamics) at a resolution of 50 μm and a 16-bit color depth. Quantitation of the hybridization signals and background subtractions were carried out with ArrayVision software (version 5.1; Imaging Research, St. Catherines, Ontario, Canada) after direct import of the Phosphor-Imager files. Calculation of normalized intensity values of individual spots was performed by using the overall-spot normalization function of ArrayVision (see reference 68).
For each of the four growth and strain conditions (condition 1 = wild type [BR16], exponential growth; condition 2 = wild type [BR16], 10 min of norvaline; condition 3 = relA mutant [BR17], exponential growth; and condition 4 = relA mutant [BR17], 10 min of norvaline), mRNA was prepared from two independent cultivations (experiments 1 and 2 [replicates]) and then used for independent cDNA synthesis. cDNA from experiments 1 and 2 was hybridized with one of two different array batches. cDNA obtained from experiment 1 was additionally hybridized with the second array batch. In all, 12 array hybridizations were performed.
To avoid extreme intensity ratios for genes close to or below the detection limit, the average of the normalized intensity of these low values was arbitrarily set to a value corresponding to a signal-to-noise ratio of 1.0. Further analysis was carried out by using GeneSpring 3.2.12 software (Silicon Genetics). Thereby, the normalized (artifact removed) volume (nARVOL) values of significantly expressed genes should be greater than the threefold nARVOL value corresponding to signal-to-noise ratio of 1.0 in at least one condition of one experiment (described above). The average of the normalized intensity values of the duplicate spots of each gene was then used to calculate the expression level ratios in comparisons of the following categories: (i) the ratio of the expression level in norvaline-treated versus exponentially growing wild-type cells; (ii) the ratio of expression in norvaline-treated versus exponentially grown relA mutant cells. Subsequently, expression level ratios from the two different hybridizations of experiment 1 were averaged and only open reading frames (ORFs) or genes showing at least a threefold difference in their expression levels in both replicate experiments were considered. Dual-channel images were generated as described for protein gels (8). Images resulting from exponential growth (control) were green, and those resulting from norvaline treatment were red. Green images (control) and red images (10 min of norvaline) were compared by using an overlay of the two images.
Northern blot analysis.
Northern blot analysis was performed as described previously (90) with 5 μg of total RNA per lane. Digoxigenin-labeled tufA-, yvyD-, gabP-, and ureA-specific RNA probes were synthesized by in vitro transcription with T7 RNA polymerase and specific PCR products as templates. Northern hybridization was carried out with RNA prepared from wild-type and relA mutant cells in control and norvaline experiments (as described above). Synthesis of the tufA, yvyD, gabP, and ureA templates by PCR was performed by using the following oligonucleotide primers: tufA-for (5′-TCTTCGAACTTATGGATGCG-3′), tufA-rev (5′-TAATACGACTCACTATAGGGAGA/ACGTTGGATTTCTTCACGAG-3′), yvyD-for (5′-TTTGACCATAGCGTGGATG-3′), yvyD-rev (5′-CTAATACGACTCACTATAGGGAGA/CGGTACACGACATTTGTAAG-3′), gabP-for (5′-ATGAACCAGTCTCAATCAGGA-3′), gabP-rev (5′-CTAATACGACTCACTATAGGGAGA/AGGCAGGATTACGGGTTGC-3′), ureA-for (5′-ATGAAACTGACACCAGTTGAAC-3′), and ureA-rev (5′-CTAATACGACTCACTATAGGGAGA/TGACTTCACCTCCGCAGAAA-3′).
RESULTS
Differential patterns of protein synthesis in wild type versus the relA mutant after exposure to norvaline.
Addition of dl-norvaline to B. subtilis cultures limits the aminoacylation of tRNAIle and tRNALeu and, hence, induces the stringent response by mimicking isoleucine and leucine starvation (36). Stringent response induction by norvaline, which involves a rapid accumulation of (p)ppGpp (36), was verified by analysis of [3H]uridine incorporation. As shown in Fig. 1A, synthesis of stable RNA (rRNA and tRNA) was strongly inhibited after the addition of norvaline to a wild-type culture but continued in the isogenic relA mutant, whereas growth and l-[35S]methionine incorporation was inhibited in both strains (Fig. 1B and C).
FIG. 1.
(A) [3H]uridine incorporation into RNA after norvaline stress (0.05% [wt/vol]) in B. subtilis BR16 (wild type) and BR17 (relA). Control refers to RNA synthesis without norvaline addition (indicated by squares); RNA synthesis before and after norvaline addition is indicated by circles. The point of addition of norvaline (NV) is indicated by arrows. (B) Growth of B. subtilis BR16 (wild type) and BR17 (relA mutant) under control conditions (squares) and after norvaline addition (circles); solid symbols refer to BR16 (wild type), and open symbols refer to BR17 (relA mutant). (C) Percent incorporation of l-[35S]methionine as measured in cultures grown with norvaline. B. subtilis BR16 (shaded columns) and BR17 (relA) (open columns) were compared.
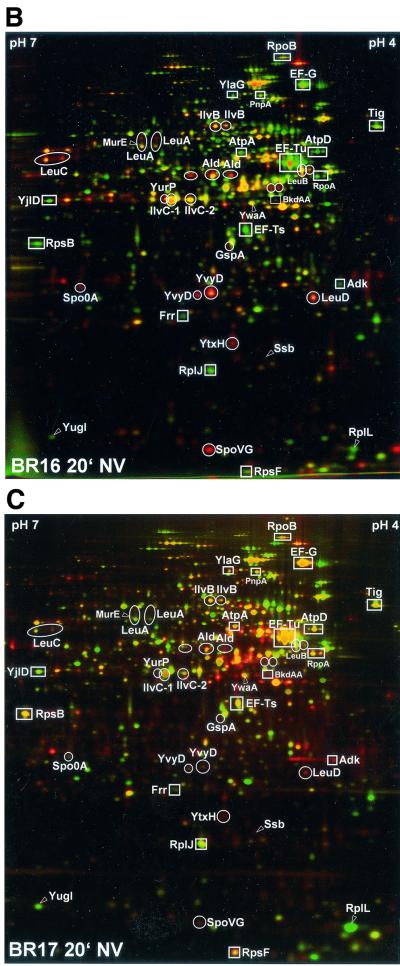
In order to examine the effects of the stringent control on the protein pattern, 2D protein gel electrophoresis of equal amounts of radioactively labeled protein extracts of exponentially grown and norvaline-treated wild-type and relA mutant cells was carried out. The created 2D gels were silver stained, dried, and exposed to PhosphorImager screens delivering autoradiograms (see Materials and Methods). The resulting autoradiograms reflect the instantaneous synthesis rates of individual proteins at the time of labeling, and the silver-stained gels represent the actual level of proteins accumulated until the time of cell harvesting. By using the dual-channel imaging technique, the synthesis rates of single proteins can be compared with their actual protein level on a single gel (8). False color images of the silver-stained gel (green channel) and the corresponding autoradiogram (red channel) (described above) were created by an overlay and matching procedure by using the DECODON Delta2D software (see Materials and Methods) (Fig. 2). The resulting red and green images reflect new synthesis (red spots = not yet accumulated), both synthesis and accumulation (yellow spots = as a combination of green and red), and repression (green spots). Green spots represent proteins that are still present in the cell but whose synthesis has been switched off.
FIG. 2.
Differential protein synthesis patterns in B. subtilis wild type (BR16) versus a relA mutant (BR17) after norvaline treatment (indication of proteins belonging to the “RelA regulon”). Dual-channel images, constructed by a combination of the silver-stained gel (green channel) and the corresponding autoradiogram (red channel), of B. subtilis BR16 (wild type) before (control [co]) (A) and after 20 min of norvaline stress (B) and of BR17 (relA strain) after 20 min of norvaline stress (C) are shown. The resulting red-green images show the whole set of newly synthesized proteins at the point of radioactive labeling (red channel), as well as the accumulated protein (green channel). Only proteins whose induction (red or orange spots) (circles) or repression (green spots) (squares) is dependent on RelA are indicated. Proteins are indicated by arrows if the RelA dependence was only demonstrated by DNA macroarray analysis (see Table 1).
Comparison of the dual-channel images of protein gels of the wild-type (stringent) strain BR16 and of the relA mutant (relaxed) strain BR17 before and after treatment with norvaline allowed the identification of proteins whose synthesis is affected by the stringent response (Fig. 2). Whereas in exponentially growing cells of both strains (shown for BR16, Fig. 2A) accumulation and synthesis are nearly in steady state (yellow color dominant), significant differences were found after norvaline treatment (Fig. 2B and C). Proteins affected by the stringent response were assigned to two classes: class I proteins were negatively controlled by the stringent response and were switched off only in the wild type (green color); class II proteins were positively regulated (orange to red color only in the wild type) (Fig. 2B and C). The autoradiograms (Fig. 3) were used to quantitate the protein synthesis rates of the proteins of classes I and II (Table 1).
FIG. 3.
Differential protein synthesis patterns in B. subtilis wild type (BR16) versus a relA mutant (BR17) after norvaline treatment (indication of proteins belonging to the “RelA regulon”). 2D protein gels (autoradiograms) of l-[35S]methionine-labeled proteins (pH 4 to 7 and basic sections from pH 3 to 10) isolated from exponentially growing B. subtilis BR16 (A) and BR17 (C), from BR16 after 20 min of norvaline stress (0.05% [wt/vol]) (B), and from BR17 after 20 min of norvaline stress (0.05% [wt/vol]) (D) are shown. These autoradiograms show only proteins synthesized during the period of the l-[35S]methionine labeling. Proteins whose induction (circles) or repression (squares) is dependent on RelA are indicated. Proteins are indicated by arrows if the RelA dependence was only demonstrated by DNA macroarray analysis (see Table 1).
TABLE 1.
Genes or proteins negatively (class I) or positively (class II) controlled by the stringent response provoked by norvaline addition, as revealed by DNA macroarray and proteome analysisa
| Class and cellular function affected | Gene (synonym)/protein | Identity/similarity/ function | (Putative) transcriptional unit | Transcriptional repression factors
|
Translational repression factors
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type | relA mutant | Wild type
|
relA mutant
|
||||||
| 10 min | 10 min | 10 min | 20 min | 10 min | 20 min | ||||
| Class I: RelA-dependent repression (negative regulation) | |||||||||
| Protein synthesis | rplK (relC) | Ribosomal protein L11 | rplK-rplA | 7.7 | 1.6 | ||||
| Protein synthesis | rplA/RplA** | Ribosomal protein L1 | rplK-rplA | 12.2 | 1.2 | ND | ND | ND | ND |
| Protein synthesis | rplJ/RplJ | Ribosomal protein L10 | rplJ-rplL-ybxB-rpoB-rpoC | 16.8 | 1.1 | 1.6 | 1.7 | 0.8 | 0.6 |
| Protein synthesis | rplL/RplL | Ribosomal protein L12 | rplJ-rplL-ybxB-rpoB-rpoC | 9.1 | 0.7 | ||||
| RNA synthesis | rpoB/RpoB | RNA polymerase (beta subunit) | rplJ-rplL-ybxB-rpoB-rpoC | 7.0 | 1.5 | 1.8 | 1.6 | 0.7 | 0.8 |
| Unknown | ybxF (ybaB) | Similar to ribosomal protein L7AE family/unknown | ybxF-rpsL-rpsG-fus-tufA | 12.1 | 1.5 | ||||
| Protein synthesis | rpsL | Ribosomal protein S12 | ybxF-rpsL-rpsG-fus-tufA | 23.2 | 1.2 | ||||
| Protein synthesis | rpsG | Ribosomal protein S7 | ybxF-rpsL-rpsG-fus-tufA | 9.3 | 1.2 | ||||
| Protein synthesis | fus/EF-G | Elongation factor G | ybxF-rpsL-rpsG-fus-tufA | 9.3 | 1.1 | 2.8 | 5.7 | 0.6 | 0.5 |
| Protein synthesis | tufA/EF-Tu* (four spots) | Elongation factor Tu | ybxF-rpsL-rpsG-fus-tufA | 7.3 | 1.1 | 2.6 | 3.7 | 0.9 | 1.0 |
| Protein synthesis | rpsJ | Ribosomal protein S10 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 10.6 | 1.4 | ||||
| Protein synthesis | rplC | Ribosomal protein L3 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 30.6 | 1.5 | ||||
| Protein synthesis | rplD | Ribosomal protein L4 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 16.3 | 1.9 | ||||
| Protein synthesis | rplW | Ribosomal protein L23 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 47.7 | 1.2 | ||||
| Protein synthesis | rplB | Ribosomal protein L2 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 38.2 | 2.5 | ||||
| Protein synthesis | rpsS | Ribosomal protein S19 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 19.1 | 1.0 | ||||
| Protein synthesis | rplV | Ribosomal protein L22 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 25.2 | 0.7 | ||||
| Protein synthesis | rpsC | Ribosomal protein S3 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 18.6 | 1.0 | ||||
| Protein synthesis | rplP | Ribosomal protein L16 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 17.1 | 1.1 | ||||
| Protein synthesis | rpmC | Ribosomal protein L29 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 20.6 | 0.5 | ||||
| Protein synthesis | rpsQ | Ribosomal protein S17 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 13.2 | 1.2 | ||||
| Protein synthesis | rplN | Ribosomal protein L14 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 16.2 | 1.1 | ||||
| Protein synthesis | rplX | Ribosomal protein L24 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 9.0 | 0.9 | ||||
| Protein synthesis | rplE/RplE** | Ribosomal protein L5 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 13.7 | 1.5 | ND | ND | ND | ND |
| Protein synthesis | rpsN | Ribosomal protein S14 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 7.1 | 1.0 | ||||
| Protein synthesis | rpsH | Ribosomal protein S8 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 6.2 | 0.8 | ||||
| Protein synthesis | rplF/RplF** | Ribosomal protein L6 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 10.6 | 0.9 | ND | ND | ND | ND |
| Protein synthesis | rplR | Ribosomal protein L18 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 6.7 | 0.6 | ||||
| Protein synthesis | rpsE | Ribosomal protein S5 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 4.0 | 0.7 | ||||
| Protein synthesis | rpmD | Ribosomal protein L30 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 4.9 | 0.8 | ||||
| Protein synthesis | rplO | Ribosomal protein L15 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 4.2 | 0.6 | ||||
| Protein synthesis | rpsE | Ribosomal protein S5 | rpsJ-rplC-D-W-B-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-X-E-rpsN-H-rplF-R-rpsE-rpmD-rplO-secY-adk-map | 4.0 | 0.7 | ||||
| Protein synthesis | rpmD | Ribosomal protein L30 | 4.9 | 0.8 | |||||
| Protein synthesis | rplO | Ribosomal protein L15 | 4.2 | 0.6 | |||||
| Protein secretion | secY | Preprotein translocase subunit | 5.6 | 0.5 | |||||
| Metabolism of nucleotides and nucleic acids | adk/Adk | Adenylate kinase | 10.3 | 0.7 | 2.6 | >6.0 | 0.8 | 0.7 | |
| Protein modification | map | Methionine aminopeptidase | 12.0 | 0.8 | |||||
| Protein synthesis | infA | Initiation factor IF-1 | infA-rpmJ-rpsM-rpsK-rpoA-rplQ | 5.6 | 1.0 | ||||
| Protein synthesis | rpmJ | Ribosomal protein L36 | infA-rpmJ-rpsM-rpsK-rpoA-rplQ | 4.7 | 0.7 | ||||
| Protein synthesis | rpsM | Ribosomal protein S13 | infA-rpmJ-rpsM-rpsK-rpoA-rplQ | 3.0 | 0.8 | ||||
| RNA synthesis | rpoA/RpoA | RNA polymerase (alpha subunit) | infA-rpmJ-rpsM-rpsK-rpoA-rplQ | 3.7 | 0.9 | 1.1 | 4.9 | 0.8 | 0.5 |
| Protein synthesis | rplQ | Ribosomal protein L17 | infA-rpmJ-rpsM-rpsK-rpoA-rplQ | 4.6 | 0.6 | ||||
| Protein synthesis | rplU | Ribosomal protein L21 | rplU-ysxB | 6.3 | 0.8 | ||||
| Unknown | ysxB | Similar to unknown proteins/unknown | rplU-ysxB | 4.2 | 0.8 | ||||
| Protein synthesis | rpsB/RpsB | Ribosomal protein S2 | rpsB | 3.6 | 0.9 | 2.1 | 4.4 | 1.0 | 0.9 |
| Protein synthesis | tsf/EF-Ts | Elongation factor Ts | tsf | 9.9 | 2.5 | 5.3 | 6.2 | 1.5 | 1.4 |
| Pyrimidine biosynthesis | pyrH (smbA) | Uridylate kinase | pyrH-frr | 9.5 | 2.5 | ||||
| Protein synthesis | frr/Frr | Ribosome recycling factor | pyrH-frr | 7.4 | 2.4 | >5.0 | >6.0 | 0.8 | ND |
| Protein synthesis | rpsD | Ribosomal protein S4 | rpsD | 9.8 | 0.8 | ||||
| Protein synthesis | rpmE | Ribosomal protein L31 | rpmE | 5.8 | 0.8 | ||||
| Protein synthesis | rpsO | Ribosomal protein S15 | rpsO | 7.5 | 1.1 | ||||
| Protein synthesis | rpsF/RpsF | Ribosomal protein S6 | rpsF-ssb-rpsR | 21.2 | 3.2 | 1.4 | 2.0 | 0.8 | 0.8 |
| DNA replication | ssb/Ssb | Single-stranded DNA-binding protein | rpsF-ssb-rpsR | 6.7 | 1.0 | ND | ND | ND | ND |
| Protein synthesis | tig/Tig* | Trigger factor (peptidyl prolyl isomerase) | tig | 6.1 | 2.2 | >5.0 | 2.0 | 1.2 | 0.7 |
| Unknown | ylaG/YlaG* | Similar to GTP-binding elongation factor/unknown | ylaG | 4.8 | 1.3 | 2.1 | 3.2 | ND | 0.7 |
| Unknown | ylxS (ymxA) | Similar to unknown proteins/unknown | ylxS-nusA-ylxR-ylxQ-infB-ylxP-rbfA-polC | 4.0 | 1.4 | ||||
| RNA synthesis | nusA | Transcription termination | ylxS-nusA-ylxR-ylxQ-infB-ylxP-rbfA-polC | 11.7 | 1.6 | ||||
| Unknown | ylxR (ymxB) | Similar to unknown proteins/unknown | ylxS-nusA-ylxR-ylxQ-infB-ylxP-rbfA-polC | 6.8 | 1.8 | ||||
| Protein synthesis | ylxQ (ymxC) | Similar to ribosomal protein L7AE family/unknown | ylxS-nusA-ylxR-ylxQ-infB-ylxP-rbfA-polC | 7.9 | 1.5 | ||||
| Protein synthesis | infC | Initiation factor IF-3 | infC-rpmI-rplT | 13.0 | 1.3 | ||||
| Protein synthesis | rpmI | Ribosomal protein L35 | infC-rpmI-rplT | 7.7 | 1.7 | ||||
| Protein synthesis | rplT | Ribosomal protein L20 | infC-rpmI-rplT | 6.5 | 1.1 | ||||
| Protein synthesis | rplS | Ribosomal protein L19 | rplS | 4.4 | 1.0 | ||||
| Protein synthesis | rpmB | Ribosomal protein L28 | rpmB | 5.4 | 1.4 | ||||
| Protein synthesis | rpsP | Ribosomal protein S16 | rpsP | 4.2 | 1.4 | ||||
| Unknown | ylbN | Unknown | ylbN-rpmF | 11.2 | 2.2 | ||||
| Protein synthesis | rpmF | Ribosomal protein L32 | ylbN-rpmF | 4.6 | 1.1 | ||||
| RNA synthesis | nusB (yqhZ) | Probably transcription termination factor | nusB | 8.8 | 1.9 | ||||
| RNA modification | truA | Pseudouridylate synthase I | truA | 3.9 | 0.4 | ||||
| Protein synthesis | rplM | Ribosomal protein L13 | rplM-rpsI | 6.6 | 1.8 | ||||
| Protein synthesis | rpsI | Ribosomal protein S9 | rplM-rpsI | 3.9 | 0.7 | ||||
| RNA modification | mc (mcS) | RNase III | mc | 3.7 | 1.6 | ||||
| Unknown | yugI/YugI | Similar to polyribonucleotide nucleotidyltransferase/unknown | yugI | 10.1 | 1.9 | >5.0 | >6.0 | ND | 1.0 |
| RNA modification | trmU (yrrA) | Probable tRNA (5-methylaminomethyl-2-thiouridylate) methyltransferase | trmU | 3.2 | 1.0 | ||||
| Cell wall | gcaD (tms, tms26) | UDP-N-acetylglucosamine pyrophosphorylase | gcaD-prs | 4.0 | 2.0 | ||||
| Nucleotide biosynthesis | prs | Phosphoribosylphosphate synthetase | gcaD-prs | 5.1 | 2.3 | ||||
| Cell wall | dltA (dae, ipa-5r) | d-Alanyl-d-alanine carrier protein ligase | dltA-dltB-dltC-dltD-dltE | 3.5 | 1.0 | ||||
| Cell wall | mbI | MreB-like protein/similar to MreB morphogene of E. coli | mbl | 3.0 | 1.1 | ||||
| Cell wall | murE/MurE | UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase | murE-mraY-murD | 7.1 | 2.6 | ND | ND | ND | ND |
| Metabolism of lipids | bkdAA (bfmBAA, bkd)/BkdAA | Branched chain alpha-keto acid dehydrogenase E1 subunit (2-oxoisovalerate dehydrogenase alpha subunit) | ptb-bcd-buk-lpdV-bkdAA-bkdAB-bkdB | 3.2 | 1.1 | ND | >6.0 | ND | 1.0 |
| Metabolism of lipids | bkdB (bfmBB) | Branched chain alpha-keto acid dehydrogenase E2 subunit (lipoamide acyltransferase) | ptb-bcd-buk-lpdV-bkdAA-bkdAB-bkdB | 3.6 | 1.0 | ||||
| Metabolism of nucleotides and nucleic acids | pnpA (comR)/PnpA | Polynucleotide phosphorylase | pnpA-ylxY? | 4.2 | 1.6 | 1.1 | 2.4 | 1.0 | 0.7 |
| Membrane bioenergetics | qoxB | Cytochrome aa3 quinol oxidase (subunit I) | qoxA-qoxB-qoxC-qoxD | 3.5 | 1.8 | ||||
| Membrane bioenergetics | qoxD | Cytochrome aa3 quinol oxidase (subunit IV) | qoxA-qoxB-qoxC-qoxD | 3.7 | 0.7 | ||||
| Membrane bioenergetics | atpB | ATP synthase (subunit a) | atpI-atpB-atpE-atpF-atpH-atpA-atpG-atpD-atpC | 5.1 | 3.0 | ||||
| Membrane bioenergetics | atpF | ATP synthase (subunit b) | atpI-atpB-atpE-atpF-atpH-atpA-atpG-atpD-atpC | 4.0 | 2.2 | ||||
| Membrane bioenergetics | atpH | ATP synthase (delta subunit) | atpI-atpB-atpE-atpF-atpH-atpA-atpG-atpD-atpC | 3.9 | 2.1 | ||||
| Membrane bioenergetics | atpA/AtpA | ATP synthase (alpha subunit) | atpI-atpB-atpE-atpF-atpH-atpA-atpG-atpD-atpC | 1.2 | 1.4 | >5.0 | >6.0 | 1.4 | 1.2 |
| Membrane bioenergetics | atpG | ATP synthase (gamma subunit) | atpI-atpB-atpE-atpF-atpH-atpA-atpG-atpD-atpC | 8.2 | 2.5 | ||||
| Membrane bioenergetics | atpD/AtpD | ATP synthase (beta subunit) | atpI-atpB-atpE-atpF-atpH-atpA-atpG-atpD-atpC | 5.4 | 1.4 | 3.9 | 6.6 | 0.6 | 0.5 |
| Membrane bioenergetics | atpC | ATP synthase (epsilon subunit) | atpI-atpB-atpE-atpF-atpH-atpA-atpG-atpD-atpC | 13.1 | 1.2 | ||||
| Specific pathways | dxs (yqiE) | Probable 1-deoxyxylulose-5-phosphate synthase | dxs? | 3.6 | 1.6 | ||||
| Unknown | yjlC | Unknown | yjlC-yjlD | 6.9 | 2.3 | ||||
| Unknown | yjlD/YjlD* | Similar to NADH dehydrogenase/unknown | yjlC-yjlD | 3.9 | 1.4 | 4.1 | >6.0 | 1.6 | 1.2 |
| Unknown | yrvE | Similar to single-stranded DNA-specific exonuclease/unknown | yrvE | 3.5 | 1.4 | ||||
| Unknown | ypuH | Similar to unknown proteins/unknown | ypuG-ypuH-ypuI | 3.2 | 1.6 | ||||
| Class II: RelA-dependent induction (positive stringent control) | |||||||||
| Biosynthesis of branched chain amino acids (Ile, Val, Leu) | ilvB/IlvB* | Acetolactate synthase (large subunit) | ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD | 1.8 | 0.4 | 4.5 | 2.4 | 1.3 | 1.1 |
| Biosynthesis of branched chain amino acids (Ile, Val, Leu) | ilvN | Acetolactate synthase (small subunit) | ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD | 3.0 | 0.3 | ||||
| Biosynthesis of branched chain amino acids (Ile, Val, Leu) | ilvC/IlvC* (two spots) | Ketol-acid reductoisomerase | ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD | 2.8 | 0.3 | 2.2 | 1.8 | 0.7 | 0.4 |
| Biosynthesis of branched chain amino acids (Ile, Val, Leu) | leuA/LeuA* (four spots) | 2-Isopropylmalate synthase | ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD | 1.8 | 0.4 | 2.3 | 1.7 | 0.7 | 1.0 |
| Biosynthesis of branched chain amino acids (Ile, Val, Leu) | leuB/LeuB* (one to two spots) | 3-Isopropylmalate dehydrogenase | ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD | 1.6 | 0.3 | 3.5 | 2.7 | 1.0 | 0.7 |
| Biosynthesis of branched chain amino acids (Ile, Val, Leu) | leuC/LeuC* (two spots) | 3-Isopropylmalate dehydratase (large subunit) | ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD | 2.3 | 0.5 | 2.3 | 1.7 | 0.6 | 0.6 |
| Biosynthesis of branched chain amino acids (Ile, Val, Leu) | leuD/LeuD* | 3-Isopropylmalate dehydratase (small subunit) | ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD | 3.5 | 0.6 | 4.5 | 2.8 | 0.6 | 0.8 |
| Unknown | ywaA/YwaA* | Similar to branched-chain amino acid aminotransferase/unknown | ywaA | 4.5 | 0.6 | 1.6 | 1.5 | ND | 0.8 |
| Metabolism of amino acids and related molecules | ureA | Urease (gamma-subunit) | ureA-ureB-ureC | 3.7 | 1.1 | ||||
| Metabolism of amino acids and related molecules | epr | Minor extracellular serine protease | epr | 6.9 | 2.1 | ||||
| Metabolism of amino acids and related molecules | vpr | Minor extracellular serine protease | vpr | 5.6 | 1.0 | ||||
| Metabolism of nucleotides and nucleic acids | adeC (ade, yzaD) | Adenine deaminase | adeC | 3.0 | 1.3 | ||||
| Unknown | yrvI | Similar to unknown proteins, unknown | relA-yrvI | 4.4 | 1.8 | ||||
| Transport | appD | Oligopeptide ABC transporter (ATP-binding protein) | appD-appF-appA | 6.7 | 0.5 | ||||
| Transport | gamP (ybfS, yzfA) | Probable PTS glucosamine-specific enzyme IICBA component | gamP | 3.7 | 1.0 | ||||
| Transport | gabP (nrg-21) | Gamma-aminobutyrate permease | gabP | 3.0 | 0.6 | ||||
| Sporulation/stationary phase | ald (spoVN)/Ald* (two to three spots) | l-Alanine dehydrogenase | ald | 6.9 | 3.0 | >5.0 | >6.0 | 0.8 | 1.5 |
| Sporulation/stationary phase | rapA (gsiAA, spo0L) | Response regulation aspartate phosphatase | rapA-phrA | 3.6 | 0.5 | ||||
| Sporulation/stationary phase | phrA | Phosphatase (RapA) inhibitor | rapA-phrA | 3.6 | 1.2 | ||||
| Sporulation/stationary phase | phrC | Phosphatase (RapC) regulator/competence and sporulation stimulation factor (CSF) | rapC-phrC | 4.4 | 1.6 | ||||
| Sporulation/stationary phase | spoVG/SpoVG* | Stage V sporulation protein, required for spore cortex synthesis, inhibitor of sporulation | spoVG | 5.8 | 0.8 | 1.7 | 2.7 | 0.9 | 0.6 |
| Sporulation/stationary phase | spo0A/Spo0A | Two-component response regulator | spo0A | 2.6 | 0.8 | 1.9 | 2.5 | ND | 1.2 |
| Sporulation/stationary phase | soj (parA) | Centromere-like function involved in forespore chromosome partitioning, inhibition of Spo0A activation | soj-spo0J | 4.4 | 1.4 | ||||
| Sporulation/stationary phase | spo0F | Two-component response regulator | spo0F | 3.7 | 0.9 | ||||
| Sporulation/stationary phase | hpr (catA, scoC) | Transcriptional repressor of sporulation and extracellular protease genes | hpr | 3.7 | 0.3 | ||||
| Adaptation to atypical conditions | gspA/GspA | General stress protein | gspA | 5.0 | 0.8 | ND | ND | ND | ND |
| Adaptation to atypical conditions | yvyD/YvyD/Hst23 (two spots) | Similar to sigma54 modulating factor of gram-negative bacteria, similar to ribosomal proteins | yvyD | 15.4 | 0.9 | 4.6 | 5.8 | 1.1 | 0.9 |
| Adaptation to atypical conditions | ytxH/YtxH | Similar to general stress protein/unknown | ytxG-ytxH-ytxJ | 3.1 | 0.8 | 2.1 | 2.0 | ND | 1.2 |
| Adaptation to atypical conditions | ytxJ | Similar to general stress protein/unknown | ytxG-ytxH-ytxJ | 4.1 | 1.2 | ||||
| Folate biosynthesis | pabB (pab) | Para-aminobenzoate synthase (subunit A) | pabB-pabA-pabC | 3.3 | 0.9 | ||||
| Mobility and chemotaxis | flgM | Flagellin synthesis regulatory protein (anti-sigma factor) | comFA-comFB-comFC-yvyF-flgM-yvyG-flgK-flgL | 3.7 | 0.7 | ||||
| Protein secretion | tepA (ylxl, ymfB) | Translocation-enhancing protein required for efficient preprotein translocation | tepA | 6.0 | 0.9 | ||||
| Protein secretion | tatAC (ynzA) | Putative component of the twin-arginine translocation pathway | tatAC-cotC | 4.3 | 1.1 | ||||
| Unknown | ytzE | Similar to transcriptional regulator (DeoR family)/unknown | ytzE | 7.9 | 1.0 | ||||
| Unknown | ydaF | Similar to acetyltransferase/ unknown | ydaF | 4.8 | 0.9 | ||||
| Unknown | yvdF | Similar to glucan 1,4-alpha-maltohydrolase/unknown | yvdF-? | 4.5 | 1.2 | ||||
| Unknown | yurP/YurP* | Similar to glutamine-fructose-6-phosphate transaminase/unknown | ?-yurP-? | 4.0 | 0.6 | ND | ND | ND | ND |
| Unknown | yrhL | Similar to acyltransferase/ unknown | yrhL-yrhK | 3.8 | 1.3 | ||||
| Unknown | ybxI (ybdS) | Similar to beta-lactamase/ unknown | ybxI | 3.0 | 1.4 | ||||
| Unknown | ywmF | Similar to unknown proteins/unknown | ywmF-csbD | 3.8 | 1.4 | ||||
| Unknown | csbD (ywmG) | SigmaB-controlled gene/unknown | ywmF-csbD | 3.8 | 1.3 | ||||
| Adaptation to atypical conditions | ydaG (yzzA) | Similar to general stress protein/unknown | ydaG | 3.4 | 0.6 | ||||
| Unknown | yxbC (yxaQ) | Similar to unknown proteins/unknown | yxbC-yxbD? | 5.7 | 0.5 | ||||
| Unknown | ytdI | Similar to unknown proteins/unknown | ytdl | 5.3 | 0.8 | ||||
| Unknown | yneR | Similar to unknown proteins/unknown | yneS?-yneR | 4.4 | 1.0 | ||||
| Unknown | yczJ | Similar to unknown proteins/unknown | yczJ | 4.4 | 1.1 | ||||
| Unknown | ybaJ | Similar to unknown proteins/unknown | ybaJ-ybaK? | 4.1 | 0.7 | ||||
| Unknown | ywpF | Similar to unknown proteins/unknown | ywpF | 4.0 | 0.9 | ||||
| Unknown | yabA | Similar to unknown proteins/unknown | ?-yabA-? | 3.9 | 1.6 | ||||
| Unknown | yjjA | Similar to unknown proteins/unknown | yjjA | 3.9 | 0.8 | ||||
| Unknown | ypiB | Similar to unknown proteins/unknown | ypiA?-ypiB-ypiF? | 3.0 | 0.7 | ||||
| Unknown | ytzB | Similar to unknown proteins/unknown | ytzB | 3.5 | 0.7 | ||||
| Unknown | yetH | Similar to unknown proteins/unknown | yetH | 3.3 | 1.1 | ||||
| Unknown | yhdX | Unknown | yhdX | 7.4 | 1.3 | ||||
| Unknown | ybdN | Unknown | ybdN | 5.3 | 1.0 | ||||
| Unknown | yscB | Unknown | yscA?-yscB | 5.2 | 1.5 | ||||
| Unknown | ykzF | Unknown | ?-ykzF-? | 3.6 | 1.2 | ||||
| Unknown | yvdC | Unknown | yvdC | 3.2 | 0.6 | ||||
| Unknown | ybyB | Unknown | ybyB | 3.1 | 0.8 | ||||
If the corresponding protein was identified on the 2D gel, the protein symbol is given in addition to the gene symbol (e.g., “tufA/EF-Tu”). For the DNA macroarray analysis, the average transcription level ratios (10 min after norvaline addition) from two different hybridizations of cDNA obtained from experiment 1 (see Materials and Methods) are indicated. Transcriptional repression factors were determined as follows: normalized intensitycontrol/normalized intensitynorvaline. Transcriptional induction factors were determined as follows: normalized intensitynorvaline/normalized intensitycontrol. For this analysis, we considered only genes showing at least a threefold difference in their expression. For proteome analysis, translation level ratios (10 and 20 min after norvaline addition; see Materials and Methods) are also included. Translational repression factors were determined as follows: % quantitycontrol/% quantitynorvaline. Translational induction factors were determined as follows: % quantitynorvaline/% quantitycontrol. ND, not defined (i.e., no separation of protein in the pH range from 4 to 7 or no spot detection by the DECODON Delta2D software under the used parameter). The “>[highest calculated expression level ratio]” value was set if an expression level ratio could not be calculated because of an extremely low (not detectable) spot in the control or after norvaline treatment. Genes or proteins that showed at least a twofold difference in their translational expression were also included even if their transcriptional expression level ratio was < 3. If the transcription data reflect the protein synthesis data, the gene or protein symbol is given in boldface. Transcriptional units (predicted and validated), with data obtained from the Subtilist database at http://genolist.pasteur.fr/SubtiList/ (59, 60), are also indicated.
Asterisks: identified by MALDI-TOF MS (this study)
Asterisks: identified by MALDI-TOF MS (66). Other proteins were reallocated from the Sub2D 2D database (http://microbio2.biologie.uni-greifswald.de:8880/sub2d.htm [8]).
Synthesis of proteins of class I was strongly decreased only in the wild type (negative stringent control).
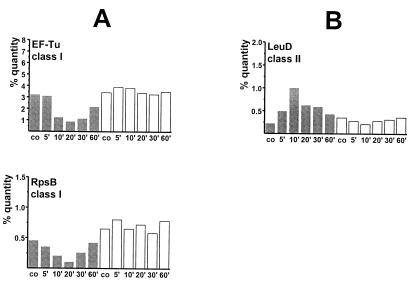
As shown in Fig. 2 and 3, the synthesis rates of translation factors (EF-Tu, EF-G, EF-Ts, and YlaG [a protein similar to GTP-binding elongation factors]), the trigger factor Tig, ribosomal proteins (r-proteins) (RpsB and -F; RplA, -E, -F, and -J), the adenylate kinase (Adk), the α- and β-subunits of RNAP (RpoA and -B), and the α- and β-subunits of ATP synthase (AtpA and -D) were strongly reduced in the wild type (Fig. 2B and 3B) but were still synthesized at a high rate in the relA mutant (Fig. 2C and 3D) after the stringent response was provoked. As demonstrated by dual-channel imaging, the stringently controlled proteins were still present but no longer synthesized (green color) (Fig. 2B). Quantitation of relative synthesis rates at different time points after norvaline addition demonstrates that the synthesis was switched off 5 to 10 min after induction of the stringent response in the wild type, whereas synthesis continued in the relA mutant (shown for RpsB and EF-Tu in Fig. 4A; see also Table 1).
FIG. 4.
Quantitation of the relative synthesis rates of EF-Tu and RpsB (members of class I) (A) and LeuD (member of class II) (B) after norvaline stress (0.05% [wt/vol]) in percent quantity as determined with the software DECODON Delta2D. B. subtilis BR16 (shaded columns) and BR17 (relA) (open columns) were compared.
Positive stringent control (proteins of class II).
Proteins that were induced only in the wild type or exhibited a higher induction rate in the wild type than in relA mutant cells were referred to as positively regulated by the stringent response. It has been shown previously that the general stress protein YvyD is the most abundant RelA-dependent protein induced after norvaline addition or in the course of amino acid starvation (21, 23). Here we demonstrate that the enzymes encoded by the ilv-leu operon which are involved in the synthesis of branched chain amino acids are also induced in a RelA-dependent manner (Fig. 2 and 3, Table 1). Norvaline, a leucine analogon, functions as an inhibitor of isoleucyl- and leucyl-tRNA synthetase and thus provokes isoleucine and/or leucine starvation (36). Mupirocin, likewise an inhibitor of isoleucyl-tRNA synthetase (28), also induces the ilv-leu operon in a RelA-dependent manner (not shown). The ilv-leu operon, a member of the T-box regulon, is regulated by transcriptional attenuation (29, 55). Because of the absence of valine, isoleucine, and leucine in this growth medium, this operon was already derepressed before norvaline addition, but it is further enhanced in the wild type only after norvaline treatment (Fig. 2A and B; Fig. 3A and B).
YurP and Ald, a sporulation-specific l-alanine dehydrogenase (also known as SpoVN), represent further members of this class (Fig. 2 and 3; Table 1). In some cases the induction of synthesis resulted in a significant accumulation of proteins only in the wild type (see Fig. 2 [e.g., LeuD, orange color]) and not in the relA mutant. For LeuD, a representative member of this class, a kinetic analysis of the protein synthesis rates is provided in Fig. 4B.
Proteins whose synthesis is induced or repressed independently of the stringent response (classes III and IV).
Some proteins are induced or repressed in both the wild type and the relA mutant in response to norvaline treatment and are referred to classes III and IV (indicated in Fig. 5, Table 2). Synthesis of enzymes involved in arginine biosynthesis (CarA and -B; ArgB, -C, -D, -F, -G, and -H) were induced in both strains, but this induction seemed to be delayed in the relA mutant (see Table 2). Enzymes maybe involved in methionine or cysteine biosynthesis (encoded by genes of the S-box regulon (e.g., yicI) (31) were induced in both strains (class III).
FIG. 5.
Sections of 2D protein gels (autoradiograms) of l-[35S]methionine-labeled proteins separated on immobilized pH gradient (IPG) strips in the pH ranges from 4 to 7 and from 4.5 to 5.5. Proteins were isolated from exponentially growing B. subtilis BR16 before (A) and 20 min after (B) norvaline addition and from B. subtilis BR17 20 min after norvaline addition (C). Only proteins induced (circles; arrows) or repressed (squares) independently of RelA are indicated.
TABLE 2.
Genes or proteins induced (class III) or repressed (class IV) independently of the stringent response, which was provoked by norvaline addition, as revealed by DNA macroarray and proteome analysisa
| Class and cellular function affected | Gene (synonym)/ protein | Identity/similarity/ function | (Putative) transcriptional unit | Transcriptional repression factors
|
Translational repression factors
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type | relA mutant | Wild type
|
relA mutant
|
||||||
| 10 min | 10 min | 10 min | 20 min | 10 min | 20 min | ||||
| Class III: RelA-independent induction | |||||||||
| Metabolism of nucleotides and nucleic acids | guaB/GuaB* (two spots) | Inosine-monophosphate dehydrogenase | guaB | 3.1 | 1.4 | 1.7 | 2.2 | 1.0 | 2.4 |
| Metabolism of amino acids and related proteins | yjcI/YjcI* (two spots) | Similar to cystathione γ-synthase/probable part of the S-box regulon/unknown | yjcI-yjcJ | 3.7 | 3.6 | 4.3 | 4.5 | >2.0 | >6.0 |
| Metabolism of amino acids and related proteins | yjcJ/YjcJ* (two spots) | Similar to cystathione β-lyase/probable part of the S-box regulon/unknown | yjcI-yjcJ | 3.0 | 4.3 | 6.0 | 4.5 | 2.7 | 3.9 |
| Specific pathways | yoaD | Similar to unknown proteins/probable part of the S-box regulon/unknown | yoaD-yoaC-yoaB? | 3.2 | 2.6 | ||||
| Specific pathways | yoaC | Similar to unknown proteins/probable part of the S-box regulon/unknown | yoaD-yoaC-yoaB? | 3.8 | 3.0 | ||||
| Transport | yheI | Similar to ABC transporter (ATP-binding protein)/unknown | yheJ-yheI-yheH | 4.0 | 3.3 | ||||
| Transport | yheH | Similar to ABC transporter (ATP-binding protein)/unknown | yheJ-yheI-yheH | 4.0 | 4.9 | ||||
| Unknown | ykrT/YkrT* | Similar to unknown proteins/probable part of the S-box regulon/unknown | ykrT-ykrS | 3.7 | 3.4 | 4.2 | 3.2 | 1.5 | 3.0 |
| Unknown | ykrS/YkrS* | Similar to eukaryotic initiation factor eIF-2B (alpha subunit)/probable part of the S-box regulon/unknown | ykrT-ykrS | 3.9 | 5.0 | >5.0 | >6.0 | >2.0 | >6.0 |
| Unknown | yitJ/YitJ* (two spots) | Similar to unknown proteins/probable part of the S-box regulon/unknown | yitJ | 5.7 | 3.3 | >5.0 | >6.0 | >2.0 | 1.8 |
| Unknown | ykrX | Similar to unknown proteins/probable part of the S-box regulon/unknown | ykrX-ykrY-ykrZ | 3.4 | 3.1 | ||||
| Unknown | ykrZ/YkrZ | Similar to unknown proteins/probable part of the S-box regulon/unknown | ykrX-ykrY-ykrZ | 4.1 | 2.8 | ND | ND | ND | ND |
| Arginine biosynthesis | argG/ArgG* (two spots) | Argininosuccinate synthase | argG-argH-ytzD | 7.0 | 4.4 | 5.0 | >6.0 | 1.5 | 4.1 |
| Arginine biosynthesis | argH/ArgH* (two spots) | Argininosuccinate lyase | argG-argH-ytzD | 7.0 | 4.4 | ND | ND | ND | ND |
| Unknown | ytzD | Unknown | argG-argH-ytzD | 14.2 | 5.2 | ||||
| Transport | yqiX/YqiX* | Similar to amino acid ABC transporter/unknown | yqiX | 3.3 | 1.6 | 2.4 | 2.1 | 1.3 | 2.9 |
| Arginine biosynthesis | argC/ArgC* | N-Acetylglutamate γ-semialdehyde dehydrogenase | argC-argJ-argB-argD-carA-carB-argF | 10.8 | 3.0 | 6.2 | 6.0 | 2.0 | 7.1 |
| Arginine biosynthesis | argJ | Omithine acetyltransferase/amino acid acetyltransferase | argC-argJ-argB-argD-carA-carB-argF | 10.1 | 2.8 | ||||
| Arginine biosynthesis | argB/ArgB* | N-Acetylglutamate 5-phosphotransferase | argC-argJ-argB-argD-carA-carB-argF | 10.3 | 4.5 | 5.7 | 4.7 | 1.5 | 6.0 |
| Arginine biosynthesis | argD/argD* | N-Acetylomithine aminotransferase | argC-argJ-argB-argD-carA-carB-argF | 10.9 | 4.3 | 4.0 | 5.5 | ND | 6.7 |
| Arginine biosynthesis | carA/CarA* | Carbamoyl-phosphate transferase-arginine (subunit A) | argC-argJ-argB-argD-carA-carB-argF | 5.6 | 3.4 | >5.0 | 2.4 | ND | 2.0 |
| Arginine biosynthesis | carB/carB* | Carbamoyl-phosphate transferase-arginine (subunit B) | argC-argJ-argB-argD-carA-carB-argF | 3.5 | 3.1 | 1.9 | 9.0 | 1.2 | 7.1 |
| Arginine biosynthesis | argF/ArgF* | Ornithine carbamoyltransferase | argC-argJ-argB-argD-carA-carB-argF | 11.6 | 3.9 | ND | ND | ND | ND |
| Class IV: RelA-independent repression | |||||||||
| Main glycolytic pathways | pgk/Pgk* | Phosphoglycerate kinase | cggR-gapA-pgk-tpi-pgm-eno | 3.0 | 2.5 | 1.3 | 1.5 | 2.8 | ND |
| Main glycolytic pathways | tpi/Tpi* | Triose phosphate isomerase | cggR-gapA-pgk-tpi-pgm-eno | 3.8 | 3.6 | 1.3 | 1.9 | ND | 1.5 |
| Main glycolytic pathways | pgm/Pgm* | Phosphoglycerate mutase | cggR-gapA-pgk-tpi-pgm-eno | 3.3 | 6.0 | 2.4 | 3.2 | 1.5 | ND |
| Main glycolytic pathways | eno/Eno* | Enolase | cggR-gapA-pgk-tpi-pgm-eno | 5.3 | 3.9 | 1.5 | 1.8 | 1.5 | 1.8 |
| Main glycolytic pathways | pfk/Pfk* | 6-Phosphofructokinase | pfk | 2.3 | 2.4 | >5.0 | >6.0 | >3.0 | 2.1 |
| Main glycolytic pathways | pdhA/PdhA | Pyruvate dehydrogenase (E1 alpha subunit) | pdhA-pdhB-pdhC-pdhD | 5.5 | 3.8 | >5.0 | >6.0 | 3.0 | 2.4 |
| Main glycolytic pathways | pdhB/PdhB | Pyruvate dehydrogenase (E1 beta subunit) | pdhA-pdhB-pdhC-pdhD | 18.2 | 8.1 | 11.2 | 10.3 | 3.3 | 4.6 |
| Main glycolytic pathways | pdhC/PdhC | Pyruvate dehydrogenase (dihydrolipoamide acetyltransferase E2 subunit) | pdhA-pdhB-pdhC-pdhD | 23.7 | 6.4 | 2.7 | 3.1 | 5.2 | 3.2 |
| Main glycolytic pathways | pdhD/PdhD* | Pyruvate dehydrogenase/2-oxoglutarate DH | pdhA-pdhB-pdhC-pdhD | 6.6 | 3.3 | >5.0 | 5.3 | 2.7 | 2.2 |
| Protein synthesis | pheS | Phenylalanyl-tRNA synthetase (alpha subunit) | pheS-pheT | 4.0 | 3.2 | ||||
| Protein synthesis | pheT/PheT* | Phenylalanyl-tRNA synthetase (beta subunit) | pheS-pheT | 2.6 | 1.8 | 4.8 | 4.0 | 2.0 | 2.5 |
| Thiamin biosynthesis | thiA (thiC)/ThiA | Biosynthesis of the pyrimidine moiety of thiamine | thiA | 4.9 | 2.7 | >5.0 | >6.0 | 2.0 | ND |
| Transport | fhuD | Ferrichrome ABC transporter (ferrichrome-binding protein) | fhuD | 4.0 | 4.2 | ||||
| Transport | cysP (ylnA) | Sulfate permease | cysP-prkC | 5.1 | 3.3 | ||||
| Transport | prkC (yloP) | Probable membrane-linked protein kinase | cysP-prkC | 4.0 | 3.1 | ||||
| Long chain fatty acid biosynthesis | accB (fabE, yqhW) | Acetyl-CoA carboxylase (biotin carboxyl carrier subunit) | accB-accC | 4.0 | 3.1 | ||||
| Long chain fatty acid biosynthesis | accC | Acetyl-CoA carboxylase (biotin carboxylase subunit) | accB-accC | 4.5 | 3.1 | ||||
| Histidine biosynthesis | hisD | Histidinol dehydrogenase | hisZ-hisG-hisD-hisB-hisH-hisA-hisF-hisI | 3.1 | 7.3 | ||||
| Histidine biosynthesis | hisB | Imidazoleglycerol-phosphate dehydratase | hisZ-hisG-hisD-hisB-hisH-hisA-hisF-hisI | 3.3 | 6.8 | ||||
| Histidine biosynthesis | hisH | Amidotransferase | hisZ-hisG-hisD-hisB-hisH-hisA-hisF-hisI | 2.7 | 4.4 | ||||
| Histidine biosynthesis | hisA | Phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase | hisZ-hisG-hisD-hisB-hisH-hisA-hisF-hisI | 2.0 | 4.3 | ||||
| Histidine biosynthesis | hisF | HisF cyclase-like protein | hisZ-hisG-hisD-hisB-hisH-hisA-hisF-hisI | 4.0 | 8.0 | ||||
| Histidine biosynthesis | hisI | Phosphoribosyl-AMP cyclohydrolase/phosphoribosyl-ATP pyrophospho-hydrolase | hisZ-hisG-hisD-hisB-hisH-hisA-hisF-hisI | 5.7 | 5.7 | ||||
| Purine biosynthesis | purE | Phosphoribosylaminoimidazole carboxylase I | purE-K-B-C-L-Q-F-purM-purN-H-D | 3.8 | 5.1 | ||||
| Purine biosynthesis | purK | Phosphoribosylaminoimidazole carboxylase II | purE-K-B-C-L-Q-F-purM-purN-H-D | 3.6 | 4.3 | ||||
| Purine biosynthesis | purB/PurB | Adenylosuccinate lyase | purE-K-B-C-L-Q-F-purM-purN-H-D | 7.9 | 4.6 | >5.0 | >6.0 | 3.1 | >3.0 |
| Purine biosynthesis | purC/PurC | Phosphoribosylaminoimidazole succinocarboxamide synthetase | purE-K-B-C-L-Q-F-purM-purN-H-D | 9.0 | 4.6 | 3.4 | 3.0 | 1.7 | ND |
| Purine biosynthesis | purL/PurL* | Phosphoribosylformylglycinamidine synthetase II | purE-K-B-C-L-Q-F-purM-purN-H-D | 6.8 | 4.2 | >5.0 | >6.0 | >3.0 | >3.0 |
| Purine biosynthesis | purQ | Phosphoribosylformylglycinamidine synthetase I | purE-K-B-C-L-Q-F-purM-purN-H-D | 3.6 | 3.0 | ||||
| Purine biosynthesis | purF | Phosphoribosylpyrophosphate amidotransferase | purE-K-B-C-L-Q-F-purM-purN-H-D | 3.6 | 2.6 | ||||
| Purine biosynthesis | purM/PurM | Phosphoribosylaminoimidazole synthetase | purE-K-B-C-L-Q-F-purM-purN-H-D | 1.9 | 2.6 | >5.0 | >6.0 | 2.1 | >3.0 |
| Purine biosynthesis | purN | Phosphoribosylglycinamide formyltransferase | purE-K-B-C-L-Q-F-purM-purN-H-D | 18.5 | 5.5 | ||||
| Purine biosynthesis | purH/PurH* | Phosphoribosylaminoimidazole carboxy formyl formyltransferase | purE-K-B-C-L-Q-F-purM-purN-H-D | 10.8 | 2.7 | 1.5 | 1.7 | ND | 1.5 |
| Purine biosynthesis | purD/PurD* | Phosphoribosylglycinamide synthetase | purE-K-B-C-L-Q-F-purM-purN-H-D | 3.5 | 1.3 | >5.0 | >6.0 | ND | >3.0 |
| Purine biosynthesis | purS (yexA) | Required for phosphoribosylformylglycinamidine synthetase activity | purS | 3.5 | 4.2 | ||||
| Purine biosynthesis | purA | Adenylosuccinate synthetase | purA | 2.1 | 5.4 | ||||
| Pyrimidine biosynthesis | pyrR | Transcriptional attenuation of the pyrimidine operon/uracil phosphoribosyltransferase activity | pyrR-pyrP-pyrB-C-pyrAA-pyrAB-DII-D-pyrF-E | 4.3 | 10.2 | ||||
| Pyrimidine biosynthesis | pyrB | Aspartate carbamoyltransferase | pyrR-pyrP-pyrB-C-pyrAA-pyrAB-DII-D-pyrF-E | 6.6 | 8.0 | ||||
| Pyrimidine biosynthesis | pyrC | Dihydrooratase | pyrR-pyrP-pyrB-C-pyrAA-pyrAB-DII-D-pyrF-E | 8.6 | 8.5 | ||||
| Pyrimidine biosynthesis | pyrAA/PyrAA* | Carbamoyl-phosphate synthetase | pyrR-pyrP-pyrB-C-pyrAA-pyrAB-DII-D-pyrF-E | 5.3 | 11.5 | >5.0 | >6.0 | >3.0 | >3.0 |
| Pyrimidine biosynthesis | pyrAB/PyrAB | Carbamoyl-phosphate synthetase (catalytic subunit) | pyrR-pyrP-pyrB-C-pyrAA-pyrAB-DII-D-pyrF-E | 2.1 | 2.0 | >5.0 | >6.0 | >3.0 | >3.0 |
| Pyrimidine biosynthesis | pyrF | Orotidine 5′-phosphate decarboxylase | pyrR-pyrP-pyrB-C-pyrAA-pyrAB-DII-D-pyrF-E | 3.2 | 2.7 | ||||
| Pyrimidine biosynthesis | pyrE | Orotate phosphoribosyltransferase | pyrR-pyrP-pyrB-C-pyrAA-pyrAB-DII-D-pyrF-E | 11.6 | 4.8 | ||||
| Unknown | yclP | Similar to ferrichrome ABC transporter (ATP-binding protein)/unknown | yclP | 4.3 | 5.1 | ||||
| Unknown | yclQ | Similar to ferrichrome ABC transporter (binding protein)/unknown | yclQ | 3.0 | 4.8 | ||||
| Unknown | yukC | Similar to unknown proteins/unknown | yukC | 4.2 | 4.6 | ||||
| Unknown | yumC/YumC* | Similar to thioredoxin reductase/unknown | yumC | 1.8 | 2.1 | >5.0 | >6.0 | >3.0 | >3.0 |
| Unknown | yaaD/YaaD* | Similar to superoxide-inducible protein/unknown | yaaD | 1.0 | 1.4 | 2.4 | 3.0 | 1.7 | 1.9 |
For the complete legend, see Table 1. Note that the repression of YumC and YaaD synthesis was not reflected at the mRNA level. CoA, coenzyme A.
The synthesis of proteins of class IV was repressed independently of (p)ppGpp. The synthesis of PheT, for example, the β-subunit of phenylalanyl-tRNA synthetase, was repressed in the wild type, as well as in the relA mutant. Furthermore, enzymes of glycolysis (e.g., 6-phosphofructokinase; Table 2), purine (e.g., PurD and PurL) and pyrimidine biosynthesis (PyrAB) showed a decreased synthesis after norvaline addition in both strains (Fig. 5 and Table 2). Because the RelA protein, isolated from the relaxed mutant BR17 (84), possesses ca. 2% residual (p)ppGpp-synthetase activity compared to the wild-type protein (80), the RelA-independent effects could be caused by residual synthesis of (p)ppGpp in the relA point mutant. Therefore, we also examined a relA null mutant (89). All of the proteins mentioned above were also induced or repressed in the relA deletion mutant BCE16, thus underlining their RelA-independent regulation (data not shown).
Transcriptome analysis by using DNA macroarrays.
The repression or induction of protein synthesis modulated by the stringent response almost certainly mirrors changes in transcription. Therefore, proteome data were compared to and complemented by global transcriptional studies by using DNA macroarrays. For a complete analysis of stringently controlled genes, we used DNA macroarrays containing all currently known protein-coding B. subtilis ORFs or genes (n = 4,107). Hybridizations were carried out with cDNA obtained from two independently isolated RNA preparations (two experiments) of each condition (condition 1 = wild type [BR16], exponential growth; condition 2 = wild type [BR16], 10 min of norvaline; condition 3 = relA mutant [BR17], exponential growth; condition 4 = relA mutant [BR17], 10 min of norvaline) by using two different array batches (see Materials and Methods). Expression intensities (nARVOL values) above a signal-to-noise ratio of 3.0 in at least one condition were obtained for 2,611 genes. For these significantly expressed genes, the expression level ratios were calculated. All genes showing at least threefold induction or repression factors in both experiments were considered.
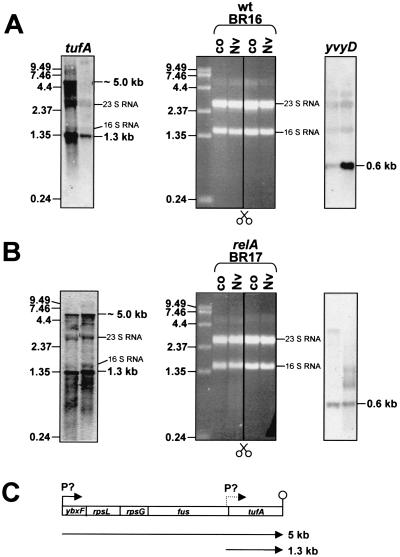
In order to guarantee high RNA quality, total RNA extracted from exponentially growing or norvaline-treated wild-type and relA mutant cells was checked for induction of model genes by Northern blot analysis first. yvyD transcription was analyzed as an example of a RelA-dependently induced gene (21, 23) and tufA as an example of RelA-dependent repressed transcription. As shown in Fig. 6, yvyD transcription was indeed induced only in the wild type. Using a tufA probe, strong transcriptional repression of a 5-kb mRNA and a 1.3-kb mRNA was detected in the wild type but not in the relA mutant. The 1.3-kb transcript corresponds to a monocistronic tufA mRNA, indicating a promoter immediately upstream from tufA. The large transcript of ∼5 kb may correspond to the transcriptional unit ybxF-rpsL-rpsG-fus-tufA (Fig. 6C). The same structures of the str operon of B. subtilis and Bacillus stearothermophilus and a promoter in front of the tufA gene from B. stearothermophilus were established by Krasny et al. (47). RNA checked in this way was used to generate cDNA for hybridization with DNA macroarrays. For a first overview, dual-channel images (as shown for protein gels [8]; described above) were created from the resulting autoradiograms. DNA macroarray images obtained from control experiments (exponential growth) were colored green, and images obtained from norvaline induction experiments were colored red. After overlaying of both arrays, newly induced genes not transcribed before the imposition of norvaline appeared as red double spots. Genes that were induced by norvaline but already transcribed before at a lower rate appeared as more or less orange. Downregulated genes no longer transcribed after the imposition of norvaline resulted in green spots (results are shown for sections in Fig. 7). Finally, genes that were transcribed with a similar intensity before and after norvaline resulted in yellow spots. The spot intensities were quantitated, and the transcription level ratios were determined (for a complete analysis, see Table 1 and 2). In most cases there was a good correlation between the data at the transcriptional and translational levels, but expression level ratios seemed to be higher at the transcriptional level. Only in a few cases (i.e., yumC and yaaD) did the transcriptional expression data not reflect the protein synthesis data (see Tables 1 and 2).
FIG. 6.
Quality check of RNA used for DNA macroarray analysis. A total of 5 μg of RNA isolated from exponentially growing cells before and 10 min after addition of norvaline from the wild type (A) and the relA mutant (B), respectively, was separated through a 1.2% agarose gel and stained with ethidium bromide (middle); the bands for 16S and 23S RNA are indicated. These ethidium bromide-stained gels were used for Northern blot analysis. To check the transcription of yvyD and tufA, the blotted membrane was divided as indicated in the figure. As suggested from previous studies and from protein data, transcriptional induction of yvyD, as well as transcriptional repression of tufA, occurred only in B. subtilis wild-type BR16. The positions of the RNA molecular size standard bands and the sizes of the yvyD or tufA transcripts are indicated. (C) Transcriptional organization of the str operon (which contains tufA).
FIG. 7.
Sections of dual-channel images generated by DNA macroarray analysis (Sigma-Genosys). PCR-derived DNA samples for each B. subtilis gene were spotted onto nylon membranes as recommended by the manufacturer (Sigma-Genosys). Each gene or ORF is represented by one double spot. A transcriptome comparison of B. subtilis strains BR16 and BR17 in response to norvaline treatment is shown. Dual-channel images were generated by combining macroarray image 1 (exponential growth; green channel) and macroarray image 2 (10 min of norvaline stress, red channel) (see Materials and Methods). Red spots indicate transcriptional induction by norvaline treatment. Orange- and red-bordered yellow spots represent genes whose transcription was already strong under control conditions but was further enhanced by norvaline treatment. Green spots represent genes whose transcription is switched off. Some prominently induced or repressed genes are indicated. (A) RelA-dependent transcriptional repression of genes encoding r-proteins and ybxF and map (class I). (B) RelA-dependent induced genes of the ilv-leu operon (class II). (C) RelA-independent induced genes (class III). (D) RelA-independent repressed genes encoding enzymes of purine biosynthesis.
Detection of genes that are negatively controlled by the stringent response (class I).
In accordance with the proteome data, a strong RelA-dependent repression of genes coding for components of the translational apparatus was found. Thus, transcription of 48 of 54 genes encoding r-proteins or proteins with similarity to r-proteins, four of six translation factors (e.g., tufA, tsf, fus, and ylaG), and two of six initiation factors (e.g., infA and infC) was switched off more than threefold only in the wild type (green spots) but continued in the relA mutant (yellow or orange spots) (Table 1; for examples, see Fig. 7A). For example, wild-type repression factors of genes of the str operon (described above) varied from 7.3-fold (tufA) to 23.2-fold (rpsL), whereas no or only little repression (1.1- to 1.5-fold) of this transcriptional unit was observed in the relA mutant (Table 1). Besides these components of the translational apparatus, transcription of genes whose products are involved in some other processes typical of growing cells seemed to be negatively controlled by (p)ppGpp. As shown in Table 1, transcription of a few genes functioning in RNA synthesis (e.g., rpoA and -B, nusA- and -B), DNA replication (ssb), protein modification (map; see also Fig. 7A), RNA modification (truA, rnc, and trmU), nucleotide metabolism (adk, pnpA, and pyrH), cell wall synthesis (gcaD, dltA, mbl, and murE), lipid metabolism (e.g., bkdAA and bkdB), and energy metabolism (atpG, -D, and -C; qoxD) was significantly repressed in the wild type only (3-fold [mbl] to 13.1-fold [atpC]). Curiously, other genes involved in these processes did not change their expression (e.g., dnaA) or repression was <3-fold (e.g., dnaC and rpoC) under these conditions and/or escaped detection because of a signal-to-noise intensity of <3 in all growth and strain conditions (see Materials and Methods; also data not shown).
Detection of genes that are positively controlled by the stringent response (class II).
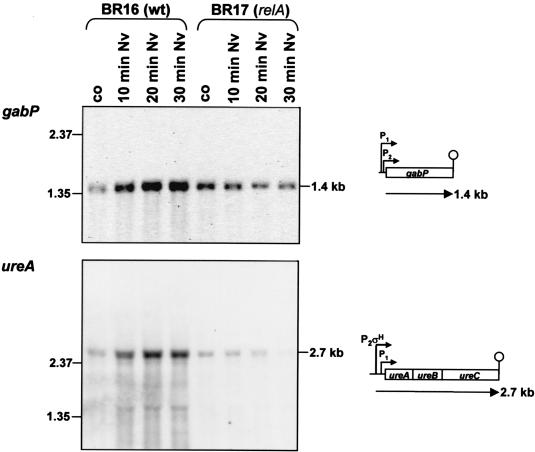
RelA-dependent induction of all of the proteins identified by proteomics was confirmed by transcriptional analysis (Table 1). For example, a threefold induction of the ilv-leu-operon was found (Fig. 7B). Furthermore, the RelA-dependent induction of the σH-dependent transcriptional units yvyD, ytxGHI, spoVG, and spo0A (23) was confirmed by DNA array analysis. In addition to these genes, the known σH-dependent genes spo0F (69) and phrA and phrC (57) were induced only in wild-type cells. Surprisingly, we also found an induction of the σB-dependent gene gspA (2) without induction of sigB itself (see also Fig. 2). Furthermore, (i) gabP coding for γ-aminobutyrate permease (10), (ii) ureA (91), (iii) genes coding for the extracellular serine proteases vpr (79) and epr (78), (iv) adeC (64), and (v) many genes of as-yet-unknown function were significantly induced in the wild type only. The gene ald was also induced in the relA mutant but at a lower rate (Table 1). The positive control of the genes gabP and ureA, which are under CodY control, was confirmed by Northern analysis (see Discussion; Fig. 8).
FIG. 8.
Northern blot analysis of the induction of the CodY-dependent gene gabP and the ure operon in response to the treatment of wild type and relA mutant with norvaline. Total RNA was isolated from strains BR16 and BR17 (relA) before (control [co]) and 10, 20, and 30 min after the addition of norvaline. A total of 5 μg of RNA was applied to each lane. The RNA probes gabP and ureA were used. The locations of RNA size markers, the size of the transcripts, and the transcriptional organization are indicated.
Detection of genes whose transcriptional induction or repression is scarcely influenced by the stringent response (classes III and IV).
As also observed at the protein level, the transcription of arginine biosynthetic genes was increased in both the wild type and the relA mutant (Fig. 7C and Table 2), but the wild-type expression level ratios of genes of the argC-F operon were significantly higher than in the relA mutant. Our data, however, indicate that transcriptional induction of this operon is delayed only in the relA mutant (not shown). In agreement with the proteome data, the transcription of genes which are probably part of the S box regulon was increased in both the wild type and the relA mutant (Fig. 7C; Table 2).
Transcription of genes involved in glycolysis (e.g., eno, tpi, and pfk) was inhibited in both the wild type and the relA mutant (Fig. 7D). Whereas the synthesis rates of proteins involved in purine and pyrimidine biosynthesis decreased in both the wild type and the relA mutant (Fig. 5), the repression level ratios of some pur and pyr genes was similar in both strains, but for other pur and pyr genes the repression level ratios differed in wild-type and relA mutant cells (Table 2).
DISCUSSION
A comprehensive analysis of the B. subtilis stringent response was carried out by using high-resolution 2D protein gel electrophoresis and DNA macroarray techniques. (p)ppGpp functions, as in E. coli, both as a negative and a positive regulator in B. subtilis.
A comparison of wild-type and relA mutant proteome and transcriptome patterns showed that 20 proteins and ca. 40 transcriptional units were negatively controlled by the stringent response, whereas 13 proteins and 50 transcriptional units seemed to be positively controlled. In E. coli in most cases (p)ppGpp functions at the level of transcriptional initiation and elongation, but our data do not provide information on the mechanism of (p)ppGpp action. Our data show that the changes (repression or induction) measured at the transcriptional level were also found at the protein synthesis level, although in most cases at lower ratios.
Our results confirm the proteome data of Wendrich and Marahiel (89) but add a large number of new genes affected by the stringent response in B. subtilis (see Table 1). Most of these new genes, whose expression seems to be controlled by (p)ppGpp, were found only by transcriptome analysis. Failure to detect certain gene products by 2D gel analysis may be due to the fact that certain subproteomic fractions, e.g., those with membrane-spanning domains, extremely alkaline proteins (66), or secreted proteins (3) were not considered in our analysis.
Negative stringent control.
As already known from E. coli, the hallmark of the stringent response consists of the negative regulation of components of the translational apparatus including rRNAs, tRNAs, ribosomal proteins (r-proteins), and translation factors (12). It has been known for a long time that transcription of stable RNA after amino acid starvation is only repressed in the wild type and not in a relA mutant of B. subtilis (80) (see Fig. 1). Several groups observed a negative effect of (p)ppGpp on elongation of transcription which was explained by the “pausing” of RNA polymerase (RNAP) at specific sites (e.g., reference 48). Recently, it has been suggested for E. coli that the effector molecule (p)ppGpp binds to the β- and β′-subunits of RNAP (13, 86), causing a rapid reduction of the transcription of rrn operons, probably by reducing the stability of the open promoter-RNAP complexes at rrn P1 promoters (4, 6). Data concerning the transcriptional regulation of rrn operons by (p)ppGpp in B. subtilis are still missing. Genes for tRNAs and rRNAs are neither represented on the macroarrays that we used nor transcribed into cDNA and therefore escaped detection by array hybridization with cDNA.
This study clearly shows that transcription of almost all genes encoding r-proteins (results are shown for 48 genes, 2 of which show similarity to r-proteins, e.g., the S10-spc-α-region [52, 83] and the rif cluster [18]) and their translation (results are shown for six r-proteins) were switched off by more than threefold after norvaline addition only in the wild type and not in the relA mutant. Only 6 of the ca. 50 known ribosomal proteins (see the Subtilist database [http://genolist.pasteur.fr/Subtilist/]) (59, 60) were identified on our 2D gels (pH 4 to 7 and pH 3 to 10) because the majority of these proteins have very basic pI values (see also reference 66). Another ribosomal protein, RplL, contains no methionine and was therefore not visualized on autoradiograms.
In E. coli, r-protein synthesis is regulated at the level of translation by an autogenous feedback mechanism (15, 16). Furthermore, it could be demonstrated in vitro that transcription of some r-protein genes is also repressed by (p)ppGpp (20, 46, 71, 72). The mechanism of the stringent response of ribosomal protein synthesis in B. subtilis is completely unknown.
Finally, other components of the translation apparatus, such as the trigger factor (tig), initiation (infA, infB, and infC), and elongation factors (fus, tufA, and tsf), were strongly repressed at the level of transcription, which resulted in decreased synthesis of their gene products only in the wild type. This is not surprising because these genes (except tsf) are cotranscribed with genes encoding ribosomal proteins (Table 1). It is interesting that tig encoding the trigger factor belongs to the negatively controlled genes in B. subtilis. In E. coli, the ribosome-bound trigger factor tig is part of the translational apparatus and participates in folding of newly synthesized proteins (11, 53). A new protein of B. subtilis similar to GTP-binding elongation factors was identified as a product of the gene ylaG, which, like the known elongation factors, is negatively controlled by the stringent response. Negative stringent control of elongation factors (9, 58, 65, 74) or the initiation factor 3 (InfC) (22) was demonstrated in E. coli but only for EF-Ts in B. subtilis (89).
Besides the genes encoding translational proteins, many genes involved in RNA synthesis (e.g., nusA, rpoA, and rpoB) are also located within operons encoding proteins of the translational apparatus (Table 1), which explains their repression in a RelA-dependent manner. rpoC coding for the β′-subunit of RNAP that might be also located in a ribosomal protein gene cluster was repressed only 1.8-fold in the wild type but not in the relA mutant (not shown). The repression of RpoB and -C (β- and β′-subunits of RNAP) has been described in E. coli (76). As in E. coli (9, 63), aminoacyl-tRNA synthetases were not regulated by the stringent response or by norvaline treatment in B. subtilis (except PheT).
In addition to genes of the translational machinery, genes involved in other processes typical of growing cells (nucleotide biosynthesis, synthesis of lipids, energy metabolism, RNA modification, protein modification, cell wall synthesis, and DNA replication) appeared to be subject to negative stringent control in B. subtilis, as shown by DNA macroarray analysis. It should be mentioned, however, that not all genes involved in anabolic reactions, such as cell wall or DNA synthesis, showed a noticeable expression pattern in growing cells or in response to norvaline. We cannot exclude, however, that some of these genes are also stringently controlled but were not considered in our list because they had a repression level ratio lower than threefold. For genes encoding as-yet-unknown proteins whose expression is under negative stringent control, one preliminary prediction of their function is possible: their products might also be involved in cellular reactions typical of growing cells.
Promoters of E. coli genes under negative stringent control are characterized by GC-rich sequences between the −10 box and the potential transcriptional start point (GC discriminator) (e.g., rrnB P1 [44, 93]). Another similarity of stringent promoters is the relative instability of the open promoter complexes and the requirement of high concentrations of the initiating nucleotides (ATP or GTP) for maximal transcription in vivo (26). The reduction in ATP and GTP pools that occurs during the stringent response (54) might cause inhibition of rRNA transcription and probably of the other stringent promoters. However, the most essential mechanism is probably the destabilization of open promoter-RNAP complexes by the (p)ppGpp-bound RNAP, which resulted in decreased transcription initiation (4). GC-rich discriminators were not found in rrn promoters of B. subtilis or of other negatively controlled genes (not shown) or in the known σA-dependent promoter of rpsD (30).
Positive stringent control.
In agreement with related data for E. coli and other eubacteria, our results demonstrate that (p)ppGpp also functions as a positive effector of gene expression in B. subtilis. We found 50 transcriptional units whose expression level seemed to be stimulated >3-fold only in the wild type. Twelve of the positively controlled genes have already been already identified by our proteomics approach. In this context, RelA-dependent induction of the ilv-leu operon in norvaline-treated cells is of particular interest and might explain the results of Wendrich and Marahiel (89), who noted that the relA deletion mutant is auxotrophic for valine, leucine, and isoleucine. In E. coli, the transcription of many operons encoding enzymes of amino acid biosynthesis pathways requires (p)ppGpp, and cells lacking (p)ppGpp (relA spoT double mutants) show a polyauxotroph phenotype (92). The physiological significance of this positive control could be to escape from amino acid starvation in norvaline-treated cells. This could also be the physiological role of the induction of γ-aminobutyrate permease (gabP), urease (ure operon), and two extracellular serine proteases (vpr and epr). Very recently, a RelA/DegU-dependent induction of the alkaline protease gene aprE of B. subtilis was described (32). aprE is also induced in a relA-dependent manner in response to norvaline, but the induction ratio of 2.7-fold in the wild type was below our threshold level. Surprisingly, we did not find transcriptional elevation of genes involved in amino acid transport, which had been suggested to be under positive stringent control in E. coli (12). However, we cannot exclude that some of the genes with an as-yet-unknown function code for potential amino acid transporters. Very recently, it was shown by DNA microarray analysis that acivicin, which is also an amino acid antagonist and inhibitor of glutamine amidotransferase, probably triggers the stringent response in E. coli that resulted in the induction of amino acid biosynthetic genes and only one amino acid transport gene. Otherwise, the transcription of 49 genes involved in translation was repressed (81).
The ilv-leu operon is transcribed from a σA-dependent promoter (29) like other amino acid biosynthetic operons in B. subtilis. ald, coding for l-alanine dehydrogenase and functioning in sporulation, is also transcribed from a σA-dependent promoter (43) and induced in a RelA-dependent manner, as revealed by both proteomic and transcriptional analysis.
In addition to these σA-dependent genes, the four σH-dependent transcriptional units yvyD, ytxGHI, spo0A, and spoVG were induced as shown previously, leading to a more effective sporulation in the wild type than in the relA mutant (23). In addition, the σH-dependent genes spo0F, phrA, and phrC were induced in response to norvaline, as revealed by transcriptome analysis.
There are many examples of positive control in other bacteria (12, 14), such as rpoS of E. coli (27) or RpoS-dependent promoters (50), the his promoter of Salmonella enterica serovar Typhimurium (17), the Pseudomonas-derived σ54-dependent promoter of the dmp operon (85), and promoters from different amino acid biosynthetic genes in E. coli (5). Furthermore, (p)ppGpp seems to be involved in the regulation of developmental processes and antibiotic production of gram-positive bacteria (for a review, see reference 14).
The mechanism of transcriptional activation by (p)ppGpp is unknown. Recent data from several groups indicate that free functional RNA polymerase is a limiting factor for transcriptional initiation, suggesting a real competition of sigma factors for the core enzyme of RNA polymerase (25, 41, 45). Barker et al. (4, 5) suggested that E. coli genes whose expression is induced by (p)ppGpp require relatively high concentrations of RNA polymerase for their expression. Their genes might not be saturated with RNA polymerase during growth but will be induced when more RNA polymerase is available for their transcription as a result of the stringent control. This “passive model” relying on the redistribution of RNA polymerase in amino acid-starved cells (see also reference 50) could also explain the positive stringent control in B. subtilis. Such a passive model, however, may not be the only reason for an induction of >10-fold (yvyD). Additional mechanisms directly or only indirectly dependent on (p)ppGpp should be taken into consideration.
Ratnayake-Lecamwasan et al. (73) suggest that the stringent response, which triggers a decrease in the GTP pool (54), is involved in the derepression of CodY. CodY is a GTP-sensing protein and might function as a repressor under high-GTP-level conditions (73). The positively stringently controlled genes gabP and ure operon (described above) are repressed by CodY in the presence of a preferred N and C source (24, 91). Their RelA-dependent induction, which was confirmed by Northern analysis (see Fig. 8), supports the idea of Ratnayake-Lecamwasan et al. (73). However, because the CodY-dependent dpp operon (75, 77) was not induced by norvaline in a RelA-dependent manner (not shown), the relationship between CodY and RelA seemed to be more complex. The elucidation of this interplay awaits further studies.
Finally, some genes are induced in norvaline-treated cells of both strains. Genes belonging to the S-box regulon and genes involved in arginine biosynthesis are induced in the wild type, as well as in the relA mutant (class III). These results indicate the involvement of specific regulators independently of RelA. The arg operons are not induced in response to lysine starvation, and their induction in response to norvaline addition seems to be specific. Motyl and coworkers (61) found that norvaline functions as a competitive inhibitor of ornithine carbamoyltransferase, and this observation may explain the RelA-independent induction of the arg operons. Proteins involved in glycolysis, as well as purine and pyrimidine biosynthesis, are downregulated independently of RelA. This is in contrast to E. coli, in which the transcription of some genes involved in purine and pyrimidine biosynthesis is repressed by the stringent response (19, 87).
Acknowledgments
This work was supported by grants of the DFG, the BMBF, the EU consortium (GLG2-CT-1999-01455), and the Fonds der Chemischen Industrie to M.H.
We thank R. Wagner and U. Völker for the critical reading of the manuscript.
Footnotes
Dedicated to K. Altendorf, Osnabrück, Germany, on the occasion of his 60th birthday.
REFERENCES
- 1.Antelmann, H., J. Bernhardt, R. Schmid, H. Mach, U. Völker, and M. Hecker. 1997. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18:1451-1463. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann, H., J. Bernhardt, R. Schmid, and M. Hecker. 1995. A gene at 333 degrees on the Bacillus subtilis chromosome encodes the newly identified σB-dependent general stress protein GspA. J. Bacteriol. 177:3540-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 5.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:699-702. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett, M. S., T. Gaal, W. Ross, and R. L. Gourse. 1998. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J. Mol. Biol. 279:341-345. [DOI] [PubMed] [Google Scholar]
- 7.Bernhardt, J., U. Völker, A. Völker, H. Antelmann, R. Schmid, H. Mach, and M. Hecker. 1997. Specific and general stress proteins in Bacillus subtilis: a two-dimensional protein electrophoresis study. Microbiology 143:1009-1017. [DOI] [PubMed] [Google Scholar]
- 8.Bernhardt, J., K. Büttner, C. Scharf, and M. Hecker. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20:2225-2240. [DOI] [PubMed] [Google Scholar]
- 9.Blumenthal, R. M., P. G. Lemaux, F. C. Neidhardt, and P. P. Dennis. 1976. The effects of the relA gene on the synthesis of aminoacyl-tRNA synthetases and other transcription and translation proteins in Escherichia coli. Mol. Gen. Genet. 149:291-296. [DOI] [PubMed] [Google Scholar]
- 10.Brechtel, C. E., and S. C. King. 1998. 4-Aminobutyrate (GABA) transporters from the amine-polyamine-choline superfamily: substrate specifity and ligand recognition profile of the 4-aminobutyrate permease from Bacillus subtilis. Biochem. J. 333:565-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukau, B., E. Deuerling, C. Pfund, and E. A. Craig. 2000. Getting newly synthesized proteins into shape. Cell 101:119-122. [DOI] [PubMed] [Google Scholar]
- 12.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 13.Chatterji, D., N. Fujita, and A. Ishihama. 1998. The mediator for stringent control, ppGpp, binds to the β-subunit of Escherichia coli RNA polymerase. Genes Cells 8:279-287. [DOI] [PubMed] [Google Scholar]
- 14.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 15.Cole, J. R., and M. Nomura. 1986. Translational regulation is responsible for growth-rate dependent and stringent control of the synthesis of ribosomal proteins L11 and L1 in Escherichia coli. Proc. Natl. Acad. Sci. USA 83:4129-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole, J. R., and M. Nomura. 1986. Changes in the half-life of ribosomal protein messenger RNA by translational repression. J. Mol. Biol. 188:383-392. [DOI] [PubMed] [Google Scholar]
- 17.Dabbs, E. R. 1984. Order of ribosomal protein genes in the Rif cluster of Bacillus subtilis is identical to that of Escherichia coli. J. Bacteriol. 159:770-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Da Costa, X. J., and S. W. Artz. 1997. Mutations that render the promoter of the histidine operon of Salmonella typhimurium insensitive to nutrient-rich medium repression and amino acid downshift. J. Bacteriol. 179:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies, I. J., and W. T. Drabble. 1996. Stringent and growth-rate-dependent control of the gua operon of Escherichia coli. Microbiology 142:2429-2437. [DOI] [PubMed] [Google Scholar]
- 20.Dennis, P. P., and M. Nomura. 1974. Stringent control of ribosomal protein gene expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 71:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drzewiecki, K., C. Eymann, G. Mittenhuber, and M. Hecker. 1998. The yvyD gene of Bacillus subtilis is under dual control of σB and σH. J. Bacteriol. 180:6674-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elhardt, D., R. Wirth, and A. Bock. 1982. Regulation of formation of threonyl-tRNA synthetase and protein synthesis initiation factor 3 from Escherichia coli in vivo and in vitro. Eur. J. Biochem. 123:477-482. [DOI] [PubMed] [Google Scholar]
- 23.Eymann, C., G. Mittenhuber, and M. Hecker. 2001. The stringent response, σH-dependent gene expression and sporulation in Bacillus subtilis. Mol. Gen. Genet. 4:913-923. [DOI] [PubMed] [Google Scholar]
- 24.Ferson, A. E., L. V. Wray, Jr., and S. H. Fisher. 1996. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol. Microbiol. 22:693-701. [DOI] [PubMed] [Google Scholar]
- 25.Fujita, M., and Y. Sadaie. 1998. Promoter selectivity of the Bacillus subtilis RNA polymerase σA and σH holoenzymes. J. Biochem. 124:89-97. [DOI] [PubMed] [Google Scholar]
- 26.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rrnA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 27.Gentry, D. R., V. J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor σS is positively regulated by ppGpp. J. Bacteriol. 175:7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbart, J., C. R. Perry, and B. Slocombe. 1993. High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob. Agents Chemother. 37:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grandoni, J. A., S. A. Zahler, and J. M. Calvo. 1992. Transcriptional regulation of the ilv-leu operon of Bacillus subtilis. J. Bacteriol. 174:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grundy, F. J., and T. Henkin. 1992. Characterization of the Bacillus subtilis rpsD regulatory target site. J. Bacteriol. 174:6763-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundy, F. J., and T. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:747-749. [DOI] [PubMed] [Google Scholar]
- 32.Hata, M., M. Ogura, and T. Tanaka. 2001. Involvement of stringent factor RelA in expression of the alkaline protease gene aprE in Bacillus subtilis. J. Bacteriol. 183:4648-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hauser, N. C., M. Vingron, M. Scheideler, B. Krems, K. Hellmuth, K.-D. Entian, and J. D. Hoheisel. 1998. Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast 14:1209-1221. [DOI] [PubMed] [Google Scholar]
- 34.Hecker, M., A. Schroeter, and F. Mach. 1983. Replication of pBR322 DNA in stringent and relaxed strains of Escherichia coli. Mol. Gen. Genet. 190:355-357. [DOI] [PubMed] [Google Scholar]
- 35.Hecker, M., A. Schroeter, K. Trader, and F. Mach. 1986. Role of relA mutation in the survival of amino acid-starved Escherichia coli. Arch. Micobiol. 143:400-402. [DOI] [PubMed] [Google Scholar]
- 36.Hecker, M., A. Richter, A. Schroeter, L. Wölfel, and F. Mach. 1987. Synthesis of heat shock proteins following amino acid or oxygen limitation in Bacillus subtilis relA+ and relA strains. Z. Naturforsch. Teil C 42:941-947. (In German.) [PubMed]
- 37.Hecker, M., and U. Völker. 1998. Non-specific, general and multiple stress resistence of growth-restricted Bacillus subtilis cells by the expression of the σB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 38.Hecker, M., and S. Engelmann. 2000. Proteomics, DNA arrays, and the analysis of still unknown regulons and unknown proteins of Bacillus subtilis and pathogenic gram-positive bacteria. Int. J. Med. Microbiol. 190:123-134. [DOI] [PubMed] [Google Scholar]
- 39.Hecker, M., and U. Völker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 40.Hengge-Aronis, R. 1999. Interplay of global regulators and the cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 41.Hicks, K. A., and A. D. Grossman. 1996. Altering the level and regulation of the major sigma subunit of RNA polymerase affects gene expression and development in Bacillus subtilis. Mol. Microbiol. 20:211-212. [DOI] [PubMed] [Google Scholar]
- 42.Homuth, G., S. Masuda, A. Mogk, Y. Kobayashi, and W. Schumann. 1997. The dnaK operon of Bacillus subtilis is heptacistronic. J. Bacteriol. 179:1153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaacks, S. K., K. Ireton, and A. D. Grossman. 1993. Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J. Bacteriol. 175:6789-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Josaitis, C. A., T. Gaal, W. Ross, and R. L. Gourse. 1995. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc. Natl. Acad. Sci. USA 92:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ju, J., T. Mitchell, H. Peters III, and W. G. Haldenwang. 1999. Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol. 181:4969-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kajitani, M., and A. Ishihama. 1984. Promoter selectivity of Escherichia coli RNA polymerase. Differential stringent control of the multiple promoters from ribosomal RNA and protein operons. J. Biol. Chem. 10:1951-1957. [PubMed] [Google Scholar]
- 47.Krasny, L., T. Vaxik, V. Fucik, and J. Jonak. 2000. Cloning and characterization of the str operon and elongation factor Tu expression in Bacillus stearothermophilus. J. Bacteriol. 182:6114-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krohn, M., and R. Wagner. 1996. Transcriptional pausing of RNA polymerase in the presence of guanosine tetraphosphate depends on the promoter and gene sequence. J. Biol. Chem. 271:23884-23894. [DOI] [PubMed] [Google Scholar]
- 49.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 50.Kvint, K., A. Farewell, and T. Nyström. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σS. J. Biol. Chem. 275:14795-14798. [DOI] [PubMed] [Google Scholar]
- 51.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σSsubunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, X., L. Lindahl, Y. Sha, and J. M. Zengel. 1997. Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-α cluster. J. Bacteriol. 179:7046-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lill, R., E. Crooke, B. Guthrie, and W. Wickner. 1988. The “trigger factor cycle” includes ribosomes, presecretory proteins, and the plasma membrane. Cell 54:1013-1018. [DOI] [PubMed] [Google Scholar]
- 54.Lopez, J., C. Marks, and E. Freese. 1981. The decrease in guanosine nucleotides initiates the sporulation in Bacillus subtilis. Biochim. Biophys. Acta 587:238-252. [DOI] [PubMed] [Google Scholar]
- 55.Marta, P. T., R. D. Ladner, and J. A. Grandoni. 1996. A CUC triplet confers leucine-dependent regulation of the Bacillus subtilis ilv-leu operon. J. Bacteriol. 178:2150-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maul, B., U. Völker, S. Riethdorf, S. Engelmann, and M. Hecker. 1995. σB-Dependent regulation of gsiB in response to multiple stimuli in Bacillus subtilis. Mol. Gen. Genet. 248:114-120. [DOI] [PubMed] [Google Scholar]
- 57.McQuade, R. S., N. Comella, and A. D. Grossman. 2001. Control of a family of phosphatase regulatory genes (phr) by the alternate sigma factor sigma-H of Bacillus subtilis. J. Bacteriol. 183:4905-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mizushima-Sugano, J., and Y. Kaziro. 1985. Regulation of the expression of the tufB operon by guanosine-5′-diphosphate-3′-diphosphate. EMBO J. 4:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moszer, I., P. Glaser, and A. Danchin. 1995. Subtilist: a relational database for the Bacillus subtilis genome. Microbiology 141:261-268. [DOI] [PubMed] [Google Scholar]
- 60.Moszer, I. 1998. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 430:28-36. [DOI] [PubMed] [Google Scholar]
- 61.Motyl, T., A. Orzechowski, and S. Pierzynowski. 1988. The relationship between ureA and pyrimidine de novo synthesis in ruminant liver. Q. J. Exp. Physiol. 73:1-6. [DOI] [PubMed]
- 62.Nishino, T., J. Gallant, P. Shalit, L. Palmer, and T. Wehr. 1979. Regulatory nucleotides involved in the Rel function of Bacillus subtilis. J. Bacteriol. 140:671-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ny, T., J. Thomale, K. Hjalmarsson, G. Nass, and G. R. Bjork. 1980. Non-coordinate regulation of enzymes involved in transfer RNA metabolism in Escherichia coli. Biochim. Biophys. Acta 607:77-84. [DOI] [PubMed] [Google Scholar]
- 64.Nygaard, P., P. Duckert, and H. H. Saxild. 1996. Role of adenine deaminase in purine salvage and nitrogen metabolism and characterization of the ade gene in Bacillus subtilis. J. Bacteriol. 178:846-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nyström, T. 1994. Role of guanosine tetraphosphatein gene expression and the survival of glucose or seryl-tRNA starved cells of Escherichia coli K12. Mol. Gen. Genet. 245:355-362. [DOI] [PubMed] [Google Scholar]
- 66.Ohlmeier, S., C. Scharf, and M. Hecker. 2000. Alkaline proteins of Bacillus subtilis: optimization of methods and first steps towards a two-dimensional alkaline master gel. Electrophoresis 21:3701-3709. [DOI] [PubMed] [Google Scholar]
- 67.Otto, A., B. Thiede, E. C. Mueller, C. Scheler, B. Wittmann-Liebhold, and P. Jungblut. 1996. Identification of human myocardial proteins separated by two-dimensional electrophoresis using an effective sample preparation for mass spectrometry. Electrophoresis 17:1643-1650. [DOI] [PubMed] [Google Scholar]
- 68.Petersohn, A., M. Brigulla, S. Haas, J. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Predich, M., G. Nair, and I. Smith. 1992. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing sigma H. J. Bacteriol. 174:2771-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria, p. 179-197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 71.Raghavan, A., and D. Chatterji. 1998. Guanosine tetraphosphate-induced dissociation of open complexes at the Escherichia coli ribosomal protein promoters rplJ and rpsA P1: nanosecond depolarization spectroscopic studies. Biophys. Biochem. 75:21-32. [DOI] [PubMed] [Google Scholar]
- 72.Raghavan, A., D. B. Kameshwari, and D. Chatterji. 1998. The differential effects of guanosine tetraphosphate on open complex formation at the Escherichia coli ribosomal protein promoters rplJ and rpsAP1. Biophys. Biochem. 75:7-19. [DOI] [PubMed] [Google Scholar]
- 73.Ratnayake-Lecamwasan, M., P. Serror, K. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reeh, S., S. Pedersen, and J. D. Friesen. 1976. Biosynthetic regulation of individual proteins in relA+ and relA strains of Escherichia coli during amino acid starvation. Mol. Gen. Genet. 149:279-289. [DOI] [PubMed] [Google Scholar]
- 75.Serror, P., and A. L. Sonenshein. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20:843-852. [DOI] [PubMed] [Google Scholar]
- 76.Shakulov, R. S., E. V. Kliachko, and O. I. Ponomarenko. 1985. The role of ppGpp in the coordination of transcription of the ribosomal protein genes rplKAJL and RNA-polymerase rpoBC genes in Escherichia coli cells. Mol. Biol. 19:1603-1609. [PubMed] [Google Scholar]
- 77.Slack, F., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689-702. [DOI] [PubMed] [Google Scholar]
- 78.Sloma, A., A. Ally, D. Ally, and J. Pero. 1988. Gene encoding a minor extracellular protease in Bacillus subtilis. J. Bacteriol. 170:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sloma, A., G. A. Rufo, Jr., K. A. Theriault, M. Dwyer, S. W. Wilson, and J. Pero. 1991. Cloning and characterization of the gene for an additional extracellular serine protease of Bacillus subtilis. J. Bacteriol. 173:6889-6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith, I., P. Paress, K. Cabane, and E. Dubnau. 1980. Genetics and physiology of the rel system of Bacillus subtilis. Mol. Gen. Genet. 178:271-279. [DOI] [PubMed] [Google Scholar]
- 81.Smulski, D. R., L. L. Huang, M. P. McCluskey, M. J. Reeve, A. C. Vollmer, T. K. Van Dyk, and R. A. LaRossa. 2001. Combined, functional genomic-biochemical approach to intermediary metabolism: interaction of acivicin, a glutamine amidotransferase inhibitor, with Escherichia coli K-12. J. Bacteriol. 183:3353-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of β-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041-2045. [DOI] [PubMed] [Google Scholar]
- 83.Suh, J. W., S. A. Boylan, S. H. Oh, and C. W. Price. 1996. Genetic and transcriptional organization of the Bacillus subtilis spc-alpha region. Gene 169:17-23. [DOI] [PubMed] [Google Scholar]
- 84.Swanton, M., and G. Edlin. 1972. Isolation and characterization of an RNA relaxed mutant of Bacillus subtilis. Biochem. Biophys. Res. Commun. 46:583-588. [DOI] [PubMed] [Google Scholar]
- 85.Sze, C. C., and V. Shingler. 1999. The alarmone (p)ppGpp mediates physiological-responsive control at the sigma 54-dependent Po promoter. Mol. Microbiol. 31:217-228. [DOI] [PubMed] [Google Scholar]
- 86.Toulokhonov, I. I., I. Shulgina, and V. J. Hernandez. 2001. Binding of the effector ppGpp to E. coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the β subunit. J. Biol. Chem. 276:1220-1225. [DOI] [PubMed] [Google Scholar]
- 87.Turnbough, C. L., Jr. 1983. Regulation of Escherichia coli aspartate transcarbamylase synthesis by guanosine tetraphosphate and pyrimidine ribonucleoside triphosphates. J. Bacteriol. 153:998-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Völker, U., A. Völker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate SigB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26:65-79. [DOI] [PubMed] [Google Scholar]
- 90.Wetzstein, M., U. Völker, J. Dedio, S. Löbau, U. Zuber, M. Schiesswohl, C. Herget, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, and molecular analysis of the dnaK locus of Bacillus subtilis. J. Bacteriol. 174:3300-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can eleminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 93.Zhang, X., and H. Bremer. 1995. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 270:11181-11189. [DOI] [PubMed] [Google Scholar]