Abstract
Superoxide dismutase (SOD) profiles of clinical isolates of Staphylococcus aureus and coagulase-negative staphylococci (CoNS) were determined by using whole-cell lysates and activity gels. All S. aureus clinical isolates exhibited three closely migrating bands of activity as previously determined for laboratory strains of S. aureus: SodM, SodA, and a hybrid composed of SodM and SodA (M. W. Valderas and M. E. Hart, J. Bacteriol. 183:3399-3407, 2001). In contrast, the CoNS produced only one SOD activity, which migrated similarly to SodA of S. aureus. Southern analysis of eight CoNS species identified only a single sod gene in each case. A full-length sod gene was cloned from Staphylococcus epidermidis and determined to be more similar to sodA than to sodM of S. aureus. Therefore, this gene was designated sodA. The deduced amino acid sequence of the S. epidermidis sodA was 92 and 76% identical to that of the SodA and SodM proteins of S. aureus, respectively. The S. epidermidis sodA gene expressed from a plasmid complemented a sodA mutation in S. aureus, and the protein formed a hybrid with SodM of S. aureus. Both hybrid SOD forms as well as the SodM and SodA proteins of S. aureus and the S. epidermidis SodA protein exist as dimers. These data indicate that sodM is found only in S. aureus and not in the CoNS, suggesting an important divergence in the evolution of this genus and a unique role for SodM in S. aureus.
The staphylococci are a diverse group of species that are routinely categorized in the clinical setting as either coagulase positive or coagulase negative (3). In most cases, coagulase-positive staphylococci isolated from humans are Staphylococcus aureus, while the coagulase-negative staphylococci (CoNS) may include any of the remaining 32 species that constitute the genus Staphylococcus (34). A notable exception to this axiom is Staphylococcus intermedius, which is coagulase positive (33). However, only about 15 of the coagulase-negative species are indigenous to humans, with Staphylococcus epidermidis being the species most frequently isolated from bloodstream infections (27, 34).
S. aureus has always been considered a human pathogen with a wide array of disease syndromes, ranging from minor skin abscesses to life-threatening endocarditis, osteomyelitis, and pneumonia (51). Contributing to this array of diseases is the capacity of this organism to produce numerous proteins with cytotoxic and immunogenic properties as well as surface-associated factors that promote adherence and evade host defenses (28, 45). In contrast, the CoNS have been regarded as apathogenic commensals residing on human skin and frequent contaminants of clinical samples (27). However, the increasing use of invasive medical devices in recent years has made the CoNS the pathogens most commonly isolated from bloodstream infections in intensive care units (11). While the determination of virulence factors has not been pursued as vigorously for CoNS as for S. aureus, it is evident that capsular polysaccharides are a major factor contributing to attachment to foreign bodies (27). Therefore, a more complete understanding of the mechanisms of disease caused by staphylococci is drastically needed.
The staphylococci reside primarily on the skin and mucous membranes of warm-blooded animals (33). In humans, approximately 30% of healthy individuals and up to 90% of health care workers are carriers of staphylococci (51). Once the bacteria enter the human body through a break in the skin or mucous membranes, they are confronted by the professional phagocytes (50). These host immune cells utilize reactive oxygen intermediates (ROIs), such as superoxide, hydrogen peroxide, and hydroxyl ions, to aid in the killing of phagocytosed bacteria (17, 32, 44). In addition, bacteria must also prevent damage to nucleic acids, proteins, and cell membranes from ROIs that arise from incomplete reduction of oxygen during aerobic respiration (reviewed in references 22 and 47). Most microorganisms that utilize aerobic respiration produce a number of enzymes that counteract the deleterious effects of ROIs (19, 29). For example, superoxide dismutase (SOD) converts superoxide to hydrogen peroxide and oxygen and catalase converts hydrogen peroxide to water and oxygen (22, 47).
SOD has been shown to be important in several bacteria for defense against killing by professional phagocytes of vertebrate hosts. Extracellular SODs, such as those from Mycobacterium tuberculosis and Nocardia asteroides, as well as the periplasmically located Cu/Zn SOD from Escherichia coli protect these microorganisms from phagocytic killing (2, 5, 6, 7). Inactivation of the cytoplasmically located SODs of Shigella flexneri and E. coli K-12 results in increased sensitivity to killing by serum and neutrophils (21, 38). In addition, sod mutations in Streptococcus pneumoniae, Campylobacter coli, Yersinia enterocolitica, and Haemophilus influenzae result in attenuation of virulence, reduced colonization of the chicken stomach, decreased survival in the spleens and livers of mice, and the inability to colonize the rat nasopharynx, respectively (15, 41, 42, 53).
Apart from earlier conflicting reports (31, 37) regarding the importance of staphylococcal SOD in disease, the role of SOD in S. aureus has only recently been addressed. This organism contains two genes, sodM and sodA, that account for three SOD activities (13, 40, 48). The sodM and sodA gene products are important for the viability of S. aureus when grown under oxidative stress conditions (13, 48). In addition, the ability to survive amino acid starvation during aerobic growth is reduced in a S. aureus sodA mutant (13, 52). However, the sodA mutation did not affect the organism's ability to recover from starvation (13, 52). While a sodA mutation in S. aureus was demonstrated to have no effect on virulence in a mouse abscess model (13), the effect of sodM and sodM sodA mutations on virulence has not been determined.
Because S. aureus produces three SODs, a characteristic unique among the gram-positive bacteria, the purpose of the present study was to determine if SOD activities are different among species of Staphylococcus. Results from this study indicate that unlike S. aureus, the CoNS produced only one SOD activity, which is most closely related to SodA of S. aureus with respect to migration on activity gels, Southern analysis, and amino acid similarities. Because S. aureus is considered a primary pathogen and the CoNS are typically recognized as opportunistic pathogens, the presence of a second SOD (namely, SodM) in S. aureus may be related to this organism's ability to cause disease.
MATERIALS AND METHODS
Staphylococcal strains and growth conditions.
Staphylococcal strains used in this study are listed in Table 1. Strains were routinely grown overnight (15 to 18 h) in tryptic soy broth (Difco Laboratories, Detroit, Mich.) at 37°C with rotary aeration (180 rpm) or on tryptic soy agar plates (tryptic soy broth containing 1.5% agar). Clinical isolates were provided by Larry Kemp of the Osteopathic Medical Center of Texas, Fort Worth, Tex., and by Ken Waites, Division of Laboratory Medicine, University of Alabama at Birmingham. Gram-positive cocci possessing catalase activity were categorized as coagulase positive or negative by inoculating 0.5 ml of reconstituted rabbit plasma (Difco Laboratories) with a single isolated colony and incubating at 37°C for 3 h. Tubes were observed for the presence of a fibrin clot. Species identification and characterization were carried out by using positive combination type 6 panels (Dade International, Inc., West Sacramento, Calif.) read after a 16- to 24-h incubation at 35°C with a Microscan Walkaway automated instrument (Dade). These panels use the results of 18 separate biochemical tests and the susceptibility to 18 different antibiotics to identify and characterize Staphylococcus species (Dade).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristics | Sourcea |
|---|---|---|
| S. aureus | ||
| RN6390 | Prototypic strain | M. S. Smeltzer, University of Arkansas for Medical Sciences |
| ATCC 25923 | ATCC | |
| HAC1 | Blood | OMCT |
| HAC2 | Skin abscess | OMCT |
| HAC3 | Peritoneal fluid | OMCT |
| HAC4 | Respiratory secretions | OMCT |
| HAC6 | Surgical wound | OMCT |
| UAB1 | Sputum | UAB |
| UAB2 | Sputum | UAB |
| UAB4 | Sputum | UAB |
| UAB5 | Sputum | UAB |
| CoNS | ||
| S. epidermidis ATCC 12228 | ATCC | |
| S. epidermidis HAC33 | Blood | OMCT |
| S. epidermidis HAC36 | Surgical wound | OMCT |
| S. epidermidis HAC94 | Unknown | OMCT |
| S. epidermidis HAC111 | Triple lumen catheter | OMCT |
| S. epidermidis HAC112 | Wound isolate | OMCT |
| S. epidermidis HAC113 | Blood | OMCT |
| S. epidermidis HAC114 | Triple lumen catheter | OMCT |
| S. epidermidis HAC115 | Blood | OMCT |
| S. auricularis HAC140 | Unknown | OMCT |
| S. auricularis HAC145 | Deep tissue wound | OMCT |
| S. capitis HAC138 | Tissue wound | OMCT |
| S. carnosus KSI2019 | Food isolate | J. J. Iandolo, University of Oklahoma Health Science Center |
| S. haemolyticus HAC146 | Arterial line catheter | OMCT |
| S. hominis HAC79 | Blood | OMCT |
| S. hominis HAC139 | Tissue wound | OMCT |
| S. hominis HAC142 | Urine | OMCT |
| S. hominis HAC144 | Blood | OMCT |
| S. lugdunensis HAC51 | Abscess | OMCT |
| S. lugdunensis HAC141 | Deep tissue wound | OMCT |
| S. simulans HAC44 | Deep tissue wound | OMCT |
| S. simulans HAC143 | Blood | OMCT |
ATCC, American Type Culture Collection, Manassas, Va.; OMCT, Osteopathic Medical Center of Texas; UAB, University of Alabama at Birmingham.
Preparation of cell lysates and SOD activity assay.
Whole-cell lysates of staphylococcal strains were prepared by using the procedure of Valderas and Hart (48). Total protein of whole cell lysates was determined by the Bradford assay (Bio-Rad Laboratories, Richmond, Calif.). Cell protein (5 or 50 μg) was loaded onto 15% (wt/vol) nondenaturing polyacrylamide gels and separated by electrophoresis in buffer lacking sodium dodecyl sulfate. SOD activity was determined by the nitroblue tetrazolium negative staining method of Beauchamp and Fridovich (8).
SOD subunit composition.
Whole-cell lysates of S. aureus RN6390, S. epidermidis ATCC 12228, and the sodA mutant of S. aureus RN6390 containing the S. epidermidis sod gene on a plasmid (pCL15 epi-sod) were prepared as described above. Total protein (5 μg) from each strain along with standard proteins of known molecular weights (Sigma) were loaded on nondenaturing polyacrylamide gels of various concentrations (12, 15, 18, and 21%). Proteins were separated by electrophoresis and stained for SOD activity as previously described (8). The relative mobility (Rf) of each band of activity was determined. Gels were then rinsed in water overnight and stained with Coomassie brilliant blue (Fisher) (4), and the Rf values for the standard proteins were determined. Rf values for standard proteins and SOD activity bands were used to generate Ferguson plots (20) as per Sigma technical bulletin no. MKR-137. The correlation (r) of slopes versus the molecular weights of standard proteins was 0.987.
Chromosomal DNA analysis, cloning, and complementation.
Chromosomal DNA was isolated from staphylococci by the guanidine-HCl-CsCl method described by Dyer and Iandolo (16). DNA was digested with either EcoRI or HindIII, resolved by agarose gel electrophoresis, and transferred by passive diffusion to neutral nylon membranes (MagnaGraph; Micron Separations Inc., Westborough, Mass.). Membranes were hybridized overnight (18 to 24 h) at 65°C with PCR products containing either sodM, sodA, or S. epidermidis ATCC 12228 sodA labeled with digoxigenin-11-UTP (Roche Molecular Biochemicals, Indianapolis, Ind.) as described by Smeltzer et al. (46) and Hart et al. (25). Hybridizing probes were detected by autoradiography with alkaline phosphatase-conjugated antidigoxigenin F(ab′)2 antibody fragments (Roche Molecular Biochemicals) and the chemiluminescent substrate CDP-Star (Roche Molecular Biochemicals).
Sequence from the 5′ end of the putative sod gene reported by Heidrich et al. (26) for S. epidermidis BN280 (GenBank accession no. X97011) was used to perform a BLAST search of the genomic DNA database (in progress) for S. epidermidis RP62A maintained by The Institute for Genomic Research (TIGR) (http://www.tigr.org). The oligonucleotide primers 5′-AGGCCATTGGTCGTATTT-3′ and 5′-GCAAATCATCTAAGGGCTATG-3′ were designed and used to amplify an approximately 0.9-kbp region containing the sodA gene from S. epidermidis ATCC 12228 by PCR. The PCR product was ligated into pCR2.1 (Invitrogen, Carlsbad, Calif.) and used to transform E. coli INVαF′, as recommended by the manufacturer (Invitrogen). Plasmid DNA from antibiotic-resistant transformants was isolated using a plasmid miniprep kit (Bio-Rad Laboratories) and digested with EcoRI to verify the presence of an approximately 0.9-kbp insert. Plasmid DNA containing the S. epidermidis sodA gene (pCR2.1 epi-sod) was sequenced at the University of Arkansas for Medical Sciences DNA Sequencing Core Facility (Little Rock) with a DNA sequencer (Perkin-Elmer Biosystems, Foster City, Calif.).
The EcoRI fragment containing the S. epidermidis sodA gene was also ligated into the shuttle vector pCL15 (kindly provided by Chia Lee at the University of Kansas Medical Center) and transformed into E. coli HB101. Plasmid DNA (pCL15 epi-sod) isolated from antibiotic-resistant colonies was used to transform the S. aureus RN4220 sodA mutant (48) by electroporation (35). Plasmid isolation and Southern analysis were used to confirm the presence of pCL15 epi-sod in chloramphenicol-resistant transformants. Plasmid pCL15 was also transformed into the sodA mutant as a vector control.
Nucleotide sequence accession number.
The full-length sodA gene from S. epidermidis ATCC 12228 has been assigned GenBank accession number AF410177.
RESULTS AND DISCUSSION
SOD subunit composition.
Most procaryotic SOD proteins studied thus far exist as either dimers or tetramers (10). Genetic evidence from a previous study demonstrated that at least the middle band of activity found in cell lysates of S. aureus consists of a multimeric form composed of SodM and SodA (48). In the present study, we compared the relative mobility of each band of SOD activity to that of proteins of known molecular weight by nondenaturing polyacrylamide gel electrophoresis (PAGE) and SOD staining. These values were used to generate Ferguson plots (20), which allowed the calculation of the molecular weight of each band of SOD activity. Results indicate that the S. aureus SodM and SodA, the S. epidermidis SodA and the hybrids composed of SodM and SodA exist as dimers (Table 2).
TABLE 2.
Subunit composition of staphylococcal SODs
| SOD | Predicted mol wta | Mol wt of active enzymeb | No. of subunits/active enzymec |
|---|---|---|---|
| S. aureus SodM | 23,015 | 45,512 | 1.98 |
| S. aureus SodA | 22,686 | 38,572 | 1.70 |
| S. epidermidis SodA | 22,679 | 38,572 | 1.70 |
| S. aureus SodM/SodA | 41,953 | 1.82/1.85 | |
| S. aureus SodM/S. epidermidis SodA | 41,953 | 1.82/1.85 |
The predicted molecular weight of each SOD was calculated from the deduced amino acid composition of the respective gene.
The molecular weight of each active enzyme was estimated by Ferguson plot (20) as described in Materials and Methods.
Calculated by dividing the molecular weight of the active enzyme by the predicted molecular weight. Values for the hybrid forms were calculated using the predicted molecular weights of SodM and SodA, respectively.
SOD profiles among clinical isolates of S. aureus.
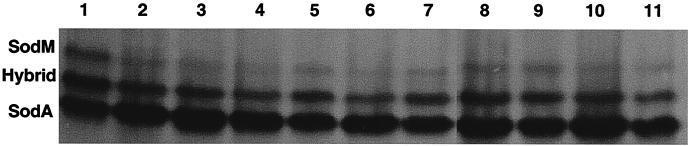
Because the SOD profile of laboratory strains of S. aureus appears to be unique among the gram-positive bacteria (23, 24, 30, 39, 48, 49, 53), we examined the SOD profiles of other staphylococci, in particular, clinical isolates of S. aureus and CoNS. Clinical S. aureus strains isolated from two different geographical locations were analyzed for SOD activity using nondenaturing PAGE and SOD staining (Fig. 1). Cell lysates from all eight S. aureus isolates (Fig. 1, lanes 2 to 6 and 8 to 11) as well as ATCC 25923 (lane 7) demonstrated a SOD profile similar to that of S. aureus RN6390 (lane 1). Originally we loaded 5 μg of total protein from each clinical strain and observed only two bands of SOD activity that exhibited a migration pattern identical to that of the SodA and SodM/SodA hybrid proteins of S. aureus RN6390. Analysis of 50 μg of total protein revealed, in each case, a third band of activity that migrated to a position similar to that of SodM of S. aureus RN6390 (Fig. 1). Interestingly, the level of activity for SodM ranged from detectable to approximately half of that observed for S. aureus RN6390. In addition, in all S. aureus strains (including laboratory strains) examined thus far, the SodM homodimer band of activity has always been less than the heterodimeric hybrid band composed of SodM and SodA (48). It is not known at present why the SOD heterodimer would exhibit more activity than the homodimer SodM protein. Perhaps the heterodimer is more stable than the SodM homodimer in cell lysates or the SodM homodimer is either secreted or associated with the cytoplasmic membrane, which has been observed with some bacterial SODs (1, 7, 14, 18, 24). However, we have compared SOD activity from S. aureus spent media to that of cell lysates and determined that the specific activity for all three SODs is approximately the same for both preparations, thereby suggesting that the SOD activity associated with spent media is not the result of secretion (data not shown).
FIG. 1.
Activity gel analysis of S. aureus SOD. Lane 1, RN6390; lane 2, HAC1; lane 3, HAC2; lane 4, HAC3; lane 5, HAC4; lane 6, HAC6; lane 7, ATCC 25923; lane 8, UAB1; lane 9, UAB2; lane 10, UAB4; lane 11, UAB5. Each lane contains approximately 50 μg of protein (see Results and Discussion). Stained gels were scanned with an AlphaImager 2000 (Alpha Innotech Corp.) imaging system, and the inverse image was generated with NIH Image software.
SOD profiles among CoNS.
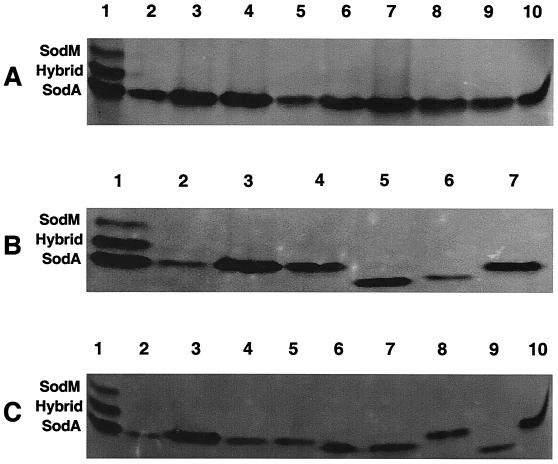
Given the S. aureus characteristic three bands of SOD activity, we decided to determine whether CoNS have similar SOD activities. Cell lysate total protein (5 μg) from each of 22 CoNS encompassing eight different species was resolved by nondenaturing PAGE and stained for SOD activity (Fig. 2). All 22 CoNS exhibited a single band of SOD activity migrating to a position similar to that of SodA of S. aureus RN6390 (Fig. 2, lanes 1). Activity of the single band ranged from barely detectable to equaling the SodA band of S. aureus RN6390 (Fig. 2). Levels of activity among the nine S. epidermidis strains (Fig. 2A, lanes 2 to 10), which included ATCC 12228, were approximately the same. These data indicate that CoNS contain only one SOD activity with a migratory pattern similar to that of the S. aureus SodA protein. While we were unable to isolate sufficient amounts of total protein from all CoNS strains, for those that we were able to isolate, no additional bands of activity were observed when 50 μg of protein was resolved by nondenaturing PAGE and stained for SOD activity (data not shown).
FIG. 2.
Activity gel analysis of CoNS. (A) S. epidermidis isolates. Lane 1, S. aureus RN6390; lane 2, HAC33; lane 3, HAC36; lane 4, HAC94; lane 5, HAC111; lane 6, HAC112; lane 7, HAC113; lane 8, HAC114; lane 9, HAC115; lane 10, ATCC 12228. (B and C) Other CoNS. (B) Lane 1, S. aureus RN6390; lane 2, S. auricularis HAC140; lane 3, S. auricularis HAC145; lane 4, S. capitis HAC138; lane 5, S. carnosus KSI2019; lane 6, S. haemolyticus HAC146; lane 7, S. epidermidis ATCC 12228. (C) Lane 1, S. aureus RN6390; lane 2, S. hominis HAC79; lane 3, S. hominis HAC139; lane 4, S. hominis HAC142; lane 5, S. hominis HAC144; lane 6, S. lugdunensis HAC51; lane 7, S. lugdunensis HAC141; lane 8, S. simulans HAC44; lane 9, S. simulans HAC143; lane 10, S. epidermidis ATCC 12228. Each lane contains approximately 5 μg of protein (see Results and Discussion). The gels were analyzed and the image was generated as described for Fig. 1.
Southern analysis of clinical isolates of CoNS.
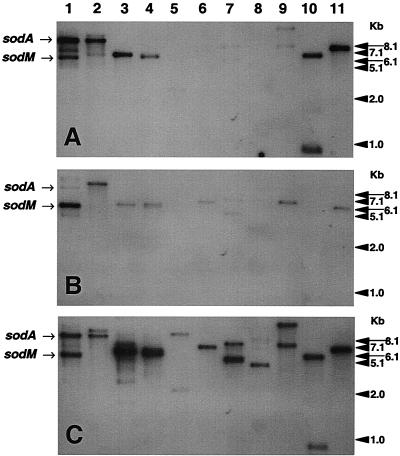
To determine whether the lack of additional bands of SOD activity among the CoNS was due to the absence of a S. aureus sodM gene equivalent, we isolated chromosomal DNA from a representative strain of each of the eight species of CoNS examined. The DNA was digested with either EcoRI (Fig. 3) or HindIII (data not shown) and hybridized with probes generated from the S. aureus RN6390 sodM and sodA genes and from the sodA gene isolated from S. epidermidis ATCC 12228. The nucleic acid sequences of all three sod genes are 74% identical (data not shown), and as expected, at a hybridization temperature of 65°C, some cross-hybridization occurred. This is particularly evident with S. aureus strains RN6390 and UAB1 (Fig. 3, lanes 1 and 2). All three probes were able to hybridize with EcoRI fragments containing the sodA and sodM genes, although the fragment containing the sodA gene was barely detectable when probed with sodM (Fig. 3, lanes 1 and 2). Nevertheless, Southern analysis using these probes indicates that the CoNS examined in this study contain only one sod gene. A single EcoRI fragment-hybridizing band was observed for S. epidermidis ATCC 12228 and clinical isolates, Staphylococcus carnosus, Staphylococcus simulans, and Staphylococcus auricularis, while Staphylococcus hominis, Staphylococcus capitis, Staphylococcus lugdunensis, and Staphylococcus haemolyticus exhibited two EcoRI fragment-hybridizing bands (Fig. 3). In all cases, the hybridizing bands were identical in size regardless of which probe was used, and the intensity of the hybridizing bands was always greater with the S. aureus sodA or S. epidermidis sod probe than with the sodM probe. In addition, species exhibiting two hybridizing bands with EcoRI-digested DNA exhibited only one band when digested with HindIII and hybridized with the sod probes, indicating only one sod gene in these species (data not shown). Chromosomal DNA hybridized with the sodM probe and in some cases those probed with sodA required extended exposure times in order to detect hybridizing bands. No bands were detected for S. auricularis and S. haemolyticus when hybridized with the sodM probe (Fig. 3B, lanes 8 and 10). While the clinical isolate of S. epidermidis (Fig. 3C, lane 11) exhibited a single hybridizing band when probed with the S. epidermidis sod, S. epidermidis ATCC 12228 exhibited a number of less intense bands (Fig. 3C, lane 3). These hybridizing bands may share a level of nucleic acid relatedness to the S. epidermidis sod probe. However, it is unlikely that these fragments represent additional sod genes due to the appearance of only one band of SOD activity (Fig. 2A, lane 10) and the absence of additional genes identified through searches of the DNA database. In addition, the sodA probe also hybridized to additional EcoRI chromosomal fragments of S. aureus, albeit less intensely than the EcoRI fragments containing either the sodM or sodA genes (Fig. 3A and C, lanes 1 and 2). Again, it is unlikely that these hybridizing fragments represent additional sod genes, since SOD activity is undetectable in a sodM sodA mutant of S. aureus (48). However, it is possible that additional sod genes were not expressed due to the growth conditions employed in our study or that the genes contained mutations. In addition, the products of these genes could be unstable in cell lysates. The failure to find the E. coli Cu/Zn SOD until recently has been attributed to the loss of the protein during isolation primarily due to its periplasmic location but also due to instability during the isolation procedures employed (9).
FIG. 3.
Southern analysis of S. aureus and CoNS chromosomal DNA hybridized with probes specific for S. aureus sodA (A), S. aureus sodM (B), or S. epidermidis sod (C). Lane 1, S. aureus RN6390; lane 2, S. aureus UAB1; lane 3, S. epidermidis ATCC 12228; lane 4, S. simulans HAC143; lane 5, S. hominis HAC 79; lane 6, S. carnosus KSI2019; lane 7, S. capitis HAC138; lane 8, S. auricularis HAC140; lane 9, S. lugdunensis HAC141; lane 10, S. haemolyticus HAC146; lane 11, S. epidermidis HAC33. Arrows show the positions of the S. aureus RN6390 EcoRI fragment-hybridizing bands that contain the sodM and sodA genes.
sod gene from S. epidermidis.
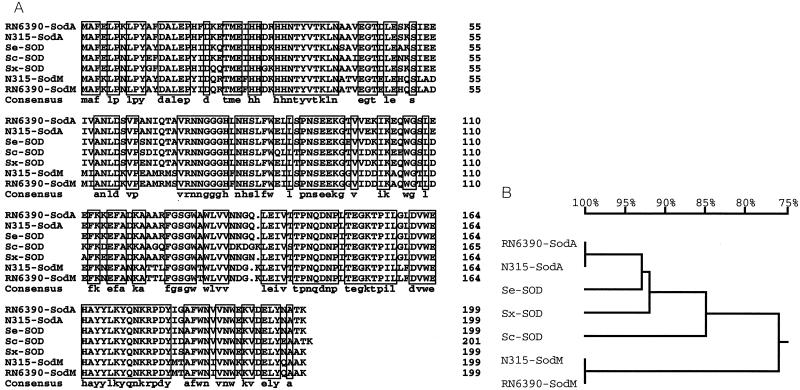
To assess the relatedness of the sod gene from S. epidermidis ATCC 12228 to the sodM and sodA genes of S. aureus, the nucleic acid sequence of the 5′ end of the putative sod gene of S. epidermidis BN280 (26) (GenBank accession no. X97011) was used to search the S. epidermidis RP62A genomic sequence maintained by TIGR (http://www.tigr.org) to find the entire open reading frame of the sod gene. PCR primers designed from this sequence were used to amplify an approximately 0.9-kbp fragment from chromosomal DNA of S. epidermidis ATCC 12228, which was cloned and sequenced. The deduced amino acid sequence of the ORF was 100% identical to the sequence reported for S. epidermidis RP62A by TIGR (data not shown). The predicted amino acid sequence of the S. epidermidis sod ORF is 92 and 76% identical to the amino acid sequences of SodA and SodM of S. aureus RN6390, respectively (Fig. 4A). In addition, the predicted SOD amino acid sequences from S. carnosus (AJ295150) and Staphylococcus xylosus (AJ276960) and the recently reported sequences from S. aureus N315 (AP003129 and AP003134 for sodM and sodA, respectively) (36) were included for comparison (Fig. 4A). The homologs share 67.8% identity as a group.
FIG. 4.
(A) Amino acid sequence alignments of staphylococcal SODs. The S. aureus RN6390 SodA (48) and SodM (48) (AF273269) proteins, the S. aureus N315 SodA and SodM proteins (36) (AP003129 and AP003134, respectively), and the Sod proteins of S. epidermidis (Se) ATCC 12228 (AF410177), S. carnosus (Sc) (CAC14833), and S. xylosus (Sx) (CAB95744) are shown. Consensus sequences are boxed. A gap at position 136 of the SodM sequences was manually inserted. (B) Phylogenetic tree. Amino acid sequences were analyzed with DNAMAN (Lynnon BioSoft, Vaudreuil, Quebec, Canada) software, which uses the neighbor-joining method described by Saitou and Nei (43).
These sequences were also used to generate a phylogenetic tree (Fig. 4B) to determine their relatedness. Data from this analysis suggest that the SODs in S. epidermidis, S. carnosus, and S. xylosus are closely related to SodA of S. aureus, whereas these proteins along with SodA of S. aureus are only distantly related to SodM, exhibiting 51% similarity to it.
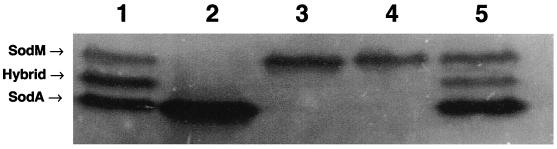
In addition, the sod gene from S. epidermidis ATCC 12228 was cloned into the staphylococcal shuttle vector pCL15 and transformed into the sodA mutant of S. aureus (Fig. 5). Cell lysates from the parental strain, S. aureus RN4220 (Fig. 5, lane 1), and S. epidermidis ATCC 12228 (Fig. 5, lane 2) exhibited the expected patterns of SOD activity. Only the SodM band of activity was observed in the sodA mutant (48) (Fig. 5, lane 3) and the sodA mutant containing pCL15 (Fig. 5, lane 4). However, the sodA mutant containing pCL15 epi-sod exhibited a band of SOD activity comparable to that of S. epidermidis and a hybrid band of activity similar to that observed for the parental S. aureus strain (48) (Fig. 5, lane 5). These data demonstrate that the S. epidermidis sodA gene expressed in S. aureus and the gene product formed a hybrid with SodM of S. aureus and, like the hybrid SOD band seen with wild-type S. aureus, exist as a heterodimer (Table 2). The formation of a hybrid of two SOD proteins is seen not only in S. aureus (48) but also in E. coli (12). In E. coli the hybrid is a dimeric protein consisting of one subunit each of SodA (Mn-containing enzyme) and SodB (Fe-containing enzyme). In E. coli as well as in S. aureus, it is not known whether the formation of a hybrid SOD protein has physiological relevance or is the result of subunit exchange between two related proteins.
FIG. 5.
SOD activity gel of S. aureus sodA mutant complemented with S. epidermidis sod. Lane 1, S. aureus RN4220; lane 2, S. epidermidis ATCC 12228; lane 3, S. aureus RN4220 sodA mutant; lane 4, S. aureus RN4220 sodA mutant containing pCL15; lane 5, S. aureus RN4220 sodA mutant containing pCL15 epi-sod. The gel was analyzed and the image was generated as described for Fig. 1.
Originally, it was reported that sodM from S. aureus RN6390 encoded a 187-amino-acid protein with 76% identity to SodA (48) (GenBank accession no. AF273269). We have now determined that the sodM sequence that was reported earlier contained an incorrect base that resulted in a 12-amino-acid truncation of the predicted sodM ORF. The correct base was verified by sequencing the region containing the base in question, and we now report that the S. aureus RN6390 sodM ORF encodes a protein of 199 amino acids with a predicted molecular mass of 22.7 kDa (Fig. 4A).
In summary, we have determined that eight representative species of the CoNS contain only one sod gene that yields one band of SOD activity as determined by nondenaturing PAGE and staining for SOD activity. This is in contrast to S. aureus, which contains two genes responsible for three bands of activity (48). The putative amino acid sequence from three CoNS sod genes indicates that these genes are more similar to the sodA gene than the sodM gene of S. aureus. Therefore, the S. epidermidis gene isolated in this study is designated sodA. Furthermore, the CoNS SOD proteins migrate on activity gels to a position similar to that of the S. aureus SodA protein. Whether the differences observed with the SOD profiles between S. aureus and the CoNS represent an important divergence in the evolution of the staphylococci is not known at present. However, the origin of the S. aureus sodM gene is an intriguing question. We recently demonstrated that the sodM gene is important in maintaining viability under oxidative stress conditions in an S. aureus strain containing a sodA mutation (48). Expression of sodM increased as cells entered the postexponential and stationary phases of growth (48) similar to that observed for sodA (13). However, while SodA is the most abundant of the three SOD activities observed, the increase in total SOD activity as cells entered the postexponential and stationary phases of growth is attributed to the increased production of SodM (48). These data suggest a regulatory mechanism for sodM independent of sodA and a unique role for SodM. Results of the present study support a unique role for sodM in that all S. aureus isolates, including those isolated from the clinical environment, contain sodM while the CoNS do not. As a pathogen, S. aureus is certainly better equipped than the CoNS, as it produces numerous toxins, enzymes, and cell wall-associated proteins, which concertedly cause a wide variety of disease syndromes in humans (28). Perhaps the S. aureus SodM protein is yet another important factor that contributes to the disease-causing ability of this organism. Studies addressing this question are in progress.
ADDENDUM
During the review of this article, a paper that described the use of PCR to amplify an internal fragment of the sodA gene in 40 CoNS type strains was published (40a). In that study, protein (50 μg) from cell lysates of the 40 CoNS strains as well as 25 unrelated clinical strains of S. aureus was resolved by nondenaturing PAGE and stained for SOD activity. All CoNS type strains exhibited a single band of SOD activity while all of the S. aureus isolates exhibited three closely migrating bands of SOD activity. Our data are in agreement with the finding of Poyart et al. that CoNS strains express only one SOD while S. aureus strains express three.
Acknowledgments
This work was supported by Public Health Service grant AI-36934 from the National Institute of Allergy and Infectious Diseases and a Faculty Research Grant from the University of North Texas Health Science Center (UNTHSC).
We are indebted to Tony Romeo and Jerry Simecka at UNTHSC and John Iandolo at OUHSC for helpful discussions, critical reading of the manuscript, and continuous encouragement throughout this work. We are also indebted to Larry Kemp of the Osteopathic Medical Center of Texas and Ken Waites, Division of Laboratory Medicine, University of Alabama at Birmingham, for providing clinical isolates. A special thanks goes to Allen Gies of the University of Arkansas for Medical Sciences DNA Sequencing Core Facility for sequencing. Preliminary sequence data were obtained from TIGR, University of Oklahoma's Advanced Center for Genome Technology, and the National Center for Biotechnology Information.
REFERENCES
- 1.Abou-Zeid, C., I. Smith, J. M. Grange, T. L. Ratliff, J. Steele, and G. A. W. Rook. 1988. The secreted antigens of Mycobacterium tuberculosis and their relationship to those recognized by the available antibodies. J. Gen. Microbiol. 134:531-538. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, P., D. Askgaard, L. Ljungqvist, J. Bennedsen, and I. Heron. 1991. Proteins released from Mycobacterium tuberculosis during growth. Infect. Immun. 59:1905-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, G. L. 1995. Staphylococcus epidermidis and other coagulase-negative staphylococci, p. 1777-1784. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 4th ed. Churchill Livingstone, New York, N.Y.
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology, p. 10.6.1-10.6.2. John Wiley & Sons, Inc., New York, N.Y.
- 5.Battistoni, A., G. Donnarumma, R. Greco, P. Valenti, and G. Rotilio. 1998. Overexpression of a hydrogen peroxide-resistant periplasmic Cu, Zn superoxide dismutase protects Escherichia coli from macrophage killing. Biochem. Biophys. Res. Commun. 243:804-807. [DOI] [PubMed] [Google Scholar]
- 6.Beaman, B. L., C. M. Black, F. Doughty, and L. Beaman. 1985. Role of superoxide dismutase and catalase as determinants of pathogenicity of Nocardia asteroides: importance in resistance to microbicidal activities of human polymorphonuclear neutrophils. Infect. Immun. 47:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaman, B. L., S. M. Scates, S. E. Moring, R. Deem, and H. P. Misra. 1983. Purification and properties of a unique superoxide dismutase from Nocardia asteroides. J. Biol. Chem. 258:91-96. [PubMed] [Google Scholar]
- 8.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 9.Benov, L. T., and I. Fridovich. 1994. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J. Biol. Chem. 269:25310-25314. [PubMed] [Google Scholar]
- 10.Beyer, W., J. Imlay, and I. Fridovich. 1991. Superoxide dismutases. Prog. Nucleic Acid Res. Mol. Biol. 40:221-253. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 1999. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1990-May 1999, issued June 1999. Am. J. Infect. Control 27:520-532. [DOI] [PubMed] [Google Scholar]
- 12.Clare, D. A., J. Blum, and I. Fridovich. 1984. A hybrid superoxide dismutase containing both functional iron and manganese. J. Biol. Chem. 259:5932-5936. [PubMed] [Google Scholar]
- 13.Clements, M. O., S. P. Watson, and S. J. Foster. 1999. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J. Bacteriol. 181:3898-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins, F. M., J. R. Lamb, and D. B. Young. 1988. Biological activity of protein antigens isolated from Mycobacterium tuberculosis culture filtrate. Infect. Immun. 56:1260-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Mello, R. A., P. R. Langford, and J. S. Kroll. 1997. Role of bacterial Mn-cofactored superoxide dismutase in oxidative stress responses, nasopharyngeal colonization, and sustained bacteremia caused by Haemophilus influenzae type b. Infect. Immun. 65:2700-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyer, D. W., and J. J. Iandolo. 1983. Rapid isolation of DNA from Staphylococcus aureus. Appl. Environ. Microbiol. 46:283-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsbach, P., and J. Weiss. 1988. Phagocytic cells: oxygen-independent antimicrobial systems, p. 445-470. In J. I. Gallin, I. M. Godstein, and R. Snyderman (ed.), Inflammation: basic principles and clinical correlates. Raven Press, New York, N.Y.
- 18.Escuyer, V., N. Haddad, C. Frehel, and P. Berche. 1996. Molecular characterization of a surface-exposed superoxide dismutase of Mycobacterium avium. Microb. Pathog. 20:41-55. [DOI] [PubMed] [Google Scholar]
- 19.Farr, S. B., and T. Kogoma. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferguson, K. A. 1964. Starch-gel electrophoresis: application to the classification of pituitary proteins and polypeptides. Metabolism 13:985-1002. [DOI] [PubMed] [Google Scholar]
- 21.Franzon, V. L., J. Arondel, and P. J. Sansonetti. 1990. Contribution of superoxide dismutase and catalase activities to Shigella flexneri pathogenesis. Infect. Immun. 58:529-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fridovich, I. 1995. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64:97-112. [DOI] [PubMed] [Google Scholar]
- 23.Gaillot, O., C. Poyart, P. Berche, and P. Trieu-Cuot. 1997. Molecular characterization and expression analysis of the superoxide dismutase gene from Streptococcus agalactiae. Gene 204:213-218. [DOI] [PubMed] [Google Scholar]
- 24.Gerlach, D., W. Reichardt, and S. Vettermann. 1998. Extracellular superoxide dismutase from Streptococcus pyogenes type 12 strain is manganese-dependent. FEMS Microbiol. Lett. 160:217-224. [DOI] [PubMed] [Google Scholar]
- 25.Hart, M. E., M. S. Smeltzer, and J. J. Iandolo. 1993. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J. Bacteriol. 175:7875-7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidrich, C., K. Hantke, G. Bierbaum, and H. G. Sahl. 1996. Identification and analysis of a gene encoding a Fur-like protein of Staphylococcus epidermidis. FEMS Microbiol. Lett. 140:253-259. [DOI] [PubMed] [Google Scholar]
- 27.Huebner, J., and D. A. Goldmann. 1999. Coagulase-negative staphylococci: role as pathogens. Annu. Rev. Med. 50:223-236. [DOI] [PubMed] [Google Scholar]
- 28.Iandolo, J. J. 1990. The genetics of staphylococcal toxins and virulence factors, p. 399-426. In B. H. Iglewski and V. L. Clark (ed.), Molecular basis of bacterial pathogenesis. Academic Press, Inc., New York, N.Y.
- 29.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1309. [DOI] [PubMed] [Google Scholar]
- 30.Inaoka, T., Y. Matsumura, and T. Tsuchido. 1998. Molecular cloning and nucleotide sequence of the superoxide dismutase gene and characterization of its product from Bacillus subtilis. J. Bacteriol. 180:3697-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanafani, H., and S. E. Martin. 1985. Catalase and superoxide dismutase activities in virulent and nonvirulent Staphylococcus aureus isolates. J. Clin. Microbiol. 21:607-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klebanoff, S. J. 1991. Myeloperoxidase: occurrence and biological function, p. 1-35. In J. Everse, K. E. Everse, and M. B. Grisham (ed.), Peroxidases in chemistry and biology. CRC Press, Inc., Boca Raton, Fla.
- 33.Kloos, W. E., and K. H. Schleifer. 1986. Staphylococcus, p. 1013-1035. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams and Wilkins Co., Baltimore, Md. [Google Scholar]
- 34.Kloos, W. E., and T. L. Bannermann. 1994. Update on the clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7:117-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373-376. [Google Scholar]
- 36.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Q. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 37.Mandell, G. L. 1975. Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. J. Clin. Investig. 55:561-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McManus, D. C., and P. D. Josephy. 1995. Superoxide dismutase protects Escherichia coli against killing by human serum. Arch. Biochem. Biophys. 317:57-61. [DOI] [PubMed] [Google Scholar]
- 39.Merkamm, M., and A. Guyonvarch. 2001. Cloning of the sodA gene from Corynebacterium melassecola and role of superoxide dismutase in cellular viability. J. Bacteriol. 183:1284-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poyart, C., P. Berche, and P. Trieu-Cuot. 1995. Characterization of superoxide dismutase genes from gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol. Lett. 131:41-45. [DOI] [PubMed] [Google Scholar]
- 40a.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purdy, D., S. Cawthraw, J. H. Dickinson, D. G. Newell, and S. F. Park. 1999. Generation of a superoxide dismutase (SOD)-deficient mutant of Campylobacter coli: evidence for the significance of SOD in Campylobacter survival and colonization. Appl. Environ. Microbiol. 65:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roggenkamp, A., T. Bittner, L. Leitritz, A. Sing, and J. Heesemann. 1997. Contribution of the Mn-cofactored superoxide dismutase (SodA) to the virulence of Yersinia enterocolitica serotype O8. Infect. Immun. 65:4705-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 44.Segal, A. W. 1989. The electron transport chain of the microbicidal oxidase of phagocytic cells and its involvement in the molecular pathology of chronic granulomatous disease. J. Clin. Investig. 83:1785-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smeltzer, M. S. 2000. Characterization of staphylococcal adhesins for adherence to host tissues, p. 411-444. In Y. H. An and R. J. Friedman (ed.), Handbook of bacterial adhesion, principles, methods and applications. Humana Press, Totowa, N.J.
- 46.Smeltzer, M. S., M. E. Hart, and J. J. Iandolo. 1993. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus. Infect. Immun. 61:919-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Touati, D. 1997. Superoxide dismutases in bacteria and pathogen protists, p. 447-493. In J. G. Scadalios (ed.), Oxidative stress and the molecular biology of antioxidant defenses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Valderas, M. W., and M. E. Hart. 2001. Identification and characterization of a second superoxide dismutase gene (sodM) from Staphylococcus aureus. J. Bacteriol. 183:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasconcelos, J. A., and H. G. Deneer. 1994. Expression of superoxide dismutase in Listeria monocytogenes. Appl. Environ. Microbiol. 60:2360-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhoef, J. 1997. Host defense against infection, p. 213-232. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 51.Waldvogel, F. A. 1995. Staphylococcus aureus (including toxic shock syndrome), p. 1754-1778. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 4th ed. Churchill Livingstone, New York, N.Y.
- 52.Watson, S. P., M. Antonio, and S. J. Foster. 1998. Isolation and characterization of Staphylococcus aureus starvation-induced, stationary-phase mutants defective in survival or recovery. Microbiology 144:3159-3169. [DOI] [PubMed] [Google Scholar]
- 53.Yesilakaya, H., A. Kadioglu, N. Gingles, J. E. Alexander, T. J. Mitchell, and P. W. Andrew. 2000. Role of the manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect. Immun. 68:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]