Abstract
The terminal enzyme of heme biosynthesis, ferrochelatase (EC 4.99.1.1), catalyzes the insertion of ferrous iron into protoporphyrin IX to form protoheme. Prior to the present work, [2Fe-2S] clusters have been identified and characterized in animal ferrochelatases but not in plant or prokaryotic ferrochelatases. Herein we present evidence that ferrochelatases from the bacteria Caulobacter crescentus and Mycobacterium tuberculosis possess [2Fe-2S] clusters. The enzyme from C. crescentus is a homodimeric, membrane-associated protein while the enzyme from M. tuberculosis is monomeric and soluble. The clusters of the C. crescentus and M. tuberculosis ferrochelatases are ligated by four cysteines but possess ligand spacings that are unlike those of any previously characterized [2Fe-2S] cluster-containing protein, including the ferrochelatase of the yeast Schizosaccharomyces pombe. Thus, the microbial ferrochelatases represent a new group of [2Fe-2S] cluster-containing proteins.
The enzyme ferrochelatase (EC 4.99.1.1) catalyzes the insertion of ferrous iron into protoporphyrin IX to form protoheme IX (heme) (4, 6). This enzyme is present in essentially all eukaryotes and prokaryotes with the exception of a few obligate pathogens and some anaerobic prokaryotes. Recently a gene sequence that appears by homology to be ferrochelatase was reported for the archaeon, Thermoplasma acidophilum (17). A comparison of all currently known ferrochelatase sequences reveals the existence of three distinct segments or domains (6). The first segment (I) is present in all eukaryotes and is identifiable as an amino-terminal organelle-targeting motif that is proteolytically removed after organelle targeting (12, 16). The second (II) represents the core 330 amino acid residues of the enzyme and is present in all ferrochelatases. The third (III) is a 30- to 50-amino-acid-residue sequence found at the carboxyl terminus of some ferrochelatases. Region III of animal ferrochelatases, which is involved in the dimerization motif for these enzymes (21), contains three of the four cysteinyl ligands for a [2Fe-2S] cluster with the fourth cluster ligand present in region II (3, 5, 19, 21). Region III of plant ferrochelatases is approximately 50 residues in length, and its role is currently unknown (6). Plant ferrochelatases do not possess the cysteinyl residues in region III and do not contain [2Fe-2S] clusters. The ferrochelatase of the yeast Saccharomyces cerevisiae, which has been cloned, purified, and characterized (9, 10, 13), possesses region III, does not contain any cysteinyl residues in this region, and does not possess a [2Fe-2S] cluster. The gene for ferrochelatase of the yeast Schizosaccharomyces pombe has been sequenced (GenBank accession number AL022245) and, interestingly, revealed the presence of four cysteinyl residues in the same sequence position as is found in animal ferrochelatases. Previously, as part of a study on the carboxyl termini of eukaryotic ferrochelatases, it was determined that S. pombe ferrochelatase contains a [2Fe-2S] cluster (14).
All bacterial ferrochelatases previously biochemically characterized possessed only region II (see references 6 and 11). However, with the plethora of genome sequencing projects, there are now several putative ferrochelatases in the bacterial genome databases which also possess a region III in their derived amino acid sequences. Of the region III-possessing bacterial ferrochelatases identified to date, all contain cysteinyl residues, but none have the same cluster-ligating cysteinyl residue motif as is found in animal ferrochelatases or other [2Fe-2S]-containing proteins.
In the present work we have expressed, purified, and characterized ferrochelatases from the bacteria Caulobacter crescentus and Mycobacterium tuberculosis. Both of these possess region III. The C. crescentus enzyme is homodimeric and membrane-associated and contains [2Fe-2S] clusters. However, although the M. tuberculosis ferrochelatase contains region III and a [2Fe-2S] cluster, it is monomeric and soluble.
MATERIALS AND METHODS
Cloning of the ferrochelatase genes from C. crescentus and M. tuberculosis.
The DNA sequence coding for the putative C. crescentus ferrochelatase was found in The Institute for Genomic Research database with Drosophila ferrochelatase as the query sequence. C. crescentus genomic DNA was a gift from Michael Laub. The primers used for PCR of the full-length clone from the genomic DNA were sense, 5′-GTTTCTTGCTAGCACCCAGAAGCTCGCCGTCG-3′, and antisense, 5′-GTTTCTTAAGCTTTCACGCGGAGGCTCCTTCG-3′. The italicized portion was added to facilitate cloning (2); restriction sites for cloning are in bold; the underlined portion is homologous to the target gene. The gene was amplified with Pfu polymerase (Promega), cleaned with QiaexII (Qiagen), and cloned into pTrcHisA (Invitrogen) by using the NheI and HindIII cloning sites. Mutagenesis of C. crescentus ferrochelatase residues H104A, T133C, T133A, S132A/T133A/T134A, C158A, C328S, C332S, C339S, and C341S was accomplished by using Quikchange mutagenesis (Stratagene). The Δ328-347 truncation was accomplished with the same sense primer as for the wild-type expression plasmid and the antisense primer 5′-GTTTCTTAAGCTTTCAGGCGGAAGACACCGTCCC-3′; the coding region was amplified by PCR and cloned into pTrcHisA by using the NheI and HindIII sites as described above. Following confirmation of ferrochelatase activity, the sequence for C. crescentus ferrochelatase was submitted to GenBank.
The gene for the M. tuberculosis ferrochelatase was obtained by PCR from cosmid MTCY277 (gift of Karin Eiglmeier, GenBank accession number Z79701). The sense primer was 5′-GTTTCTTGGATCCCAATTTGATGCCGTCCTGCTG-3′; the antisense primer was 5′-GTTTCTTAAGCTTTCACGGCGATCCTGCACTCGG-3′. The gene was amplified with Pfu Turbo polymerase (Stratagene), and the resulting fragment was cloned into pTrcHisA (Invitrogen) by using the BamHI and HindIII cloning sites.
The sequences for all of the constructs described above were confirmed by the University of Georgia's Molecular Genetics Instrumentation Facility. Each of the resulting expression constructs was also transformed into Escherichia coli ΔhemH, a ferrochelatase-deficient strain (16), to assess its ability to complement the growth of these cells (7).
Expression and purification of recombinant ferrochelatases.
For expression of the proteins, a 100-ml culture of Circlegrow (Bio101, Inc.) containing 100 μg of ampicillin/ml (final concentration) was inoculated with E. coli JM109 containing an expression plasmid and grown at 30°C with shaking for 5 to 6 h. This culture was then used as an inoculum for 1 liter of Circlegrow, also containing 100 μg of ampicillin/ml, and grown for 18 to 20 h at 30°C with shaking. The cells were then harvested, resuspended in 65 ml of solubilization buffer (50 mM Tris-morpholinepropanesulfonic acid [Tris-MOPS] [pH 8.0], 0.1 M KCl, 1% [wt/vol] sodium cholate, 10 μg of phenylmethylsulfonyl fluoride/ml), sonicated four times (30 s each time) on ice, and centrifuged at 100,000 × g for 30 min at 4°C. The resulting supernatant was loaded onto a column containing 2.5 ml of a 50% slurry of Talon matrix (Clontech) previously equilibrated with solubilization buffer. The column was then washed with 25 ml of solubilization buffer, and the protein was eluted off the column matrix with solubilization buffer containing 300 mM imidazole. Enzyme assays were as described previously (18). Protein concentrations were calculated spectrophotometrically, and UV and visible light scans were acquired with a Cary 1G dual-beam spectrophotometer. The millimolar extinction coefficients at 280 nm (expressed per millimolar centimeter) employed were based upon amino acid composition and confirmed fluorimetrically by tryptophan content. The values for C. crescentus and M. tuberculosis are 53 and 50, respectively. Molecular weight determinations were made by using fast protein liquid chromatography (FPLC) in solubilization buffer. The iron content was determined spectrophotometrically with ferrozine and ascorbic acid (5) following denaturation of the enzyme at 90°C with 2.0% (wt/vol) sodium dodecyl sulfate (SDS). Triton X-114 partitioning was done as described previously (1).
Nucleotide sequence accession number.
The sequence for C. crescentus ferrochelatase was submitted to GenBank and assigned accession number AF184071.
RESULTS
Ferrochelatases of C. crescentus and M. tuberculosis were cloned and expressed as hexahistidine amino-tagged proteins in E. coli in the vector pTrcHisA. The C. crescentus enzyme was efficiently expressed by E. coli and yielded at a rate of approximately 10 mg per liter of culture. The yield of the M. tuberculosis preparations were somewhat lower, approximately 2.5 mg per liter of culture. The single Talon matrix column was sufficient to obtain homogeneous protein preparations (not shown). Unlike most animal ferrochelatases, the ferrochelatases of C. crescentus and M. tuberculosis were relatively stable upon storage at 4°C. The C. crescentus enzyme preparations remained in solution and active after over a week of storage at 4°C, and surprisingly, the characteristic spectra of the cluster remained even in the presence of 1% (wt/vol) SDS at room temperature for several minutes. In order to quantitate the iron content of this enzyme it was necessary to include 2% (wt/vol) SDS, ferrozine, and ascorbic acid in the buffer and to heat the sample to 90°C. Under these conditions, a stoichiometry of 1.6 Fe/mole of enzyme was found. The determination of subunit composition for the purified proteins was done by FPLC in the presence of 1.0% (wt/vol) sodium cholate. The C. crescentus enzyme was found to be a dimer based upon the predicted monomer molecular weight of 38,491. This observation is consistent with the suggestion based on the structure of human ferrochelatase that dimerization is mediated by carboxyl-terminal extension (21), which this ferrochelatase possesses. The enzyme required detergent for solubilization and partitioned into the detergent phase, rather than the aqueous phase, of the Triton X-114 two-phase systems, thereby demonstrating a hydrophobic nature, which is consistent with membrane association. This observation is consistent with the proposal that membrane association for ferrochelatase is attributable to the presence of a hydrophobic surface largely composed of a 12-amino-acid-residue region that is present as a hydrophobic lip on the active site pocket of human ferrochelatase (6, 21). It is present in both S. pombe (T. A. Dailey and H. A. Dailey, unpublished data) and C. crescentus ferrochelatases, but it is absent in known soluble ferrochelatases such as Bacillus subtilis ferrochelatase (11). The ferrochelatase of M. tuberculosis differs from the C. crescentus enzyme in that it does not partition into the detergent phase of the Triton X-114 two-phase system, indicating that it is not membrane associated, and by FPLC it was determined to be monomeric based upon its predicted molecular weight of 41,043.
Kinetic parameters were determined for the enzymes. For C. crescentus the apparent Kms are 6.2 μM for protoporphyrin and 30 μM for iron. The Vmax is 14.5 min−1. Interestingly, C. crescentus ferrochelatase does not use cobalt as do S. pombe (data not shown) and other animal ferrochelatases, but it does catalyze metallization with nickel with an apparent Km of 13 μM and a Vmax of 16.5 min−1. Kinetic parameters were not determined for the M. tuberculosis enzyme since the activity of that enzyme was very low. Currently we do not have an explanation for this low level of enzyme activity.
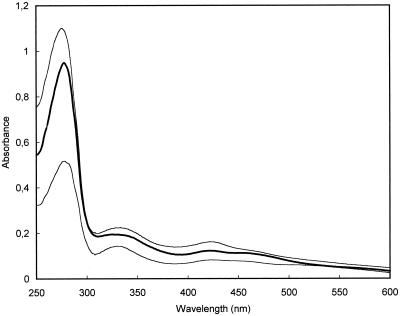
The absorbance spectra for ferrochelatases of all three organisms contain features characteristic of [2Fe-2S] cluster-containing proteins (Fig. 1). The amino acid sequence of S. pombe ferrochelatase contains cysteines analogous to the [2Fe-2S] four cluster-ligating cysteines that are found in animal ferrochelatases. However, C. crescentus ferrochelatase does not possess cysteines in these same positions, so a series of site-directed mutants were prepared and examined to determine the cluster ligands (Table 1). Since the spacing of the available cysteine residues of this protein differs dramatically from what has been found for animal ferrochelatases (Fig. 2), five cysteine residues, one histidine residue, and one serine residue were individually mutated. In addition, one triple mutation in the region from 132 to 134 was made and a carboxyl-terminal truncation was engineered. The data obtained with these mutants are consistent with a four-cysteinyl ligation involving C158, C332, C339, and C341.
FIG. 1.
UV and visible light spectra for ferrochelatases of S. pombe, C. crescentus, and M. tuberculosis. Spectra for the purified microbial ferrochelatases presented in this study are shown to illustrate the presence of spectral features of the [2Fe-2S] cluster that is present in each. The top line is for C. crescentus ferrochelatase (21 μM), the middle line (in bold) is for M. tuberculosis ferrochelatase (19 μM), and the bottom line is for S. pombe ferrochelatase (10 μM).
TABLE 1.
Site-directed mutants of C. crescentus ferrochelatase
| Mutation(s) | ΔhemH complementation | [2Fe-2S] cluster |
|---|---|---|
| H104A | + | + |
| T133C/A | + | + |
| S132A, T133A, T134A | − | Unstable |
| C158A | + | − |
| C328S | + | + |
| C332S | + | − |
| C339S | + | − |
| C341S | + | − |
| Δ328-347 | + | − |
FIG. 2.
A sequence pileup for C. crescentus, S. pombe, M. tuberculosis, and human ferrochelatases. The position of the [2Fe-2S] cluster-ligating residues for human ferrochelatase are noted with asterisks. The cysteine residues identified in the present study as ligands for C. crescentus are noted in bold italics, and putative cysteine ligands for M. tuberculosis are shown in an altered font. Conserved residues, including the carboxyl-terminal animal cysteine residue, are shown in shaded boxes.
One feature of C. crescentus ferrochelatase that is unlike any other characterized [2Fe-2S] cluster-containing ferrochelatase reported to date is that mutations which result in loss of the cluster do not cause a complete loss of enzyme activity. The C339S mutant retains approximately 5% of the wild-type activity. Although it was not possible to detect enzyme activity by the in vitro assay, the carboxyl-terminal truncation (Δ328-347) has sufficient residual activity to complement the E. coli ΔhemH mutant. Previous studies in this laboratory demonstrate that as little as 1% of residual ferrochelatase activity is sufficient to complement this mutant (7).
DISCUSSION
Prior to the present work it was generally believed that no prokaryotic ferrochelatases possessed [2Fe-2S] clusters (4, 6). The yeast S. cerevisiae (13) and plant ferrochelatase sequences do not contain the cysteine ligands involved in cluster ligation in animal ferrochelatases, and the expression of nonanimal ferrochelatases had revealed no evidence of clusters. The initially published bacterial ferrochelatase sequences did not contain the carboxyl-terminal domain III (see references 6 and 11) which is necessary for cluster formation in animal ferrochelatases (3-6, 8, 19, 21), and there was no spectral evidence of a cluster in expressed bacterial ferrochelatase proteins. Thus, the tacit assumption has been that the [2Fe-2S] cluster arose during evolution in the animal kingdom. Because of this proposition, explanations for the cluster's role became related to the evolution of multicellular animals (6, 8, 20). The data presented previously (14) and above, however, clearly show that the cluster exists not only in multicellular animals but also in at least one unicellular eukaryote and, most significantly, in some prokaryotes.
Since no [2Fe-2S] cluster has ever been reported for a bacterial ferrochelatase, the discovery of a [2Fe-2S] cluster in C. crescentus ferrochelatase was unexpected. Visual examination of the primary sequence revealed the presence of a carboxyl-terminal extension of a length similar to that of animal ferrochelatases, and this region contains three cysteine residues as are found in animal ferrochelatases (Fig. 2). However, the spacing of these residues (C-X6-C-X-C) is unlike that found in animal and S. pombe ferrochelatases (C-X2-C-X4-C) (6-9, 14, 20). More significant, however, was that the most-amino-terminal cluster-ligating cysteine residue of animal (C196 of human ferrochelatase) and S. pombe (C162) ferrochelatases is absent in C. crescentus. The residues surrounding the most-amino-terminal cysteine are also highly conserved between S. pombe and animal ferrochelatases (Y-P-Q-W/Y-S-C-A/S-T-S/T-G). Instead, there is a threonine in this position in C. crescentus (T133). While the possibility for a noncysteinyl cluster ligand exists, mutations of this threonine had no effect upon the cluster (Table 1). The identity of the amino-proximal ligand was confirmed by site-directed mutagenesis to be C158. The remaining three distal ligands were identified as C332, C339, and C341 by site-directed mutagenesis. This results in a four-cysteinyl ligated [2Fe-2S] cluster, and the spacing of its ligands is C-X170-C-X6-C-X-C, which is unique among all [2Fe-2S] cluster-containing proteins currently known. Because of the proximity of the last two cysteine residues, it may be anticipated that C339 and C341 will serve as ligands to one iron and the two amino-proximal cysteines will serve as ligands to the other iron. This arrangement would be analogous to that found in human ferrochelatase (21). Examination of the tertiary structure of human ferrochelatase reveals that the residue corresponding to C. crescentus T133, which is between β-sheet 3 and α-helix 7, is adjacent to the residue corresponding to C. crescentus C158 (human C196), which is between β-sheet 4 and α-helix 8 (21). Thus, the movement of the fourth ligand from residue 133 to residue 158 in the primary sequence may have little impact since the actual spatial position of these two residues is very close.
The M. tuberculosis ferrochelatase has another unique cysteinyl spacing in the carboxyl terminal region of the enzyme, C-X8-C-X4-C. In this ferrochelatase the putative amino-proximal cysteine ligand occurs two or three amino acid residues (depending upon the alignment parameters) distal from the corresponding cysteine of animal and S. pombe ferrochelatases.
The discovery of a [2Fe-2S] cluster in C. crescentus and M. tuberculosis raises several questions, the first of which is whether there are [2Fe-2S] clusters in other bacteria. An examination of derived amino acid sequences from public databases (GenBank and TIGR) reveals four other bacterial ferrochelatases with carboxyl-terminal extensions. One of these, Rickettsia prowazekii, possesses the same cysteine presence and spacing as is found in that of C. crescentus and would, therefore, be predicted to contain a [2Fe-2S] cluster. Those of Propionibacterium freudenreichii, Streptomyces coelicolor, and Corynebacterium diphtheriae possess carboxyl-terminal extensions and four possible ligating cysteine residues, although their spacing is unlike either animal or C. crescentus ferrochelatases and the carboxyl-terminal two cysteines are immediately adjacent residues. At present there are no known bacterial ferrochelatases with carboxyl-terminal extensions which lack cysteine residues in this region.
Another question raised by the presence of [2Fe-2S] clusters in bacterial ferrochelatase concerns the role this feature plays for the enzyme. A role as a dimerization motif (6, 21) alone is not supported by the discovery that M. tuberculosis ferrochelatase contains both this region and a [2Fe-2S] cluster but is monomeric as isolated. C. crescentus ferrochelatase may provide an important handle in addressing the question of a possible catalytic role, since the noncluster mutants produced in the present study, unlike those made previously for animal ferrochelatases, still possess minimal enzyme activity.
Acknowledgments
We thank K.-F. Wang for performing the FPLC molecular weight determinations.
This work was supported by grants from the National Institutes of Health (DK 32303) to H.A.D.
REFERENCES
- 1.Bordier, C. 1981. Phase-separation of integral membrane-proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 2.Brownstein, M. J., J. D. Carpten, and J. R. Smith. 1996. Modulation of non-templated nucleotide addition by tag DNA polymerase: primer modifications that facilitate genotyping. BioTechniques 20:1004-1010. [DOI] [PubMed] [Google Scholar]
- 3.Crouse, B. R., V. M. Sellers, M. G. Finnegan, H. A. Dailey, and M. K. Johnson. 1996. Site-directed mutagenesis and spectroscopic characterization of human ferrochelatase: identification of residues coordinating the [2Fe-2S] cluster. Biochemistry 35:16222-16229. [DOI] [PubMed] [Google Scholar]
- 4.Dailey, H. A. 1996. Ferrochelatase, p. 77-98. In R. P. Hausinger, G. L. Eichorn, and L. G. Marzilli (ed.), Mechanisms of metallocenter assembly. VCH Publishers, Inc., New York, N.Y.
- 5.Dailey, H. A., M. G. Finnegan, and M. K. Johnson. 1994. Human ferrochelatase is an iron-sulfur protein. Biochemistry 33:403-407. [DOI] [PubMed] [Google Scholar]
- 6.Dailey, H. A., T. A. Dailey, C.-K. Wu, A. E. Medlock, K.-f. Wang, J. P. Rose, and B.-C. Wang. 2000. Ferrochelatase at the millennium: structures, mechanisms, and [2Fe-2S] clusters. Cell. Mol. Life Sci. 57:1909-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dailey, H. A., V. M. Sellers, and T. A. Dailey. 1994. Mammalian ferrochelatase. Expression and characterization of normal and 2 human protoporphyric ferrochelatases. J. Biol. Chem. 269:390-395. [PubMed] [Google Scholar]
- 8.Day, A. L., B. M. Parsons, and H. A. Dailey. 1998. Cloning and characterization of Gallus and Xenopus ferrochelatases: presence of the [2Fe-2S] cluster in nonmammalian ferrochelatase. Arch. Biochem. Biophys. 359:160-169. [DOI] [PubMed] [Google Scholar]
- 9.Eldridge, M. G., and H. A. Dailey. 1992. Yeast ferrochelatase: expression in a baculovirus system and purification of the expression protein. Protein Sci. 1:271-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gora, M., E. Grzybowska, J. Rytka, and R. Labbe-Bois. 1996. Probing the active-site residues in Saccharomyces cerevisiae by directed mutagenesis. In vivo and in vitro analyses. J. Biol. Chem. 271:11810-11816. [DOI] [PubMed] [Google Scholar]
- 11.Hansson, M., and L. Hederstedt. 1994. Purification and characterization of a water-soluble ferrochelatase from Bacillus subtilis. Eur. J. Biochem. 220:201-208. [DOI] [PubMed] [Google Scholar]
- 12.Karr, S. R., and H. A. Dailey. 1988. The synthesis of murine ferrochelatase in vitro and in vivo. Biochem. J. 254:799-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labbe-Bois, R. 1990. The ferrochelatase of Saccharomyces cerevisiae. Sequence, disruption, and expression of its structural gene HEM15. J. Biol. Chem. 265:7278-7283. [PubMed] [Google Scholar]
- 14.Medlock, A. E., and H. A. Dailey. 2000. Examination of the activity of carboxyl-terminal chimeric constructs of human and yeast ferrochelatases. Biochemistry 39:7461-7467. [DOI] [PubMed] [Google Scholar]
- 15.Miyamoto, K., K. Nakahigashi, K. Nishimura, and H. Inokuchi. 1991. Isolation and characterization of visible light-sensitive mutants of Escherichia coli K12. J. Mol. Biol. 219:393-398. [DOI] [PubMed] [Google Scholar]
- 16.Prasad, A. R. K., and H. A. Dailey. 1995. Effect of cellular location on the function of ferrochelatase. J. Biol. Chem. 270:18198-18200. [DOI] [PubMed] [Google Scholar]
- 17.Ruepp, A., W. Graml, M. L. Santos-Martinez, K. K. Koretke, C. Vloker, H. W. Mewes, D. Frishman, S. Stocker, A. N. Lupas, and W. Baumeister. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:503-513. [DOI] [PubMed] [Google Scholar]
- 18.Sellers, V. M., C.-K. Wu, T. A. Dailey, and H. A. Dailey. 2001. Human ferrochelatase: characterization of substrate-iron binding and proton-abstracting residues. Biochemistry 40:9821-9827. [DOI] [PubMed] [Google Scholar]
- 19.Sellers, V. M., K.-F. Wang, M. K. Johnson, and H. A. Dailey. 1998. Evidence that the fourth ligand to the [2Fe-2S] cluster in animal ferrochelatase is a cysteine-characterization of the enzyme from Drosophila melanogaster. J. Biol. Chem. 273:22311-22316. [DOI] [PubMed] [Google Scholar]
- 20.Sellers, V. M., M. K. Johnson, and H. A. Dailey. 1996. Function of the [2Fe-2S] cluster in mammalian ferrochelatase: a possible role as a nitric oxide sensor. Biochemistry 35:2699-2704. [DOI] [PubMed] [Google Scholar]
- 21.Wu, C.-K., H. A. Dailey, J. P. Rose, A. M. Burden, and B.-C. Wang. 2001. The 2.0 angstrom structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat. Struct. Biol. 8:156-160. [DOI] [PubMed] [Google Scholar]