Abstract
The HliA protein of the cyanobacterium Synechococcus elongatus PCC 7942 is a small, thylakoid-associated protein that appears to play a role in photoprotection; its transcript rapidly accumulates in response to high-intensity light (HL) and the hli gene family is required for survival of cells in high light. In order to discover regulatory factors involved in HL acclimation in cyanobacteria, a screen was performed for chemically generated mutants unable to properly control expression of the hliA gene in response to HL. One such mutant was identified, and complementation analysis led to the identification of the affected gene, designated nblS. Based on its deduced protein sequence, NblS appears to be a membrane-bound, PAS-domain-bearing, sensor histidine kinase of two-component regulatory systems in bacteria. The nblS mutant was unable to properly control light intensity-mediated expression of several other photosynthesis-related genes, including all three psbA genes and the cpcBA genes. The mutant was also unable to control expression of the hliA and psbA genes in response to low-intensity blue/UV-A light, a response that may be related to the HL-mediated regulation of the genes. Additionally, in response to nutrient deprivation, the nblS mutant was unable to properly control accumulation of the nblA transcript and associated degradation of the light-harvesting phycobilisomes. The nblS mutant dies more rapidly than wild-type cells following exposure to HL or nutrient deprivation, likely due to its inability to properly acclimate to these stress conditions. Thus, the NblS protein is involved in the control of a number of processes critical for altering the photosynthetic apparatus in response to both HL and nutrient stress conditions.
The photosynthetic apparatus is a dynamic assemblage of activities that are strongly controlled by the environment. Environmental parameters such as light quality, light intensity, temperature, water availability, and nutrient status play critical roles in determining the activities of photosynthetic complexes and the levels of pigments and proteins associated with those complexes (17, 22). It is vital that organisms tune photosynthesis to balance the energetic requirements of the cell with the light energy absorbed by antenna pigments and the transfer of that energy to photosynthetic reaction centers. Such coordination helps prevent oxidative damage caused by overreduction of electron carriers and the accumulation of excited pigment molecules. In the short term, photosynthetic activity in response to excess absorbed light can be modulated by redistribution of excitation energy utilization between the photosystems and the quenching of excess excitation energy within antenna complexes (7, 10, 17, 22, 35). Long-term modulations involve alterations in the composition of the photosynthetic apparatus or changes in processes that modulate the assembly or disassembly of specific protein complexes that function in photosynthesis (7, 10, 17, 35). Long-term acclimation processes may reflect altered patterns of gene expression, which may be triggered by changes in light quality (through the activity of specific photoreceptors) or changes in light intensity (through the activity of redox-sensitive regulators) (2, 15, 20, 28, 31, 32, 39, 49).
Cyanobacteria possess several mechanisms for modifying the composition of the photosynthetic machinery with respect to environmental conditions. One dramatic example of this modulation is the degradation of the light-harvesting pigment-protein complex, the phycobilisome (PBS), during starvation for sulfur and nitrogen (13, 14, 52). The loss of PBS and a reduction in the level of chlorophyll per cell during nutrient-limited growth changes the appearance of cells from a deep blue-green to a chlorotic yellow, a process termed bleaching. Degradation of the PBS may provide some of the limiting nutrient and serve to reduce the light absorbed by the photosynthetic apparatus during starvation conditions, when the use of excitation energy for anabolic processes would markedly decline. One protein that appears to trigger PBS degradation during nutrient deprivation is NblA (14). The level of nblA transcript, which increases dramatically upon starvation of Synechococcus elongatus PCC 7942 for nitrogen or sulfur (14), is controlled by NblR (43). This response regulator controls activities that are critical for cell viability under a variety of stress conditions, including nutrient deprivation and high-intensity light (HL) stress.
A well-characterized response of both plants and cyanobacteria to HL is the turnover and exchange of the D1 subunits of photosystem II, the primary site of damage during photoinhibition (10). In S. elongatus PCC 7942, there are two forms of D1 that are encoded by three genes (21). The psbAI gene encodes D1, form I, which is most abundant under low light (LL) conditions. The psbAII and psbAIII genes each encode D1, form II, which becomes abundant when the cells are shifted from LL to HL, a consequence of the induction of the psbAII and psbAIII genes and a decrease in psbAI transcript levels (9). D1, form II, may provide cells with greater resistance to photooxidative damage (12). Light quality may provide part of the signal for light intensity regulation; blue light causes an increase in the accumulation of psbAII and psbAIII transcripts and a decrease in the level of the psbAI transcript (50).
Like the psbAII and psbAIII genes, expression of the hliA gene of S. elongatus PCC 7942 increases during exposure of the organism to HL or blue/UV-A light (19). The HliA protein of S. elongatus PCC 7942 has similarity to the extended chlorophyll a/b-binding family of proteins, especially to the early light-inducible proteins (ELIPs) of vascular plants, a carotenoid-binding protein (Cbr) of the alga Dunaliella, and analogous, single membrane-spanning helix proteins of other cyanobacteria (including a family of four hli genes present in Synechocystis sp. strain PCC 6803 [23]). Similar to the ELIPs, Hli polypeptides are localized in the thylakoid membrane and accumulate following exposure to HL or blue/UV-A light (19, 23). The Hli polypeptides are critical for survival in HL (23) and may be involved in the dissipation of excess absorbed excitation energy (M. Havaux, G. Guedeney, Q. He, and A. R. Grossman, unpublished data).
This paper describes the identification of an apparent sensor histidine kinase, NblS, that is involved in controlling hliA expression in response to HL. NblS was also found to be involved in the expression of nblA and degradation of the PBS during nutrient deprivation. In addition, it was found to be involved in the control of a number of other photosynthesis-related genes during exposure to HL and blue/UV-A light. Our results suggest that NblS functions as an important regulator controlling the acclimation of the photosynthetic apparatus to stress conditions.
MATERIALS AND METHODS
Culture conditions.
Culturing of Synechococcus elongatus PCC 7942 (formerly known as Anacystis nidulans R2 or Synechococcus sp. strain PCC 7942 [24]) was as previously reported (30). Antibiotics were added when appropriate (see reference 18). Cells were grown at 50 μmol photon m−2 s−1 incandescent light. The level of HL was 800 μmol photon m−2 s−1 unless otherwise indicated. The level of UV-A light from black-light blue bulbs (see reference 19) was 27 μmol photon m−2 s−1. Prior to HL or UV-A treatment, cultures were grown to an optical density at 750 nm (OD750) of approximately 1.0, diluted to an OD750 of 0.2 with fresh medium, and incubated for 18 h in LL at 10 μmol photon m−2 s−1 (unless otherwise indicated). Starvation for sulfur or nitrogen was performed as previously reported (43).
Mutagenesis and screening for hliA regulatory mutants.
The parental strain for mutagenesis was S. elongatus PCC 7942 hliNG (18). This strain carries the hliA promoter fused to both a promoterless nblA gene (14) (on an autonomously replicating plasmid) and a promoterless uidA gene (β-glucuronidase [GUS]) (incorporated into the chromosome at a neutral site). Since the presence of the NblA polypeptide in cyanobacterial cells triggers the bleaching process, a strain harboring the nblA gene fused to the hliA promoter will turn from a normal blue-green appearance to yellow upon exposure to conditions that activate the hliA promoter. Thus, the nblA reporter gene in the hliNG strain serves as a direct visual screen for hliA promoter activity. S. elongatus PCC 7942 hliNG was mutagenized with N-methyl-N′-nitrosoguanidine (18), and the mutagenized cells were screened on plates for reduced bleaching following a 12-h exposure to 500 μmol photon m−2 s−1. Putative mutants that exhibited reduced bleaching in HL were quantified for hliA activity during HL exposure by assaying GUS activity as previously reported (19). From a series of screens involving approximately 50,000 colonies, 11 isolates showed reduced bleaching and GUS activity in HL but exhibited normal bleaching under nutrient deprivation conditions and appeared to have lesions in the hliA promoter-nblA fusion. Four isolates had reduced bleaching but normal GUS activity under HL, also had reduced bleaching under nutrient deprivation conditions, and were complemented by a DNA fragment bearing the nblB gene, which encodes a polypeptide necessary for degradation of the PBS (18). A single mutant, designated nblS-1, was identified that exhibited both reduced bleaching and lowered GUS activity in HL. This mutant also was found to be nonbleaching under nutrient deprivation conditions (see below).
Complementation of the nblS-1 mutant and characterization of the nblS gene.
Molecular techniques were performed using standard protocols (42). The nblS-1 mutant was transformed (30) with a library of Sau3AI partially digested chromosomal DNA of S. elongatus PCC 7942 in pUC118 (43), and transformants were selected for ampicillin resistance, which results when a plasmid integrates into the chromosome by single homologous recombination. Because hliA promoter-driven nblA expression was reduced but not completely absent in the nblS-1 mutant, a significant level of bleaching occurred when the mutant was incubated overnight in HL. To more easily identify complemented strains, LL-adapted transformant colonies were screened for GUS activity after 3 h in HL by overspraying with the highly sensitive fluorogenic GUS substrate 4-methylumbelliferyl β-d-glucuronide (10 mg/ml dissolved in dimethyl sulfoxide) and visualizing fluorescence under long-wavelength UV light. Chromosomal DNA was extracted from one of the complemented strains, and the genomic regions flanking the inserted plasmid DNA were rescued as previously reported (43). One rescued plasmid contained a 4.5-kbp PstI segment of genomic DNA that was able to complement the nblS-1 mutant. Subclones of this fragment were used in complementation tests to define the complementing gene. The 4.5-kbp PstI fragment was sequenced by using the Applied Biosystems PRISM system (Perkin-Elmer).
RNA isolation and RNA blot hybridizations.
The wild-type and nblS-1 strains used for RNA blot hybridizations and viability assays were the hliNG strain and the nblS-1 mutant, respectively, after being cured of the hliA-nblA fusion-bearing plasmid by several passages through liquid medium devoid of antibiotics. The nblR mutant used for RNA blot analysis was the nblRΩ strain, in which the nblR gene was disrupted by insertion of a kanamycin resistance gene (43). Following the various treatments, RNA was isolated from cells (6). For all RNA blot hybridizations, equal amounts of RNA (determined spectroscopically) were resolved by electrophoresis in formaldehyde gels; ethidium bromide included in the loading buffer allowed visualization of the rRNA bands and confirmed equal loading of the RNA samples. Moreover, on one blot, equal loading of RNA samples based on ethidium bromide staining was confirmed by equal signals obtained by probing the RNA blot with an RNase P gene-specific probe as a control (see reference 41). Gene-specific probes were prepared as described for hliA (19), nblA (14), and psbAI, psbAII, and psbAIII (8). A 321-bp PCR-amplified fragment of the cpcBA operon (starting 27 bp upstream of the initial ATG of cpcB and extending into the coding region) was cloned into pGEM-T Easy (Promega). This plasmid served as the template for the preparation of an antisense RNA probe specific for transcripts bearing the cpcB gene. A 287-bp PCR-amplified fragment of glnA (starting 5 bp downstream of the initial ATG and extending into the coding region) was used to generate a DNA probe specific for that gene.
Nucleotide sequence accession number.
The sequence of the 4.5-kbp PstI fragment identified in this study was deposited in the GenBank database (accession number AF299076).
RESULTS
Characterization of a mutant with aberrant regulation of hliA and discovery of its aberrant response to nutrient deprivation.
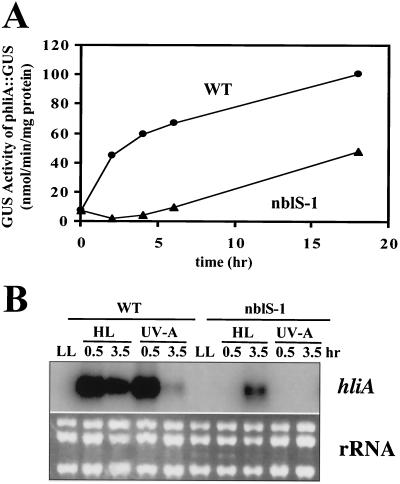
To identify regulatory factors involved in light intensity control of gene expression in cyanobacteria, we screened for chemically generated mutants unable to properly activate transcription from the hliA promoter in HL. One such mutant identified was nblS-1. Figure 1A shows activity from the hliA promoter in HL quantified in strains bearing the hliA promoter fused to the GUS gene. Wild-type cells developed high levels of GUS activity following exposure to HL, showing a rapid increase in activity during the initial 2 h of exposure, followed by a slower rise; the nblS-1 mutant exhibited a significantly lower level of activity, with the initial rapid rise in activity being absent but with the slower rise still apparent.
FIG. 1.
Characterization of expression of the hliA gene in the nblS-1 mutant. (A) Measurement of hliA promoter-driven GUS activity (phliA::GUS) during HL exposure of the hliNG wild-type (WT) strain and the nblS-1 mutant. (B) RNA blot hybridization of an hliA-specific probe to RNA isolated from the wild-type or nblS-1 strain grown for various times in HL or blue/UV-A light. For comparison, total RNA was stained with ethidium bromide (panel B, bottom).
The kinetics of HL-triggered accumulation of hliA mRNA in the nblS-1 mutant and wild-type strains agreed with the results observed using the GUS assay (Fig. 1B). A rapid increase in the level of hliA mRNA in wild-type cells in HL was followed by a gradual decrease. Similar kinetics of hliA transcript accumulation in the wild-type strain occurred in UV-A light (Fig. 1B); however, the peak at 30 min was followed by a more rapid decrease than that seen in HL, such that after 3.5 h of UV-A light there was almost no detectable hliA transcript. (This rapid decrease was not due to a general decrease in transcription during UV-A exposure, since a transcript for the glnA gene was actually more abundant following 3.5 h of UV-A light than at 30 min [data not shown].) In contrast, accumulation of hliA mRNA in the nblS-1 mutant following exposure to either HL or UV-A light was much lower than that in wild-type cells; in the mutant, accumulation of hliA mRNA was apparent only following 3.5 h of exposure to HL and no accumulation was observed in UV-A light.
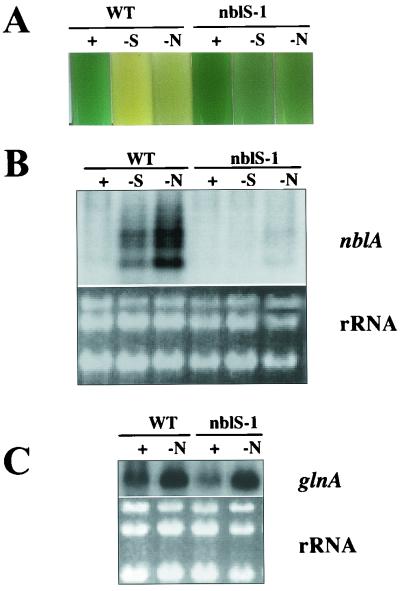
In addition to exhibiting aberrant regulation of hliA, the nblS-1 mutant showed an abnormal response to nutrient deprivation. The mutant exhibited very little of the loss of pigmentation (bleaching) that is typically found in wild-type cells during nutrient deprivation (Fig. 2A, compare lanes +, -S, and -N); the name given to the mutant, nblS (nonbleaching sensor), reflects the fact that a mutation strongly affects the pathway that controls the bleaching process in S. elongatus PCC 7942 and that the gene altered in the strain encodes a putative sensor protein (see below). The finding that the nblS-1 mutant did not bleach during nutrient deprivation suggested that this strain might be deficient in the expression of nblA, since this gene is induced during nutrient limitation and is required for bleaching to occur (14). This was confirmed by RNA blot hybridizations (Fig. 2B). On the other hand, genes critical for the uptake and assimilation of specific nutrients following nutrient deprivation were still properly regulated in the nblS-1 mutant. For example, as seen in the wild-type cells, nitrogen deprivation of the nblS-1 mutant resulted in an increased accumulation of the glnA transcript (Fig. 2C).
FIG. 2.
Characterization of the response to nutrient deprivation in the nblS-1 mutant. (A) Liquid medium cultures of wild-type cells (WT) and the nblS-1 mutant after 72 h in complete medium (+) or medium lacking sulfur (-S) or nitrogen (-N). (B) RNA blot hybridization of a riboprobe specific for nblA to RNA isolated from the wild-type or nblS-1 strain after 24 h in complete medium or medium lacking sulfur or nitrogen. (C) RNA blot hybridization of a probe specific for glnA to RNA isolated from the wild-type or nblS-1 strain grown in complete medium or in medium lacking nitrogen for 30 min. For comparison within each RNA blot, total RNA was stained with ethidium bromide (panels B and C, bottom).
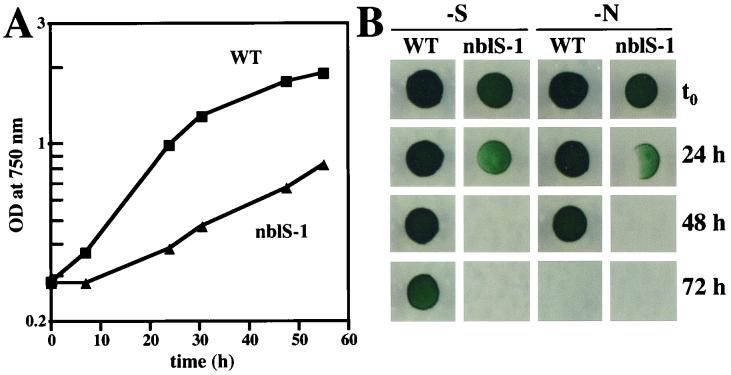
Since a mutant in the NblR response regulator that controls expression of nblA is unable to properly acclimate to stress conditions and dies upon exposure to HL or nutrient stress, we examined the nblS-1 mutant for its viability under stress conditions. Similar to the nblR mutant, the nblS-1 strain exhibited a marked decline in growth rate relative to that of wild-type cells in HL in liquid medium (Fig. 3A), with more cell death apparent in the mutant than in the wild-type strain. (After 6 h in HL, the nblS-1 mutant culture contained 3.75 × 107 CFU/ml per OD750 of 1.0, while the wild-type culture contained 1.17 × 108 CFU/ml per OD750 of 1.0; prior to HL, the cultures contained 1.60 × 108 CFU/ml per OD750 of 1.0 and 1.64 × 108 CFU/ml per OD750 of 1.0 for the mutant and wild-type strains, respectively.) Cells from the mutant strain also died more rapidly than wild-type cells during nutrient deprivation (Fig. 3B).
FIG. 3.
Viability of the nblS-1 strain during stress conditions. (A) Growth, as measured by OD750, of wild-type (WT) and nblS-1 strains in HL. (B) Wild-type (WT) and nblS-1 strains were spotted onto nutrient-replete solid medium following growth in liquid medium lacking sulfur (-S) or nitrogen (-N). Each spot consisted of 10 μl from a twofold dilution of the original culture.
Expression of other HL-regulated genes in the nblS-1 mutant.
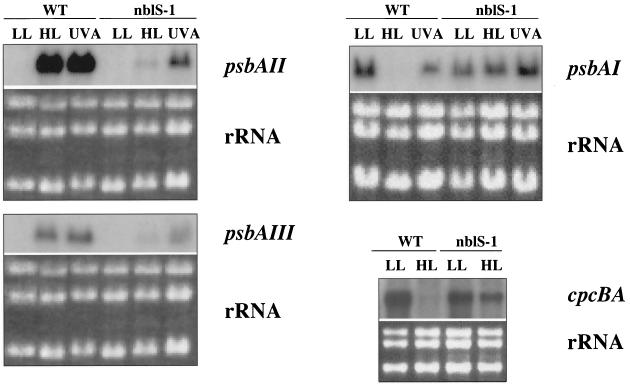
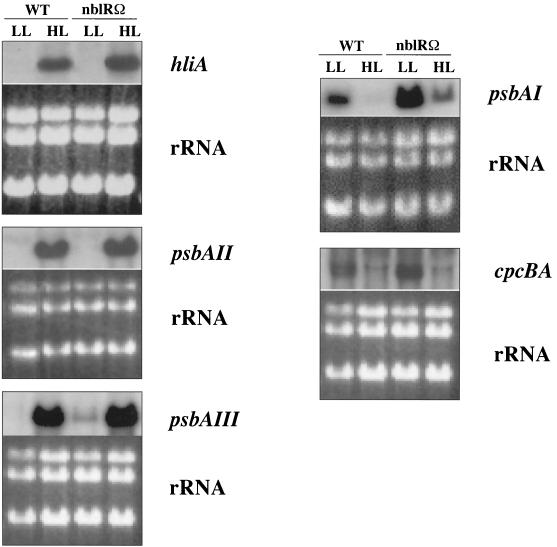
We examined expression of a number of HL-regulated genes in the nblS-1 mutant (Fig. 4). The nblS-1 mutant was aberrant with regard to the control of all three psbA genes. The mutant strain exhibited little increase in psbAII and psbAIII transcript levels and no decrease in psbAI transcript level following exposure to HL or UV-A irradiation. In fact, the levels of psbAI transcript in the mutant increased in response to UV-A exposure. Furthermore, we found in wild-type S. elongatus PCC 7942 that the cpcBA transcripts (i.e., those encoding the phycocyanin subunits, which constitute the major polypeptides of the light-harvesting PBS) declined to very low levels in HL. Such a decrease in phycocyanin levels would reduce the excitation energy absorbed by the cells during HL. However, in the nblS-1 strain there was little such decrease, with cpcBA transcript levels somewhat lower under LL and higher under HL relative to those of the wild-type strain (Fig. 4). Similarities in the light regulation of hliA and these other HL-regulated genes make it extremely likely that the altered expression of these genes in the mutant was the result of the same lesion that affects hliA expression.
FIG. 4.
RNA blot analyses of light intensity-regulated genes in the wild-type and nblS-1 strains. LL-acclimated cells were exposed for 30 min to either HL or blue/UV-A light as described in Materials and Methods. Under the usual LL conditions (10 μmol photon m−2 s−1), psbAI was not transcribed significantly. Therefore, LL conditions of 50 μmol photon m−2 s−1 were used for RNA blot analyses of psbAI expression. For comparison within each RNA blot, total RNA was stained with ethidium bromide (bottom panels).
The nblS gene encodes a PAS domain-bearing putative sensor kinase.
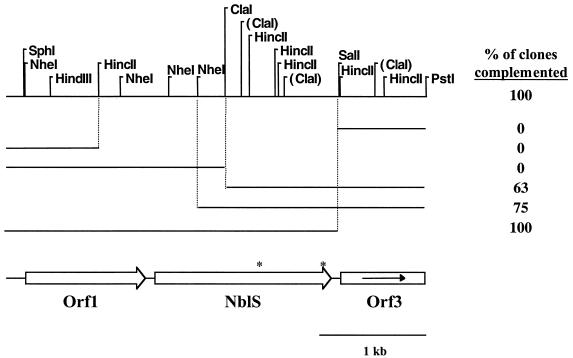
To identify the gene altered in nblS-1, the mutant was transformed with a wild-type recombinant library, and transformants were screened for complementation of the defect in hliA expression in HL as described in Materials and Methods. A 4.5-kbp PstI fragment isolated from one of the complemented strains restored hliA expression in HL. The entire fragment was sequenced, and the complementing region was localized (by using subcloning and complementation analyses) to a 664-amino acid open reading frame (ORF) that we designated NblS (Fig. 5). Another complete ORF (Orf 1) and a partial ORF (Orf 3) were found flanking NblS on the 4.5-kbp PstI fragment. Orf 1 has high sequence similarity to the family of phosphoribosylglycinamide formyltransferases (an enzyme involved in de novo purine biosynthesis), while Orf 3 shows high sequence similarity to the family of 5′-methylthioadenosine and purine-nucleoside phosphorylases (involved in purine salvage). It is not known whether these two ORFs share any functional relationship with NblS.
FIG. 5.
A physical map of the 4.5-kbp region carried by the integrative plasmid that complemented the nblS-1 mutant and fragments of this plasmid used in subcloning analysis. Also indicated are the relative locations of the two complete ORFs and one partial ORF that were discovered upon DNA sequence analysis. Indicated by asterisks on the NblS ORF are the locations of the two amino acid changes generated by the two point mutations present in the nblS-1 allele. To the right of each DNA fragment is listed the percentage of transformants bearing the integrated plasmid carrying the particular fragment that complemented hliA activity in the nblS-1 mutant. This percentage is not always 100% for those fragments that did not bear a complete nblS gene because, depending upon the location of crossover relative to the mutations in the nblS-1 allele, a complete, wild-type nblS gene was not always generated in the chromosome. The subcloning analyses localize the complementing region to the nblS gene. (The ClaI sites shown in parentheses are those sites at which the DNA was not cleaved during subcloning analyses due to methylation in the Dam+ Escherichia coli.)
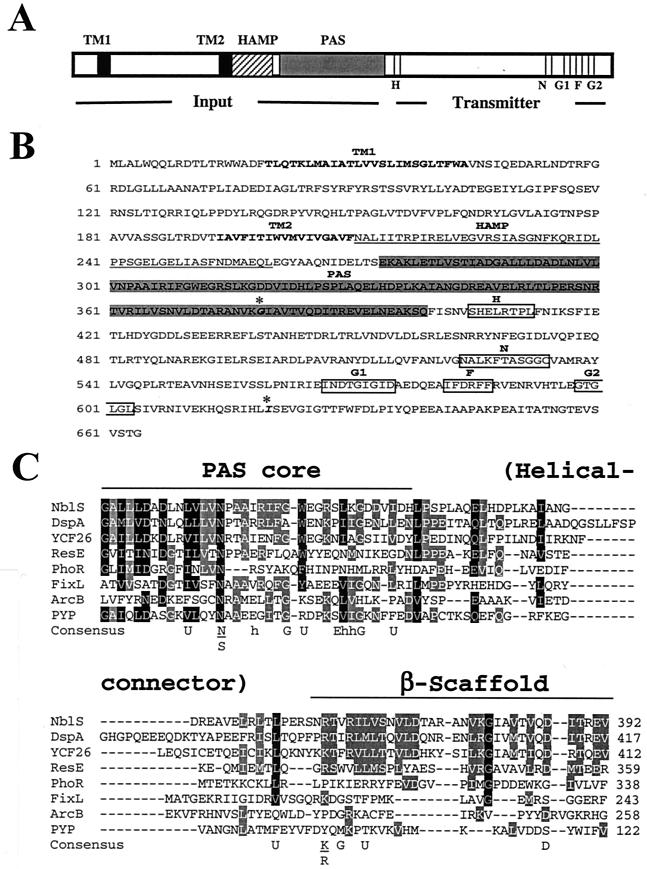
Analysis of the predicted NblS protein sequence showed that it shares strong sequence similarity with the superfamily of sensor histidine kinases of two-component regulatory systems (36, 44). It can be divided into several domains based on sequence similarity to other proteins in the databases and to characterized functional motifs (Fig. 6). The C terminus of the protein contains a typical histidine kinase transmitter module with N, G1, F, and G2 domains (37). The N-terminal putative sensory input region contains two predicted membrane-spanning regions (TM1 and TM2) followed by a region, recently termed the HAMP domain (3), with similarity to the linker region of methyl-accepting chemotaxis proteins and certain histidine kinases. HAMP domains may be involved in intramolecular interactions that control activation of the polypeptides in which they are found (3). The HAMP domain of NblS is followed by a region that bears strong similarity to PAS domains. PAS domains are involved in protein-protein interactions and the binding of redox-active cofactors that modulate protein output activity (48, 54).
FIG. 6.
Domain structure of NblS. (A) Diagrammatic representation of the NblS domain structure; (B) sequence of the NblS protein annotated with the domain structure. The two predicted transmembrane regions (TM1 and TM2), the HAMP region, and the PAS domain are shown. Also indicated are motifs characteristic of a C-terminal kinase transmitter domain (from residue 400 to the end of the protein), which are highly conserved among histidine kinases (H, N, G1, F, and G2). Indicated in boldface italic and with asterisks in panel B are the two amino acid changes (G-379 to D and I-620 to V) present in the nblS-1 mutant allele. (C) Alignment of the PAS domain with homologous regions from other proteins. Shown are PAS domains from the DspA protein (sll0698) from Synechocystis sp. strain PCC 6803 (4); the YCF26 protein from a red algal chloroplast (40); the ResE (45) and the PhoR (27) proteins, both from Bacillus subtilis; the FixL protein from Rhizobium meliloti (16); the ArcB protein from E. coli (26); and the photoactive-yellow protein (PYP) from Halochromatium salexigens (29). The consensus sequence from a published alignment of PAS domains (54) is also shown (U, bulky hydrophobic: FILMVWY; h, hydrophobic: ILMV). Residues conserved among 70% or more of the sequences are shaded in black; residues conserved among 50% or more are shaded in gray.
The nblS gene from the nblS-1 mutant was PCR amplified and sequenced. Although mutagenesis using nitrosoguanidine was performed at a rate that should generate a single mutation per cell, the gene was found to contain two mutations, both of which caused amino acid substitutions (described in the legend to Fig. 6B). This could be due to the fact that nitrosoguanidine mutagenesis commonly acts at replication forks (34), resulting in multiple mutations within a cell that are close to one another.
Several characterized histidine kinase-like proteins have high overall sequence similarity to NblS. The highest sequence similarities are between NblS and DspA of Synechocystis sp. strain PCC 6803 (58% identity overall, 54% identity within the input domain) and putative proteins encoded by the chloroplast genome of some red algae (49% identity overall, 28% identity within the input domain) (4, 40). The dspA gene product of Synechocystis sp. strain PCC 6803 was identified as a factor that confers resistance to a variety of herbicides and calmodulin antagonists (and was hypothesized to play a role in cell permeability) (4) and was later identified in a study as being involved in the response to chilling in Synechocystis sp. strain PCC 6803 (therein called Hik33) (46, 47). NblS also has high sequence similarity to ResE (45) and PhoR (27) of Bacillus subtilis (25% identity and 23% identity, respectively, overall; 19% identity and 14% identity, respectively, within the input domain). ResE is a global regulator controlling the expression of genes involved in aerobic and anaerobic respiratory growth (apparently by monitoring an intracellular signal, perhaps the reduced state of menaquinone, in controlling gene expression [33]), while PhoR is involved in controlling gene expression in response to phosphorus deprivation. (NblS also shares high sequence identity with an ResE homolog, SrrB, in Staphylococcus aureus that is involved in the global control of virulence factors in response to oxygen conditions [53].) Like NblS, all of the aforementioned proteins have TM, HAMP, and PAS domains. Figure 6C shows the PAS domain from NblS aligned with PAS domains from closely related, putative sensor proteins as well as with some of the more thoroughly characterized PAS domains.
We have attempted to generate a strain in which the nblS gene has been inactivated by insertion of an antibiotic cassette (data not shown). Despite repeated subculturing in LL, PCR analysis demonstrated that wild-type copies of the chromosome persist in this strain. The same result was seen for the Synechocystis sp. strain PCC 6803 hik33 mutant strain (47). It may be that some amount of NblS activity is essential for cell viability. Nevertheless, the 4.5-kbp PstI fragment bearing nblS was able to complement the nblS-1 mutant for the defects in HL-, UV-A-, and nutrient stress-mediated gene expression as well as in stress-related growth (data not shown). This indicates that the defects are due to the mutations in nblS and not to mutations elsewhere in the chromosome.
Are nutrient stress- and HL-regulated genes controlled through the same or different signal transduction pathways?
The NblR response regulator controls transcription of nblA and the bleaching of the cyanobacterial cells during nutrient stress (43). To determine if NblR is also important for controlling expression of the light-responsive genes, we examined the effect of light intensity on the levels of hliA, psbAI, psbAII, psbAIII, and cpcBA transcripts following exposure of an nblR mutant to HL. As shown in Fig. 7, the nblR mutant still exhibits HL induction of hliA, psbAII, and psbAIII transcripts and HL depression of psbAI and cpcBA transcripts. These results suggest that the light-regulated expression of these photosynthetic genes is not controlled through NblR. Surprisingly, in the nblR mutant, transcripts from hliA, psbA, and cpcBA genes consistently accumulate to slightly higher levels than in wild-type cells under inducing conditions, and detectable levels of psbAIII and psbAI mRNA exist in the nblR mutant under noninducing conditions (Fig. 7).
FIG. 7.
RNA blot analysis of light-intensity controlled genes in the wild-type strain and a strain in which the nblR gene was inactivated. RNA was extracted from cells grown in LL and exposed to HL (as described in the Fig. 5 legend) for 30 min (psbAII and psbAI) or 2 h (hliA, psbAIII, and cpcBA). For comparisons within each RNA blot, total RNA was stained with ethidium bromide (bottom panels).
DISCUSSION
Bacteria have evolved diverse ways for sensing and responding to changes in environmental conditions. One common mechanism for the perception and transduction of signals in bacteria is the two-component regulatory system (36, 44). This type of system typically involves a pair of proteins, namely, a histidine kinase that acts as a sensor of a signal and an associated response regulator that often is involved in binding of DNA and the regulation of gene expression. The study described here has identified, in a cyanobacterium, a putative sensor kinase, NblS, that is involved in controlling the expression of a variety of genes involved in photosystem function during HL and nutrient deprivation. NblS contains the appropriate motifs to function as a histidine kinase (including the conserved histidine residue), but it remains to be shown whether it actually functions as such at the biochemical level. We have not positively identified any response regulator(s) that interact with NblS. No gene with similarity to response-regulatory molecules was found contiguous to nblS on the genome (Fig. 5). One obvious candidate response regulator that may interact with NblS during nutrient limitation is NblR, since both NblS and NblR are involved in controlling nblA expression and PBS degradation. However, our data indicate that NblR is not directly involved in the regulation of the light-responsive genes apparently controlled by NblS. Higher-than-normal transcript levels from the light-responsive genes were observed in the nblR mutant under inducing conditions (and, in some cases, detectable levels were observed under noninducing conditions) (Fig. 7). One way this could be explained is to propose that, during changes in light intensity, NblS interacts with NblR (but not to control the light intensity-regulated genes analyzed here) as well as with one or more other response regulators. Thus, in the absence of NblR, more NblS would be free to interact with another response regulator(s), which would lead to the observed augmentation of NblS-dependent (but not NblR-dependent) light intensity-mediated responses. Of course, this hypothesis remains to be tested.
We have found that NblS is involved in controlling the expression of a number of different genes whose products are associated with photosynthesis during HL and nutrient stress. Cyanobacteria experience a variety of changes in the photosynthetic apparatus and its activities in response to HL and generalized nutrient deprivation (5, 13, 14, 25). nblS-1 mutant cells were found to die more rapidly than wild-type cells during exposure to HL and nutrient limitation. The loss in viability is likely a consequence of the inability of the cells to properly control expression of genes that are necessary for acclimation to these stress conditions (including genes whose products are involved in photosynthesis such as those identified here). Interestingly, although NblR did not appear to control the light-regulated genes analyzed here (and, indeed, is not known to control any genes other than nblA during nutrient stress), a similar loss in viability during both nutrient deprivation and HL has been observed for an nblR mutant (43). This suggests that, in addition to modulating expression of nblA under nutrient stress, in HL NblR is likely to control the expression of genes other than those studied here that are important to survival in HL.
Input signals that control potential sensor kinases such as NblS are often difficult to specifically define. At this time, we do not know what signals modulate NblS action and how such signals specifically alter the HL and nutrient stress responses. Examination of the sequence of the potential sensory input region of NblS may give clues as to how NblS could be acting as a sensor. Since PAS domains in a number of proteins appear to be involved in redox sensing (48), it is possible that the PAS domain of NblS is involved in sensing changes in photosynthetic or cellular redox during HL and nutrient stress. Absorption of light in excess of that which can be used in photosynthesis can lead to overexcitation of the photosynthetic apparatus and the generation of damaging oxygen radicals. Such an excess of light can occur during periods of HL exposure or during periods of lower light when the organism is experiencing stresses, such as nutrient limitation, that reduce anabolic processes in the cell (17). The presence of two predicted TM domains in NblS makes it likely that the protein is anchored in the cytoplasmic or thylakoid membrane. In this location it could interact with photosynthesis and/or respiration. Another potential signal input region of NblS is the area between the two TM domains, which may be located either in the periplasm or in the thylakoid lumen. Indeed, HAMP domains, such as that found in NblS, have been proposed to be linkers involved in signal transduction, transmitting conformational changes from the periplasmic region (caused by the binding of specific ligands) to the cytoplasmic regions of the molecules (3, 51). It remains to be demonstrated what role each of the domains of NblS plays in the activity of the molecule.
Our results demonstrate that NblS is in some manner involved in blue/UV-A light-mediated gene expression. Several genes in plants and cyanobacteria are responsive both to HL and to relatively low levels of blue/UV-A light (levels that would not dramatically alter cellular redox conditions) (e.g., genes encoding ELIPs [1], the hliA gene [19], and the psbA genes [50]). This suggests that a blue/UV-A photoreceptor is involved in controlling gene expression during HL acclimation of photosynthetic organisms. It is not clear what role, if any, NblS plays in actual blue/UV-A photosensing. Since some PAS domains are involved in sensing of blue light (11, 38), it is possible that NblS itself functions as a blue light photoreceptor; however, it is also possible that NblS is only indirectly involved in integrating the blue light signal and that some other system is actually responsible for perception of blue/UV-A light.
Recently, the apparent Synechocystis sp. strain PCC 6803 NblS homolog, DspA (4) (Hik33 [47]), has been shown to be involved in controlling gene expression (including expression of photosynthesis-related genes) in response to low temperature conditions (46, 47). Chilling is another condition that, like nutrient deprivation, reduces the use of photosynthate by the cell, which could result in hyperreduction of the photosynthetic electron transport chain and the accumulation of reactive oxygen species (17). This adds support to the hypothesized role of NblS as a global regulator in these cyanobacteria that integrates redox and light signals and suggests that NblS may influence other signaling pathways involved in acclimation responses.
Acknowledgments
The authors thank R. Schwarz, J. Christie, and W. Briggs for their helpful suggestions during the course of this work.
This work was supported in part by National Science Foundation Grant MCB 9727836 and U.S. Department of Agriculture grants 97-35301-4575 and 98-35301-6445 awarded to A.R.G., U.S.-Israel Bi-National Science Foundation Grant 9800146 awarded to A.R.G. and R. Schwarz, and National Science Foundation Postdoctoral Fellowship DBI 9750329 awarded to L.G.V.W.
REFERENCES
- 1.Adamska, I., K. Kloppstech, and I. Ohad. 1992. UV light stress induces the synthesis of the early light-inducible protein and prevents its degradation. J. Biol. Chem. 267:24732-24737. [PubMed] [Google Scholar]
- 2.Anderson, J. M. 1986. Photoregulation of the composition, function, and structure of thylakoid membranes. Annu. Rev. Plant Physiol. 37:93-136. [Google Scholar]
- 3.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 4.Bartsevich, V. V., and S. V. Shestakov. 1995. The dspA gene product of the cyanobacterium Synechocystis sp. strain PCC 6803 influences sensitivity to chemically different growth inhibitors and has amino acid similarity to histidine protein kinases. Microbiology 141:2915-2920. [DOI] [PubMed] [Google Scholar]
- 5.Bhaya, D., R. Schwarz, and A. R. Grossman. 2000. Molecular responses to environmental stress, p. 397-442. In B. A. Whitton and M. Potts (ed.), Ecology of cyanobacteria: diversity in time and space. Kluwer Publishers, Dordrecht, The Netherlands.
- 6.Bhaya, D., N. Watanabe, T. Ogawa, and A. R. Grossman. 1999. The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. strain PCC 6803. Proc. Natl. Acad. Sci. USA 96:3188-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorkman, O., and B. Demmig-Adams. 1994. Regulation of photosynthetic light energy capture, conversion, and dissipation in leaves of higher plants, p. 17-47. In E.-D. Schulze and M. M. Calwell (ed.), Ecophysiology of photosynthesis. Springer, Berlin, Germany.
- 8.Brusslan, J., and R. Haselkorn. 1989. Resistance to the photosystem II herbicide diuron is dominant to sensitivity in the cyanobacterium Synechococcus sp. PCC7942. EMBO J. 8:1237-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustos, S. A., M. R. Schaefer, and S. S. Golden. 1990. Different and rapid responses of four cyanobacterial psbA transcripts to changes in light intensity. J. Bacteriol. 172:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow, W. S. 1994. Photoprotection and photoinhibitory damage, p. 151-196. In J. Barber (ed.), Advances in molecular and cell biology, vol. 10. JAI Press, Greenwich, Conn. [Google Scholar]
- 11.Christie, J. M., M. Salomon, K. Nozue, M. Wada, and W. R. Briggs. 1999. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96:8779-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, A. K., V. M. Hurry, P. Gustafsson, and G. Oquist. 1993. Rapid interchange between two distinct forms of cyanobacterial photosystem II reaction-center protein D1 in response to photoinhibition. Proc. Natl. Acad. Sci. USA 90:9973-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collier, J. L., and A. R. Grossman. 1992. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J. Bacteriol. 174:4718-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collier, J. L., and A. R. Grossman. 1994. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13:1039-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danon, A., and S. P. Mayfield. 1994. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science 266:1717-1719. [DOI] [PubMed] [Google Scholar]
- 16.David, M., M. L. Daveran, J. Batut, A. Dedieu, O. Domergue, J. Ghai, C. Hertig, P. Boistard, and D. Kahn. 1988. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell 54:671-683. [DOI] [PubMed] [Google Scholar]
- 17.Demmig-Adams, B., and W. W. Adams III. 1992. Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43:599-626. [Google Scholar]
- 18.Dolganov, N., and A. R. Grossman. 1999. A polypeptide with similarity to phycocyanin α-subunit phycocyanobilin lyase involved in degradation of phycobilisomes. J. Bacteriol. 181:610-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolganov, N. A. M., D. Bhaya, and A. R. Grossman. 1995. Cyanobacterial protein with similarity to the chlorophyll a/b binding proteins of higher plants: evolution and regulation. Proc. Natl. Acad. Sci. USA 92:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escoubas, J.-M., M. Lomas, J. LaRoche, and P. G. Falkowski. 1995. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 92:10237-10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golden, S. S., J. Brusslan, and R. Haselkorn. 1986. Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J. 5:2789-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman, A. R., D. Bhaya, K. E. Apt, and D. M. Kehoe. 1995. Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu. Rev. Genet. 29:231-288. [DOI] [PubMed] [Google Scholar]
- 23.He, Q., N. Dolganov, O. Björkman, and A. R. Grossman. 2001. The high light-inducible polypeptides in Synechocystis PCC 6803. Expression and function in high light. J. Biol. Chem. 276:306-314. [DOI] [PubMed] [Google Scholar]
- 24.Herdman, M., R. W. Castenholz, I. Iteman, J. B. Waterbury, and R. Rippka. 2001. Order Chroococcales, Wettstein 1924, emend. Rippka, Deruelles, Waterbury, Herdman and Stanier 1979, p. 776. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology: the archaea and the deeply branching and phototropic bacteria, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
- 25.Hihara, Y., A. Kamei, M. Kanehisa, A. Kaplan, and M. Ikeuchi. 2001. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iuchi, S., Z. Matsuda, T. Fujiwara, and E. C. Lin. 1990. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol. Microbiol. 4:715-727. [DOI] [PubMed] [Google Scholar]
- 27.Jensen, K. K., E. Sharkova, M. F. Duggan, Y. Qi, A. Koide, J. A. Hoch, and F. M. Hulett. 1993. Bacillus subtilis transcription regulator, SpoOA, decreases alkaline phosphatase levels induced by phosphate starvation. J. Bacteriol. 175:3749-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kehoe, D. M., and A. R. Grossman. 1996. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273:1409-1412. [DOI] [PubMed] [Google Scholar]
- 29.Koh, M., G. van Driessche, B. Samyn, W. D. Hoff, T. E. Meyer, M. A. Cusanovich, and J. J. van Beeumen. 1996. Sequence evidence for strong conservation of the photoactive yellow proteins from the halophilic phototrophic bacteria Chromatium salexigens and Rhodospirillum salexigens. Biochemistry 35:2526-2534. [DOI] [PubMed] [Google Scholar]
- 30.Laudenbach, D. E., and A. R. Grossman. 1991. Characterization and mutagenesis of sulfur-regulated genes in a cyanobacterium: evidence for function in sulfate transport. J. Bacteriol. 173:2739-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, H., and L. A. Sherman. 2000. A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 182:4268-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxwell, D. P., D. E. Laudenbach, and N. P. A. Hunter. 1995. Redox regulation of light-harvesting complex II and cab mRNA abundance in Dunaliella salina. Plant Physiol. 109:787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 34.Nestmann, E. R. 1975. Mutagenesis by nitrosoguanidine, ethyl methanesulfonate, and mutator gene mutH in continuous cultures of Escherichia coli. Mutat. Res. 28:323-330. [DOI] [PubMed] [Google Scholar]
- 35.Niyogi, K. K. 1999. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:333-359. [DOI] [PubMed]
- 36.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 37.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 38.Pellequer, J. L., K. A. Wager-Smith, S. A. Kay, and E. D. Getzoff. 1998. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc. Natl. Acad. Sci. USA 95:5884-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfannschmidt, T., A. Nilsson, and J. F. Allen. 1999. Photosynthetic control of chloroplast gene expression. Nature 397:625-628. [Google Scholar]
- 40.Reith, M. E., and J. Munholland. 1995. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep. 13:333-335.
- 41.Reyes, J. C., and F. J. Florencio. 1995. Electron transport controls transcription of the glutamine synthetase gene (glnA) from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 27:789-799. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schwarz, R., and A. R. Grossman. 1998. A response regulator of cyanobacteria integrates diverse environmental signals and is critical for survival under extreme conditions. Proc. Natl. Acad. Sci. USA 95:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stock, J. B., A. M. Stock, and J. M. Mottonen. 1990. Signal transduction in bacteria. Nature 344:395-400. [DOI] [PubMed] [Google Scholar]
- 45.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, I., Y. Kanesaki, K. Mikami, M. Kanehisa, and N. Murata. 2001. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 40:235-244. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, I., D. A. Los, Y. Kanesaki, K. Mikami, and N. Murata. 2000. The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 19:1327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, W. F., and M. J. White. 1991. Physiological and molecular studies of light-regulated nuclear genes in higher plants. Annu. Rev. Plant Physiol. 42:423-466. [Google Scholar]
- 50.Tsinoremas, N. F., M. R. Schaefer, and S. S. Golden. 1994. Blue and red light reversibly control psbA expression in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Biol. Chem. 269:16143-16147. [PubMed] [Google Scholar]
- 51.Williams, S. B., and V. Stewart. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093-1102. [DOI] [PubMed] [Google Scholar]
- 52.Yamanaka, G., and A. N. Glazer. 1980. Dynamic aspects of phycobilisome structure. Phycobilisome turnover during nitrogen starvation in Synechococcus spp. Arch. Microbiol. 124:39-47. [Google Scholar]
- 53.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]