Abstract
The 3-hydroxypropionate cycle is a new autotrophic CO2 fixation pathway in Chloroflexus aurantiacus and some archaebacteria. The initial step is acetyl-coenzyme A (CoA) carboxylation to malonyl-CoA by acetyl-CoA carboxylase, followed by NADPH-dependent reduction of malonyl-CoA to 3-hydroxypropionate. This reduction step was studied in Chloroflexus aurantiacus. A new enzyme was purified, malonyl-CoA reductase, which catalyzed the two-step reduction malonyl-CoA + NADPH + H+ → malonate semialdehyde + NADP+ + CoA and malonate semialdehyde + NADPH + H+ → 3-hydroxypropionate + NADP+. The bifunctional enzyme (aldehyde dehydrogenase and alcohol dehydrogenase) had a native molecular mass of 300 kDa and consisted of a single large subunit of 145 kDa, suggesting an α2 composition. The N-terminal amino acid sequence was determined, and the incomplete gene was identified in the genome database. Obviously, the enzyme consists of an N-terminal short-chain alcohol dehydrogenase domain and a C-terminal aldehyde dehydrogenase domain. No indication of the presence of a prosthetic group was obtained; Mg2+ and Fe2+ stimulated and EDTA inhibited activity. The enzyme was highly specific for its substrates, with apparent Km values of 30 μM malonyl-CoA and 25 μM NADPH and a turnover number of 25 s−1 subunit−1. The specific activity in autotrophically grown cells was 0.08 μmol of malonyl-CoA reduced min−1 (mg of protein)−1, compared to 0.03 μmol min−1 (mg of protein)−1 in heterotrophically grown cells, indicating downregulation under heterotrophic conditions. Malonyl-CoA reductase is not required in any other known pathway and therefore can be taken as a characteristic enzyme of the 3-hydroxypropionate cycle. Furthermore, the enzyme may be useful for production of 3-hydroxypropionate and for a coupled spectrophotometric assay for activity screening of acetyl-CoA carboxylase, a target enzyme of potent herbicides.
A new autotrophic CO2 fixation cycle termed the 3-hydroxypropionate cycle has been proposed for the phototrophic green nonsulfur bacterium Chloroflexus aurantiacus (7, 10, 11, 28, 29). This bacterium grows optimally at 55°C under heterotrophic conditions, but it can also grow in mineral salt medium with CO2 as the sole carbon source (11, 20, 21, 25). In thermal springs, filamentous Chloroflexus spp . and cyanobacteria form microbial mats that probably thrive photoautotrophically (30). Indications for the operation of a similar pathway in autotrophic CO2 fixation have been obtained for acidophilic archaebacteria of the Crenarchaeota, such as Acidianus infernus, Metallosphaera sedula, and Sulfolobus metallicus (3, 13, 17).
The proposed 3-hydroxypropionate cycle is shown in Fig. 1. Each turn of the cycle results in the net fixation of two molecules of bicarbonate into one molecule of glyoxylate. Acetyl-coenzyme A (CoA) is carboxylated to malonyl-CoA by conventional ATP-dependent acetyl-CoA carboxylase. The reduction of malonyl-CoA to propionyl-CoA requires three NADPH and one MgATP and proceeds via free 3-hydroxypropionate as an intermediate. 3-Hydroxypropionate is even excreted into the medium in the late growth phase of autotrophically growing cultures (10, 29). Propionyl-CoA is carboxylated to methylmalonyl-CoA, followed by isomerization of methylmalonyl-CoA to succinyl-CoA; these reactions are conventional and are used in many organisms for propionate assimilation. Succinyl-CoA is used for malate activation by CoA transfer, forming succinate and malyl-CoA; succinate in turn is oxidized to malate by conventional enzymes. Malyl-CoA is cleaved to acetyl-CoA and glyoxylate.
FIG. 1.
Proposed 3-hydroxypropionate cycle of autotrophic CO2 fixation in the phototrophic green nonsulfur bacterium C. aurantiacus, illustrating the role of malonyl-CoA reductase. Enzyme activities: 1, acetyl-CoA carboxylase; 2, malonyl-CoA reductase (NADPH), the enzyme studied in this work, catalyzing both malonyl-CoA and malonate semialdehyde reduction; 3, 3-hydroxypropionyl-CoA synthetase; 4, 3-hydroxypropionyl-CoA dehydratase; 5, acryloyl-CoA reductase (NADPH); 6, propionyl-CoA carboxylase; 7, methylmalonyl-CoA epimerase; 8, methylmalonyl-CoA mutase; 9, succinyl-CoA:l-malate CoA transferase; 10, succinate dehydrogenase, electron acceptor unknown; 11, fumarate hydratase; 12, l-malyl-CoA lyase. Note that the different enzyme activities indicated above do not necessarily mean that all these reactions are catalyzed by different enzymes. For instance, as shown here, malonyl-CoA reductase catalyzes two reactions. [CoASH], transferred coenzyme A.
There are three characteristic processes in the proposed 3-hydroxypropionate cycle that are not present in other autotrophs. The first one is the reduction of malonyl-CoA to propionyl-CoA. This complex reaction sequence formally involves five steps (Fig. 1). The second process concerns the reformation of acetyl-CoA from malate under release of glyoxylate. Previous work showed that malate is converted to malyl-CoA with succinyl-CoA as the CoA donor, catalyzed by succinyl-CoA:l-malate CoA transferase. l-Malyl-CoA is cleaved to acetyl-CoA and glyoxylate by l-malyl-CoA lyase (9). The third characteristic feature is the unknown pathway used to assimilate glyoxylate.
The present work addressed the first problem. To date, no malonyl-CoA-reducing enzyme has been described. The reduction to 3-hydroxypropionate requires two subsequent two-electron reduction steps, with malonate semialdehyde as the likely free intermediate. In C. aurantiacus as well as in M. sedula, both partial reactions were NADPH dependent (17, 29). It was unknown whether two enzymes, a CoA-acylating aldehyde dehydrogenase and a primary alcohol dehydrogenase, or, alternatively, one bifunctional enzyme, with alcohol plus aldehyde dehydrogenase activities, catalyzes the overall reaction. Since malonyl-CoA reduction has only been studied in cell extracts, purification and characterization of this key enzyme would add strong evidence for the operation of the proposed carbon fixation cycle.
The malonyl-CoA-reducing enzyme of C. aurantiacus was purified and studied. It is a large bifunctional enzyme which catalyzes the reduction of malonyl-CoA with two NADPH to 3-hydroxypropionate. The unequivocal documentation of this new enzymatic reaction should have an impact on the acceptance of the proposed autotrophic pathway.
MATERIALS AND METHODS
Bacteria and growth conditions.
C. aurantiacus strain OK-70-fl (DSM 636) was grown in 5- or 12-liter glass fermenters to an optical density (OD) of 3.5 to 4.0 at 55°C and pH of around 8. Autotrophic growth occurred under anaerobic conditions on a minimal medium supplemented with trace elements and vitamins. The cultures were gassed with a mixture of H2 and CO2 (80%:20% [vol/vol]) as described elsewhere (29). Cells were also grown anaerobically under photoheterotrophic conditions on modified minimal medium D supplemented with 0.25% (wt/vol) Casamino Acids, 0.1% (wt/vol) yeast extract, and trace elements according to Castenholz (4). This medium was buffered with 0.05% (wt/vol) glycylglycine-Na+.
Materials.
Chemicals were obtained from Fluka (Neu-Ulm, Germany), Merck (Darmstadt, Germany), Sigma-Aldrich (Deisenhofen, Germany), or Roth (Karlsruhe, Germany); biochemicals were from Roche Diagnostics (Mannheim, Germany), Applichem (Darmstadt, Germany), or Gerbu (Craiberg, Germany). [2-14C]malonyl-coenzyme A was obtained from Amersham Pharmacia (Freiburg, Germany). Materials and equipment for fast protein liquid chromatography (FPLC) were obtained from Amersham Pharmacia or Bio-Rad (Munich, Germany).
Syntheses. (i) Malonyl-CoA.
Monothiophenylmalonate was synthesized according to published procedures (12, 18, 22) and stored under a nitrogen atmosphere at −20°C. Coenzyme A (61 μmol of) was incubated under anaerobic conditions at room temperature in 40 ml of a 0.1 M NaHCO3 solution (pH 7.5) with 122 μmol of monothiophenylmalonate (dissolved in 300 μl of dioxane and 700 μl of 0.1 M NaHCO3). After 90 min, the pH was adjusted to 3.0 by addition of 1 M HCl, and the solution was extracted twice with diethylether. The aqueous solution was lyophilized, and the powder was stored at −20°C.
(ii) Succinyl-CoA, acetyl-CoA, propionyl-CoA, and 3-hydroxypropionate.
Succinyl-CoA, acetyl-CoA, propionyl-CoA, and 3-hydroxypropionate were synthesized as described elsewhere (9), and the dry powder was stored at −20°C.
Preparation of cell extract.
Cells were suspended in a twofold volume of 50 mM Tris-HCl buffer (pH 7.8) containing 3 mM dithioerythritol (DTE) and 0.2 mg of DNase I per ml. The cell suspension was passed through a French pressure cell at 137 MPa; the lysate was ultracentrifuged (100,000 × g) at 4°C for 1 h. The supernatant was used immediately or kept frozen at −70°C.
Enzyme assays.
Malonyl-CoA reductase was tested at 55°C, routinely in the forward direction. The spectrophotometric assay was followed at 365 nm (ɛNADPH = 3.4 × 103 M−1 cm−1).
(i) Forward reaction.
The malonyl-CoA-dependent oxidation of NADPH was monitored. The assay mixture (0.5 ml) routinely contained 100 mM Tris-HCl (pH 7.8), 2 mM MgCl2, 3 mM DTE, 0.3 mM NADPH, 0.3 mM malonyl-CoA, and protein. The reaction was started by the addition of malonyl-CoA. Buffers used to determine the pH optimum were morpholinopropanesulfonic acid (MOPS)-NaOH (pH 5.9 to 7.2) and Tris-HCl (pH 6.3 to 8.4). One unit of enzyme activity refers to 2 μmol of NADPH oxidized per min, corresponding to 1 μmol of malonyl-CoA reduced to 3-hydroxypropionate per min.
(ii) Reverse reaction.
The 3-hydroxypropionate-dependent reduction of NADP+ was monitored. The assay mixture (0.5 ml) contained 100 mM Tris-HCl (pH 7.8), 2 mM MgCl2, 3 mM DTE, 0.5 mM CoA, 5 mM NADP+, and 10 mM 3-hydroxypropionate. The reaction was started by the addition of 3-hydroxypropionate.
Purification of malonyl-CoA reductase.
The purification was performed at 4°C. All buffers contained 3 mM DTE unless otherwise indicated. The preparation started with heat-precipitated cell extract from 20 g of cells (wet mass) and used four chromatographic steps; purification was performed with both heterotrophically and autotrophically grown cells.
(i) Heat precipitation.
Cell extract (100,000 × g supernatant) was incubated at 65°C for 30 min to precipitate unwanted protein, lipids, and pigments, followed by ultracentrifugation (100,000 × g) at 4°C for 40 min.
(ii) DEAE-Sepharose chromatography.
The supernatant after heat precipitation (45 ml) was applied to a DEAE-Sepharose column (Pharmacia, fast flow; diameter, 5.0 cm; volume, 80 ml) which had been equilibrated with 20 mM Tris-HCl (pH 7.8)-10% (vol/vol) glycerol (referred to as buffer A) at a flow rate of 3 ml min−1. The column was washed with 2 bed volumes of buffer A, 2 bed volumes of buffer A containing 100 mM NaCl, and 2 bed volumes of buffer A containing 140 mM NaCl. Most activity was eluted with 200 mM NaCl in buffer A in a volume of 150 ml.
(iii) Phenyl-Sepharose chromatography.
Saturated ammonium sulfate solution was added to the combined active fractions obtained from DEAE-Sepharose chromatography to a final concentration of 10% (vol/vol). This solution was applied directly to a Phenyl-Sepharose column (Pharmacia; diameter, 3.0 cm; volume, 20 ml) at a flow rate of 1 ml min−1. The column had been equilibrated with 100 mM Tris-HCl (pH 7.8)-200 mM (NH4)2SO4-5 mM MgCl2-10% (vol/vol) glycerol. After washing the column with 4 bed volumes of this buffer, the column was developed with a 250-ml decreasing linear gradient of 200 to 0 mM ammonium sulfate at 1 ml min−1. The activity was eluted between 160 and 100 mM salt, and the pooled fractions were concentrated and desalted immediately by ultrafiltration (Amicon) to a final volume of 25 ml.
(iv) Blue-Sepharose chromatography.
The combined and desalted fractions from Phenyl-Sepharose chromatography were applied to a Blue-Sepharose CL-6B column (Pharmacia; diameter, 2.2 cm; volume, 10 ml) at a flow rate of 1 ml min−1. The column had been equilibrated with buffer A without DTE. For this chromatographic step, modified buffer A with a pH of 8.5 instead of 7.8 was used (referred to as buffer B). After four washing steps (3 bed volumes of buffer A, 3 bed volumes of buffer B, 3 bed volumes of buffer B containing 200 mM NaCl, and 3 bed volumes of buffer B containing 300 mM NaCl), the activity was eluted with buffer B containing 400 mM NaCl. The pooled fractions were desalted and concentrated by ultrafiltration (Amicon) to a final volume of 10 ml. The pH was readjusted to 7.8 by addition of 0.1 M HCl.
(v) Reactive Blue 4 chromatography.
The enzyme was further purified by chromatography on a Reactive Blue 4 column (Sigma-Aldrich; diameter, 1.5 cm; volume, 5 ml). The combined and desalted fractions obtained from Blue-Sepharose chromatography were applied to the Reactive Blue 4 column at a flow rate of 1 ml min−1. The column had been equilibrated with buffer A without DTE. After three washing steps (4 bed volumes of buffer A, 4 bed volumes of buffer B, and 4 bed volumes of buffer B containing 200 mM NaCl), the activity was eluted with buffer B containing 300 mM NaCl. The active fractions were pooled and concentrated by ultrafiltration (Amicon).
Determination of native molecular mass.
The native molecular mass of the enzyme was estimated by gel filtration chromatography. Concentrated fraction obtained from Reactive Blue 4 chromatography was applied to an FPLC Superdex 200 HR 16/60 gel filtration column (Pharmacia; diameter, 1.6 cm; volume, 120 ml) which had been equilibrated with 20 mM Tris-HCl (pH 7.8) containing 100 mM NaCl and 10% (vol/vol) glycerol. The flow rate was 1 ml min−1. The column was calibrated with the following molecular mass standards: ferritin (450 kDa), catalase (240 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (67 kDa), and ovalbumin (45 kDa).
Transformation of 14C-labeled malonyl-CoA and HPLC analysis of substrate and products.
A special assay was used to follow the reduction of 14C-labeled malonyl-CoA to labeled products. The reaction mixture (250 μl) contained 100 mM Tris-HCl (pH 7.8), 2 mM MgCl2, 1 mM NADPH, and 0.15 mM [2-14C]malonyl-CoA (18.5 kBq). The reaction was started by the addition of 50 μl of enzyme (gel filtration chromatography fraction; 0.017 U, 3 μg of protein). After incubation at 55°C for different periods of time, 50-μl samples were withdrawn, and the reaction was stopped by adding 3 μl of 6 M HCl, followed by centrifugation. Then 10 μl of the supernatant was applied to an analytical high-pressure liquid chromatography (HPLC) column (Polyspher OA HY; 300 by 6.5 mm; Merck, Darmstadt, Germany) with 12.5 mM H2SO4 as the eluent at a 0.8 ml min−1 flow rate. Substrate and products were monitored simultaneously for radioactivity with a flowthrough analyzer with a solid scintillator cell and for absorption at 210 nm using a photo diode array detector. Retention times were 4.4 min for malonyl-CoA and 8.3 min for 3-hydroxypropionate.
Other methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (8% polyacrylamide) was performed as described by Laemmli (16). Proteins were visualized by Coomassie blue staining (31). Protein was determined by the method of Bradford (1) using bovine serum albumin as the standard. The N-terminal amino acid sequence of the proteins after blotting on a polyvinylidene difluoride membrane was obtained by gas- and liquid-phase sequencing with an Applied Biosystems 473A sequencer.
RESULTS
Malonyl-CoA reduction in cell extracts.
Extracts of autotrophically grown cells of C. aurantiacus catalyzed the malonyl-CoA-dependent oxidation of NADPH, corresponding to a specific activity of 0.08 μmol of malonyl-CoA reduced min−1 (mg of protein)−1 at 55°C. The specific activity of this reaction in heterotrophically grown cells was 0.03 μmol min−1 (mg of protein)−1. The optimal pH of the enzyme was 7.8. The enzyme activity was not oxygen sensitive. A stoichiometry of 2 mol of NADPH per mol of malonyl-CoA added was observed. NADPH could not be replaced by NADH. After completion of the reaction, the products formed from malonyl-CoA were 3-hydroxypropionate and CoASH.
Purification and characterization of malonyl-CoA reductase.
The malonyl-CoA-reducing enzyme was purified from autotrophically and heterotrophically grown cells. Routine investigations were done with heterotrophically grown cells because these cells were much more easily available and contained sufficient malonyl-CoA reductase activity. Cell extract was heat precipitated to remove unwanted protein, lipids, and pigments, followed by four chromatographic steps (Table 1). An almost homogeneous protein was obtained, with a 6% yield. The colorless protein eluted during gel filtration chromatography in a symmetrical peak corresponding to a native molecular mass of 300 kDa. After SDS-PAGE, one single subunit of approximately 145 kDa was observed, suggesting an α2 composition of the native enzyme (Fig. 2). A minor impurity band migrating at higher molecular mass was present, which, however, was present in larger amounts in extracts from heterotrophically grown cells compared to autotrophically grown cells. It is therefore unlikely to represent malonyl-CoA reductase.
TABLE 1.
Purification of malonyl-CoA reductase from 20 g (fresh cells) of autotrophically grown C. aurantiacusa
| Purification step | Protein (mg) | Activity (μmol min−1) | Sp act [μmol min−1 (mg of protein)−1] | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Cell extract (100,000 × g supernatant) | 616 | 99 | 0.08 | 1.0 | 100 |
| Heat precipitation | 225 | 101 | 0.225 | 2.8 | 102 |
| DEAE-Sepharose | 142 | 60 | 0.210 | 2.6 | 61 |
| Phenyl-Sepharose | 14 | 28 | 1.0 | 10 | 28 |
| Blue-Sepharose | 1.0 | 15 | 7.5 | 94 | 15 |
| Reactive Blue 4 | 0.3 | 6 | 10.0 | 125 | 6 |
The purification protocol with heterotrophically grown cells gave similar results, which differed mainly in the lower initial specific activity.
FIG. 2.
SDS-PAGE of proteins in cell extracts and in the individual enzyme fractions during purification. An 8% gel is shown that was stained with Coomassie blue. Lanes: 1, molecular mass standard proteins; 2, extract of heterotrophically grown cells (15 μg of protein); 3, extract after heat precipitation (15 μg of protein); 4, enzyme fraction after DEAE-Sepharose chromatography (10 μg of protein); 5, enzyme fraction after Phenyl-Sepharose chromatography (10 μg of protein); 6, enzyme fraction after Blue-Sepharose chromatography (5 μg of protein); 7, enzyme fraction after Reactive Blue 4 chromatography (3 μg of protein); 8, molecular mass standard proteins. The arrow on the left indicates the 145-kDa protein band which corresponds to malonyl-CoA reductase.
The enzyme was stable for weeks at −20°C in the presence of 10% (vol/vol) glycerol. The enzyme was oxygen insensitive but sensitive to repeated freezing and thawing. The UV-visible spectrum showed only the protein absorption peak at 280 nm, indicating that there is no chromophoric group present.
Sequencing of the protein revealed the following N-terminal amino acid sequence (X indicates an uncertain amino acid identification): X G T G R L A G K I A L I T G.
Catalytic properties.
The catalytic properties of the purified enzyme were studied at 55°C, which corresponds to the optimal growth temperature of this strain. The pH optimum was 7.8, and half-maximal activities were observed at pH 6.5 and 8.5. The enzyme followed Michaelis-Menten kinetics, with apparent Km values of 30 μM for malonyl-CoA and 25 μM for NADPH. A stoichiometry of 2 mol of NADPH oxidized per mol of malonyl-CoA added was observed. After completion of the reaction, the products identified were 3-hydroxypropionate, CoA, and NADP+. Hence, the enzyme catalyzed the reaction malonyl-CoA + 2 NADPH + H+ → 3-hydroxypropionate− + CoA + 2 NADP+. The specific activity of the pure enzyme was 10 μmol of malonyl-CoA reduced min−1 mg−1, corresponding to a turnover number of 50 s−1 per native enzyme.
The enzyme was highly specific for its substrates (Table 2). NADH was not oxidized, and acetyl-CoA, propionyl-CoA, and succinyl-CoA could not substitute for malonyl-CoA. Glyoxylate was not reduced. No cofactor was required; however, enzyme activity was stimulated by divalent cations in the order Fe2+ > Ca2+ ≃ Mg2+ (Table 2). When Mg2+ (routinely 2 mM) was omitted, approximately 50% residual activity was obtained. In the absence of Mg2+, the addition of EDTA (0.5 mM) resulted in 85 to 90% inhibition. Ni2+ inhibited and Zn2+ inactivated the enzyme. Since 2 μM Fe2+ had the same effect as 2 mM Mg2+, the enzyme may be activated by either cation under physiological conditions.
TABLE 2.
Molecular and catalytic properties of the new enzyme malonyl-CoA reductase from C. aurantiacus
| Property | Malonyl-CoA reductase |
|---|---|
| Substrates | Malonyl-CoA, 2 NADPH |
| Products | 3-Hydroxypropionate, 2 NADP+, CoA |
| Intermediate | Malonate semialdehyde |
| Sp act | 10 μmol min−1 (mg of protein)−1 |
| Apparent Km | 30 μM malonyl-CoA, 25 μM NADPH |
| pH optimum | 7.8 |
| Native molecular mass | 300 ± 50 kDa |
| Subunit molecular mass | 145 ± 10 kDa |
| Suggested composition | α2 |
| Catalytic number | 50 s−1 |
| Specificity | Malonyl-CoA, 100%; acetyl-CoA, <1%; succinyl-CoA, <1%; propionyl-CoA, <1%; glyoxylate, <1%; NADPH, 100%; NADH, <1% |
| Influence of divalent cations (mM)a | Mg2+ (2), 100%; Ca2+ (2), 100%; Fe2+ (0.2), 120%; Fe2+ (0.02), 110%; Fe2+ (0.002), 100%; Mn2+ (2), 40%; Co2+ (2), 40%; Ni2+ (2), 30%; Zn2+ (2), <2% |
100% = standard assay with 2 mM Mg2+.
The enzyme catalyzed the reverse reaction, i.e., 3-hydroxypropionate-dependent reduction of NADP+; NAD+ was not reduced. The rate of the reverse reaction was approximately 5% of the NADPH oxidation rate of the forward reaction at pH 7.8. Since no CoA was required for this activity, this reaction probably stops after oxidation of 3-hydroxypropionate to malonate semialdehyde: 3-hydroxypropionate + NADP+ → malonate semialdehyde + NADPH + H+.
Malonate semialdehyde as an intermediate.
Malonyl-CoA reductase seems to be a large bifunctional enzyme which has NADP+-specific aldehyde dehydrogenase (CoA-acylating) and alcohol dehydrogenase activities. Malonate semialdehyde is expected to be an intermediate, which is either released or remains enzyme bound (see Fig. 1).
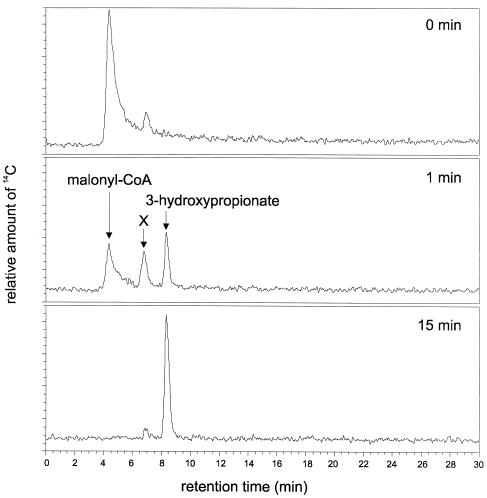
Product formation from 14C-labeled malonyl-CoA and NADPH was studied to detect a possible intermediate. When [2-14C]malonyl-CoA was used as the substrate, the transient formation of a labeled product that differed from 3-hydroxypropionate and most likely represented malonate semialdehyde was observed (Fig. 3). This intermediate was completely consumed in the course of the reaction at the expense of 3-hydroxypropionate formation. This may be interpreted to mean that malonate semialdehyde is a free intermediate of the malonyl-CoA reductase reaction.
FIG. 3.
Transformation of 14C-labeled malonyl-CoA by purified malonyl-CoA reductase and HPLC analysis of labeled substrate and products. The three panels show HPLC separation of the reaction mixture after different periods of incubation, as indicated. The 14C peak in the upper panel corresponds to 0.15 mM [2-14C]malonyl-CoA initially added to the reaction mixture. Peak X is considered to be malonate semialdehyde. The small peak at 0 min that almost comigrates with compound X is likely to be free [2-14C]malonate as a contaminant of [2-14C]malonyl-CoA. For conditions, see Materials and Methods.
To corroborate this conclusion, the reaction stoichiometry was studied in the presence and absence of the aldehyde-trapping agent semicarbazide, which forms a semicarbazone with aldehydes. The stoichiometry of 2 mol of NADPH oxidized per mol of malonyl-CoA added, which was observed in the absence of semicarbazide, was reduced to a 1:1 stoichiometry in the presence of semicarbazide, suggesting that the semialdehyde was a free intermediate. Addition of semicarbazide also resulted in 50 to 60% reduction in the initial NADPH oxidation rate. These findings indicate that the enzyme catalyzed two steps: (i) malonyl-CoA + NADPH + H+ → malonate semialdehyde + CoA + NADP+ and (ii) malonate semialdehyde + NADPH + H+ → 3-hydroxypropionate + NADP+.
DISCUSSION
New bifunctional oxidoreductase and potential application.
A new enzyme from C. aurantiacus that catalyzes the reduction of malonyl-CoA with two NADPH to 3-hydroxypropionate was described (Table 2). Malonate semialdehyde is an intermediate of the reaction. The enzyme is stimulated by Fe2+ and Mg2+. Obviously, it is a large bifunctional enzyme which harbors an aldehyde dehydrogenase (CoA-acylating) and an alcohol dehydrogenase domain. Malonyl-CoA reductase belongs to the enzyme classes EC 1.1.1 and EC 1.2.1. The enzyme may be useful for synthesis of 3-hydroxypropionate and for a coupled spectrophotometric continuous assay for activity screening of acetyl-CoA carboxylase, a target enzyme of a class of herbicides (8), which until now could only be tested by discontinuous radioisotope assay.
Role of the enzyme, required enzyme activity, and regulation.
The physiological role of malonyl-CoA reductase is to reduce malonyl-CoA to 3-hydroxypropionate. This represents the second step of the cycle, which follows after ATP-dependent carboxylation of acetyl-CoA by acetyl-CoA carboxylase. The reverse reaction, oxidation of 3-hydroxypropionate by NAD(P)+, could hardly be measured. The low apparent Km values and the high specificity for its substrates strongly favor a role of the enzyme in reduction of malonyl-CoA. Since malonyl-CoA reductase is not required in any other known bacterial pathway, it can be regarded as a characteristic enzyme of the 3-hydroxypropionate cycle. Since it is easy to measure, sufficiently active, highly specific, and reasonably stable, it is ideally suited for screening autotrophs for the operation of this pathway. The characterization of this key enzyme adds a further piece of evidence for the validity of the hypothetical carbon fixation pathway via 3-hydroxypropionate. It should be added, however, that the first half of the 3-hydroxypropionate cycle may also be suited for the assimilation of acetate.
The specific activity is high enough to meet the requirements of growing cells. More specifically, the generation time of autotrophically growing cultures was 26 h (specific growth rate, 0.027 h−1), which corresponds to a calculated CO2 fixation rate of 0.026 μmol of CO2 fixed min−1 (mg of cell protein)−1 (for calculation, see reference 9). Since two molecules of CO2 are fixed during one turn of this cycle, the minimal specific activity of the enzymes of this cycle should be 0.013 μmol min−1 (mg of protein)−1. The observed specific activity was 0.08 μmol min−1 (mg of protein)−1, which is sufficient to explain the growth rate.
Malonyl-CoA reductase activity is controlled according to its postulated physiological requirement. In heterotrophically grown cells, only about 37% of the maximal rate could be measured, although these cells grew five times faster than cells under autotrophic conditions. This downregulation effect was even more pronounced in M. sedula (17).
Evidence for two partial reactions involving malonate semialdehyde as an intermediate.
The kinetics of malonyl-CoA-dependent oxidation of NADPH seemed to be in line with normal Michaelis-Menten kinetics. No clear sign of a biphasic reaction, which might be expected when malonate semialdehyde is an intermediate that is further reduced by NADPH, was observed. Semicarbazide trapping experiments clearly showed that the semialdehyde is formed as an intermediate. The reduction of the semialdehyde to 3-hydroxypropionate seems to be fast, and the affinity for this intermediate is high. The first conclusion is derived from the observation that semicarbazide addition to the standard assay reduced the NADPH oxidation rate 50 to 60%. If semialdehyde reduction were rate limiting, this effect should be less pronounced or not observable. The second conclusion follows from the fact that the semialdehyde did not accumulate in large amounts in the course of the reaction, yet an intermediate which is likely to be the semialdehyde (malonate semialdehyde is not commercially available and therefore could not be used as a reference) was clearly detectable by HPLC methods (Fig. 3). This intermediate was rapidly transformed to 3-hydroxypropionate.
Metal requirements.
The enzyme required a divalent cation for activity, although an absolute dependence could not be shown. Addition of Mg2+, Ca2+, or Fe2+ led to a twofold stimulation, and addition of EDTA resulted in 90% inhibition. High concentrations of Mg2+ (2 mM) and low concentrations of Fe2+ (2 μM) gave similar effects. Since Mg2+ is present in much higher intracellular concentrations than Fe2+, it is not possible to fix the physiologically relevant metal cofactor. This will be discussed below. Zn2+ inactivates the enzyme, possibly by binding strongly to the metal binding site without being active in catalysis.
Comparison with similar oxidoreductases.
The oxidation of a primary alcohol via its aldehyde to the CoA thioester of the corresponding acid and the reverse reaction are common reactions in metabolism. In bacteria, such reactions play a role in the energy metabolism of many fermentative bacteria (5, 6, 26, 27). So far, reduction of malonyl-CoA or oxidation of 3-hydroxypropionate to malonyl-CoA has not been reported because such reactions do not occur in the energy metabolism of any fermenting bacteria. To our knowledge, 3-hydroxypropionate has not been tested as a growth substrate because it is not an important naturally occurring substrate. Still, the new enzyme malonyl-CoA reductase is expected to have some similarities to other alcohol and aldehyde dehydrogenases.
A database search revealed that the N-terminal amino acid sequence exactly matched the deduced N-terminal amino acid sequence (M S G T G R L A G K I A L I T G) of a putative open reading frame (ORF) in the incomplete database of the C. aurantiacus genome (http://www.jgi.doe.gov/JGI_microbial/html/chloroflexus/chloro_mainpage.html). The ORF was obviously incomplete, and the C terminus was lacking since the ORF (on contig 1090) did not contain a stop codon. The truncated ORF was highly similar to ones encoding various short-chain alcohol dehydrogenases. These proteins are in the range of 25 to 30 kDa. Since malonyl-CoA reductase is larger than 100 kDa, this finding suggests that the enzyme is likely to be composed of a short N-terminal alcohol dehydrogenase domain and a large C-terminal aldehyde dehydrogenase domain.
Since malonyl-CoA reductase represents a bifunctional enzyme with aldehyde dehydrogenase (CoA-acylating) and alcohol dehydrogenase activities, the relation to other known bifunctional oxidoreductases will be interesting. AdhE from Escherichia coli and similar enzymes, like Aad/AdhE from Clostridium acetobutylicum and AdhE from Entamoeba histolytica and Giardia lamblia, also represent bifunctional enzymes (2, 6, 14, 15, 19, 23, 24). In vivo, these enzymes catalyze the reduction of a CoA thioester of an organic acid, typically acetate, to its corresponding alcohol. These enzymes are classified in group III of alcohol dehydrogenases or metal-activated alcohol dehydrogenases, since a common property of these enzymes is activation by divalent cations (23).
AdhE from E. coli is activated by Fe2+, whereas Zn2+, Ni2+, Cu2+, and Mn2+ inhibit the enzyme (14, 15, 23). All known AdhE-type enzymes use NADH as a cofactor to reduce the CoA-ester to its alcohol and are involved in catabolism. Analysis of the protein and gene sequences suggested that these enzymes are fusion proteins resulting from fusion of an alcohol dehydrogenase gene and an aldehyde dehydrogenase gene. The N-terminal region of the protein represents the aldehyde dehydrogenase moiety, and the C-terminal region represents the alcohol dehydrogenase domain (23). The AdhE type of enzymes have subunit sizes of about 95 kDa and are believed to form homodimers. Malonyl-CoA reductase also forms a homodimer, but its subunit is much larger; the reason for this is unknown.
In contrast, the NADPH-dependent reduction of succinyl-CoA to 4-hydroxybutyrate via succinate semialdehyde in Clostridium kluyveri is catalyzed by the combined action of two separate proteins, a CoA-acylating succinate semialdehyde dehydrogenase and a 4-hydroxybutyrate dehydrogenase (26, 27). The genes coding for these enzymes constitute an operon. Under cellular conditions, two semialdehyde dehydrogenase subunits and two alcohol dehydrogenase subunits seem to form a tetrameric enzyme complex of about 200 kDa (27).
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Bonn, and the Fonds der Chemischen Industrie, Frankfurt. Special thanks are due to Richard Krieger, Institut für Organische Chemie, Universität Freiburg, for the synthesis of monothiophenylmalonate and to Nasser Gad'on, Freiburg, for assistance in growing cells.
REFERENCES
- 1.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Bruchhaus, I., and E. Tannich. 1994. Purification and molecular characterization of the NAD+-dependent acetaldehyde/alcohol dehydrogenase from Entamoeba histolytica. Biochem. J. 303:743-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton, N. P., T. D. Williams, and P. R. Norris. 1999. Carboxylase genes of Sulfolobus metallicus. Arch. Microbiol. 172:349-353. [DOI] [PubMed] [Google Scholar]
- 4.Castenholz, R. W. 1969. Thermophilic blue-green algae and the thermal environment. Bacteriol. Rev. 33:476-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Rev. 63:223-234. [DOI] [PubMed] [Google Scholar]
- 6.Dürre, P., R-J. Fischer, A. Kuhn, K. Lorenz, W. Schreiber, B. Stürzenhofecker, S. Ullmann, K. Winzer, and U. Sauer. 1995. Solventogenic enzymes of Clostridium acetobutylicum: catalytic properties, genetic organization, and transcriptional regulation. FEMS Microbiol. Rev. 17:251-262. [DOI] [PubMed] [Google Scholar]
- 7.Eisenreich, W., G. Strauss, U. Werz, G. Fuchs, and A. Bacher. 1993. Retrobiosynthetic analysis of carbon fixation in the phototrophic eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 215:619-632. [DOI] [PubMed] [Google Scholar]
- 8.Grönwald, J. W. 1994. Herbicides inhibiting acetyl-CoA carboxylase. Biochem. Soc. Trans. 22:616-621. [DOI] [PubMed] [Google Scholar]
- 9.Herter, S., J. Farfsing, N. Gad"on, C. Rieder, W. Eisenreich, A. Bacher, and G. Fuchs. 2001. Autotrophic CO2 fixation in Chloroflexus aurantiacus: study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J. Bacteriol. 183:4305-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holo, H. 1989. Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch. Microbiol. 151:252-256. [Google Scholar]
- 11.Holo, H., and R. Sirevåg. 1986. Autotrophic growth and CO2 fixation in Chloroflexus aurantiacus. Arch. Microbiol. 145:173-180. [Google Scholar]
- 12.Imamoto, T., M. Kodera, and M. Yokoyama. 1982. A convenient method for the preparation of S-esters of thio analogs of malonic acid. Bull. Chem. Soc. Jpn. 55:2303-2304. [Google Scholar]
- 13.Ishii, M., T. Miyake, T. Satoh, H. Sugiyama, Y. Oshima, T. Kodama, and Y. Igarashi. 1997. Autotrophic carbon dioxide fixation in Acidianus brierleyi. Arch. Microbiol. 166:368-371. [DOI] [PubMed] [Google Scholar]
- 14.Kessler, D., W. Herth, and J. Knappe. 1992. Ultrastructure and pyruvate formate-lyase radical quenching property of the multienzymic AdhE protein of Escherichia coli. J. Biol. Chem. 267:18073-18079. [PubMed] [Google Scholar]
- 15.Kessler, D., I. Leibrecht, and J. Knappe. 1991. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 281:59-63. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Ménendez, C., Z. Bauer, H. Huber, N. Gad'on, K. O. Stetter, and G. Fuchs. 1999. Presence of acetyl-coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for the operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J. Bacteriol. 181:1088-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohrhauer, H., K. Christiansen, M. Gan, M. Deubig, and R. T. Holman. 1968. Improved method for the preparation of malonyl coenzyme A. J. Lipid Res. 9:398-399. [PubMed] [Google Scholar]
- 19.Nair, R. V., G. E. Bennett, and E. T. Papoutsakis. 1994. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 176:871-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierson, B. K., and R. W. Castenholz. 1974. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 100:5-24. [DOI] [PubMed] [Google Scholar]
- 21.Pierson, B. K., and R. W. Castenholz. 1974. Studies of pigments and growth in Chloroflexus aurantiacus, a phototrophic filamentous bacterium. Arch. Microbiol. 100:283-305. [DOI] [PubMed] [Google Scholar]
- 22.Pollmann, W., and G. Schramm. 1964. Reactivity of metaphosphate esters prepared from P4O10 and ethylether. Biochim. Biophys. Acta 80:1-7. [Google Scholar]
- 23.Reid, M. F., and C. A. Fewson. 1994. Molecular characterization of microbial alcohol dehydrogenases. Crit. Rev. Microbiol. 20:13-56. [DOI] [PubMed] [Google Scholar]
- 24.Sánchez, L. B. 1998. Aldehyde dehydrogenase (CoA-acetylating) and the mechanism of ethanol formation in the amitochondrial protist Giardia lamblia. Arch. Biochem. Biophys. 354:57-64. [DOI] [PubMed] [Google Scholar]
- 25.Sirevåg, R., and R. Castenholz. 1979. Aspects of carbon metabolism in Chloroflexus. Arch. Microbiol. 120:151-153. [Google Scholar]
- 26.Söhling, B., and G. Gottschalk. 1993. Purification and characterization of a coenzyme-A-dependent succinate-semialdehyde dehydrogenase from Clostridium kluyveri. Eur. J. Biochem. 212:121-127. [DOI] [PubMed] [Google Scholar]
- 27.Söhling, B., and G. Gottschalk. 1996. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J. Bacteriol. 178:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauss, G., W. Eisenreich, A. Bacher, and G. Fuchs. 1992. 13C-NMR study of autotrophic CO2 fixation pathways in the sulfur-reducing archaebacterium Thermoproteus neutrophilus and in the phototrophic eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 205:853-866. [DOI] [PubMed] [Google Scholar]
- 29.Strauss, G., and G. Fuchs. 1993. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur. J. Biochem. 215:633-643. [DOI] [PubMed] [Google Scholar]
- 30.van der Meer, M. T. J., S. Schouten, J. W. de Leeuw, and D. M. Ward. 2000. Autotrophy of green non-sulphur bacteria in hot spring microbial mats: biological explanations for isotopically heavy organic carbon in the geological record. Environ. Microbiol. 2:428-435. [DOI] [PubMed] [Google Scholar]
- 31.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low-background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182:157-159. [DOI] [PubMed] [Google Scholar]