Abstract
The Bacillus subtilis competence transcription factor ComK is required for establishment of competence for genetic transformation. In an attempt to study the ComK factor further, we explored the genes regulated by ComK using the DNA microarray technique. In addition to the genes known to be dependent on ComK for expression, we found many genes or operons whose ComK dependence was not known previously. Among these genes, we confirmed the ComK dependence of 16 genes by using lacZ fusions, and three genes were partially dependent on ComK. Transformation efficiency was significantly reduced in an smf disruption mutant, although disruption of the other ComK-dependent genes did not result in significant decreases in transformation efficiency. Nucleotide sequences similar to that of the ComK box were found for most of the newly discovered genes regulated by ComK.
Bacillus subtilis cells develop competence when they enter the stationary phase of growth in response to nutrient limitation (10). During the competence development process, only a fraction of the cells differentiate into competent cells that have physiological characteristics different from those of noncompetent cells, such as an inability to synthesize DNA or undergo cell division (8, 11, 14). The ComK transcription factor plays a key role in competence development, and expression of comK is limited to competent cells (12, 14). It has been demonstrated that various signals converge during the ComK synthesis process (10, 12, 13, 42). One of the signals, cell density, is recognized by the cell through accumulation of extracellular factors, ComX and CSF, which results in phosphorylation of the ComA response regulator of the two-component regulatory system, ComP-ComA (23). In the noncompetent state, ComK is trapped in a complex consisting of ClpC, ClpP, and MecA and thus is inactive (39, 40). Phosphorylated ComA activates expression of comS, whose gene product is needed for the release of ComK from the complex, resulting in ComK activation (31, 40). Upon activation, ComK directly binds to the ComK box (17) located in the upstream regulatory regions of the target genes or operons which are involved in the synthesis of DNA uptake machinery (comE, comF, and comG operons and comC) (7, 8, 12, 42, 45). In addition, ComK regulates the genes involved in DNA metabolism and recombination, such as nucA, addBA, and recA, by binding to the ComK box (15, 16, 43). It has also been suggested that ComK participates in cellular events specific to competent cells (11, 14). Hence, further identification of ComK-regulated genes should contribute to a better understanding of the role of ComK in competent cells. In this study we analyzed a global change in transcription caused by comK deficiency by the DNA microarray technique, which has been proven to be a powerful tool for global analysis of gene transcription (20, 27, 32, 46).
MATERIALS AND METHODS
Strains and medium.
B. subtilis CU741 trpC2 leuC7, CU741c trpC2 leuC7 comK, and ODF200 hag-lacZ::Cm have been described previously (28). The lacZ fusion strains used in this study are pMutin-based disruption mutants constructed by members of the Japan (JAFAN database [http://bacillus.genome.ad.jp]) and EU (Micado database [http://locus.jouy.inra.fr/micado]) consortia of B. subtilis functional genomics. In these strains a 5′-terminal region of the genes to be studied is transcriptionally fused to the Escherichia coli lacZ gene (41). The comK mutants of the fusion strains were constructed by transformation with DNA from CU741c.
MC medium is a minimal salt-glucose medium supplemented with potassium glutamate and Casamino Acids (21).
RNA preparation, cDNA synthesis, hybridization, and microarray analysis.
Total RNAs were isolated from CU741 and CU741c cells that were grown in 100 ml of synthetic MC medium and were harvested 3 h after the end of the logarithmic growth phase. The procedures used for RNA preparation have been described previously (46). The fluorescently labeled cDNA probes used for hybridization to DNA microarrays were prepared by a two-step procedure as described previously (32). cDNAs from CU741and CU741c were labeled with Cy3 and Cy5, respectively.
DNA microarrays were prepared by the method described previously (46) and were supplied by Takara Shuzo (Shiga, Japan). A single microarray plate used in this study contained two grids, each containing 4,055 protein-encoding genes and 39 calf thymus DNA spots used as negative controls. The grids did not contain 45 genes for which PCR amplification was not successful. The hybridization and microarray analyses were performed as described previously (32, 46). The mean values for the Cy3 and Cy5 fluorescence intensities for each gene on the two grids were calculated after subtraction of the background values obtained by determining the average values for the intensities of the 39 calf thymus DNA spots, and ratios of gene expression were calculated (Table 1). transferrin receptor mRNA and the complementary human transferrin receptor primer were used as a positive, internal control (46).
TABLE 1.
Global analysis of the comK regulon
| Genea | Exptb
|
Transcription levelc
|
Descriptiond | ComK dependence on lacZ fusion | Transformation efficiency (%)
|
ComK box (figure or reference) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | Wild type | comK | Ratio | This studye | Previous studyf | ||||
| thdF | ∗ | ∗ | 1,034 | 251 | 4.1 | Thiophen and furan oxidation | Yesg | 51 | Fig. 2 | ||
| yyaF | ∗ | ∗ | ∗ | 40,637 | 509 | 79.8 | Unknown; similar to unknown proteins | Yesg | 38 | Fig. 2 | |
| rpsF | ∗ | ∗ | 13,341 | 617 | 21.6 | Ribosomal protein S6 (BS9) | |||||
| ssb | ∗ | ∗ | 22,147 | 1,292 | 17.1 | Single-stranded DNA-binding protein | |||||
| yyaN | ∗ | ∗ | 655 | 100 | 6.6 | Unknown; similar to transcriptional regulator (MerR family) | Too low | 76 | Fig. 2 | ||
| sacY | ∗ | ∗ | ∗ | 734 | 119 | 6.2 | Transcriptional antiterminator involved in positive regulation of levansucrase and sucrase synthesis | ||||
| sacX | ∗ | ∗ | ∗ | 870 | 31 | 28.1 | Negative regulatory protein of SacY | Fig. 2 | |||
| ywfM | ∗ | ∗ | ∗ | 3,038 | 180 | 16.9 | Unknown; similar to unknown proteins | Too low | 49 | ||
| ywpH | ∗ | ∗ | ∗ | 16,086 | 116 | 138.7 | Unknown; similar to single-stranded DNA-binding protein | Yesg | 20 | Fig. 2 | |
| glcR | ∗ | ∗ | ∗ | 13,759 | 3,604 | 3.8 | Transcriptional repressor involved in expression of the phosphotransferase system | Fig. 2 | |||
| comFA | ∗ | ∗ | ∗ | 54,287 | 158 | 343.6 | Late competence protein required for DNA uptake | Yesh | 0.5-0.01 (25) | 17 | |
| comFB | ∗ | ∗ | ∗ | 11,714 | 127 | 92.2 | Late competence gene | 10 (25) | |||
| comFC | ∗ | ∗ | ∗ | 25,090 | 684 | 36.7 | Late competence gene | 50 (25) | |||
| yvyF | ∗ | ∗ | ∗ | 15,554 | 1,113 | 14.0 | Unknown; similar to flagellar protein | Yesg | 41 | ||
| yvrP | ∗ | 6,326 | 456 | 13.9 | Unknown; similar to unknown proteins from B. subtilis | Yesg | 27i | Fig. 2 | |||
| argG | ∗ | ∗ | 10,555 | 172 | 61.4 | Argininosuccinate synthase | Fig. 2 | ||||
| argH | ∗ | ∗ | 2,788 | 31 | 89.9 | Argininosuccinate lyase | |||||
| ytzD | ∗ | ∗ | 606 | 34 | 17.8 | Unknown | |||||
| comC | ∗ | ∗ | ∗ | 5,315 | 155 | 34.3 | Protein required for processing and translocation of ComGC, -GD, -GE, -GG | Yesj | 0.001 (26) | 17 | |
| maf | ∗ | ∗ | ∗ | 8,218 | 912 | 9.0 | Septum formation | Fig. 2 | |||
| ysxA | ∗ | ∗ | ∗ | 1,902 | 144 | 13.2 | Unknown; similar to DNA repair protein | Yesg | 95i | Fig. 2 | |
| comER | ∗ | ∗ | ∗ | 6,540 | 27 | 242.2 | Nonessential gene for competence | 80 (18) | 17 | ||
| comEA | ∗ | ∗ | ∗ | 40,386 | 93 | 434.3 | Exogenous DNA-binding protein | 0.2-10−6 (18) | |||
| comEB | ∗ | ∗ | ∗ | 29,174 | 239 | 122.1 | Late competence operon required for DNA binding and uptake | 25-50 (18) | |||
| comEC | ∗ | ∗ | ∗ | 2,626 | 55 | 47.7 | Late competence operon required for DNA binding and uptake | Yesj | |||
| comGA | ∗ | ∗ | ∗ | 43,481 | 66 | 658.8 | Late competence gene | Yesj | <0.001 (4) | 17 | |
| comGB | ∗ | ∗ | ∗ | 6,796 | 27 | 251.7 | DNA transport machinery | <0.001 (4) | |||
| comGC | ∗ | ∗ | ∗ | 13,801 | 142 | 97.2 | Exogenous DNA binding | <0.001 (4) | |||
| comGD | ∗ | ∗ | ∗ | 40,543 | 27 | 1,501.6 | DNA transport machinery | <0.001 (4) | |||
| comGE | ∗ | ∗ | ∗ | 12,402 | 27 | 459.3 | DNA transport machinery | <0.001 (4) | |||
| comGF | ∗ | ∗ | ∗ | 15,822 | 32 | 494.4 | DNA transport machinery | <0.001 (4) | |||
| comGG | ∗ | ∗ | ∗ | 1,890 | 27 | 70.0 | DNA transport machinery | <0.001 (4) | |||
| sinI | ∗ | ∗ | ∗ | 1,855 | 61 | 30.4 | Antagonist of SinR | Fig. 2 | |||
| yneA | ∗ | ∗ | 761 | 103 | 7.4 | Unknown | Yesg | 81 | |||
| yneB | ∗ | 532 | 45 | 11.8 | Unknown; similar to resolvase | Yesg | 102 | Fig. 2 | |||
| recA | ∗ | ∗ | ∗ | 11,981 | 990 | 12.1 | Multifunctional protein involved in homologous recombination and DNA repair | Yesk | 17 | ||
| smf | ∗ | ∗ | ∗ | 6,750 | 279 | 24.2 | DNA processing Smf protein homolog | Yesg | 2 | Fig. 2 | |
| yjaZ | ∗ | ∗ | 1,696 | 114 | 14.9 | Unknown | No | 77i | Fig. 2 | ||
| argF | ∗ | ∗ | 8,647 | 175 | 49.4 | Ornithine carbamoyltransferase | |||||
| carB | ∗ | ∗ | 42,610 | 92 | 463.2 | Carbamoyl-phosphate transferase-arginine (subunit B) | |||||
| carA | ∗ | ∗ | 33,186 | 27 | 1,229.1 | Arbamoyl-phosphate transferase-arginine (subunit A) | |||||
| argD | ∗ | ∗ | 29,027 | 82 | 354.0 | N-Acetylornithine aminotransferase | |||||
| argB | ∗ | ∗ | 7,826 | 27 | 289.9 | N-Acetylglutamate 5-phosphotransferase | Fig. 2 | ||||
| yitT | ∗ | ∗ | 2,036 | 439 | 4.6 | Unknown; similar to unknown proteins | 44i | ||||
| sbcD | ∗ | 1,671 | 117 | 14.3 | Exonuclease SbcD homolog | Yesg | 9 | ||||
| yhjR | ∗ | ∗ | 2,056 | 123 | 16.7 | Unknown | Too low | 65 | Fig. 2 | ||
| yhjB | ∗ | ∗ | 4,866 | 622 | 7.8 | Unknown; similar to metabolite permease | Yesg | 22 | Fig. 2 | ||
| comK | ∗ | ∗ | ∗ | 24,507 | 33 | 742.6 | Competence transcription factor | Yesl | <0.0001 (44) | 17 | |
| yhzC | ∗ | ∗ | 512 | 85 | 6.0 | Unknown | Too low | 98 | Fig. 2 | ||
| yhcH | ∗ | ∗ | 599 | 163 | 3.7 | Unknown; similar to ABC transporter (ATP-binding protein) | Yesg | 15i | |||
| yhcG | ∗ | ∗ | 802 | 184 | 4.3 | Unknown; similar to glycine betaine-l-proline transport | Too low | 148 | |||
| yhcF | ∗ | ∗ | 2,729 | 731 | 3.7 | Unknown; similar to transcriptional regulator (GntR family) | Partialg | 98 | |||
| yhcE | ∗ | ∗ | 1,066 | 261 | 4.1 | Unknown | Partialg | 74i | |||
| yfnG | ∗ | ∗ | 220 | 43 | 5.1 | Unknown; similar to CDP-glucose 4,6-dehydratase | Too low | 114 | |||
| rapH | ∗ | ∗ | 7,146 | 603 | 11.9 | Response regulator aspartate phosphatase | Yesg | 171i | Fig. 2 | ||
| tlpC | ∗ | ∗ | 2,002 | 174 | 11.5 | Methyl-accepting chemotaxis protein | |||||
| nucA | ∗ | ∗ | ∗ | 4,152 | 27 | 153.8 | Membrane-associated nuclease | Yesm | 48 (43) | 17 | |
| nin | ∗ | ∗ | ∗ | 1,609 | 59 | 27.3 | Inhibitor of the DNA-degrading activity of NucA | ||||
| yckE | ∗ | ∗ | ∗ | 1,891 | 83 | 22.8 | Unknown; similar to beta-glucosidase | Too low | 88 | Fig. 2 | |
| cwlJ | ∗ | ∗ | ∗ | 667 | 60 | 11.1 | Cell wall hydrolase (sporulation) | ||||
| ybdK | ∗ | 1,097 | 84 | 13.1 | Unknown; similar to two-component sensor histidine kinase | Yesg | 68 | Fig. 2 | |||
Genes are arranged counterclockwise from the origin. The operon structures for the following groups of genes have been confirmed (yyaF-rpsF-ssb) or deduced (JAFAN database [http://bacillus.genome.ad.jp]): yyaF, rpsF, and ssb; sacY and sacX; comFA, comFB, and comFC; argG and argH; comER, comEA, comEB, and comEC; comGA, comGB, comGC, comGD, comGE, comGF, and comGG; yneA and yneB; argF, carB, carA, argD, and argB; yhcH and yhcG; yhcF and yhcE; and nucA and nin.
The asterisks indicate the experiments in which each gene was identified as a target candidate (see text).
The wild-type and comK transcription levels are the signal intensities of the microarray spots (in arbitrary units) observed in experiment 1 for the wild-type and comK strains, respectively. Each ratio was determined by dividing the signal intensity for the wild-type strain by the signal intensity for the comK mutant.
Data obtained from the JAFAN database.
Transformation was performed as described in Materials and Methods. Transformation efficiencies were calculated by dividing the transformation frequencies of the mutants by the transformation frequency of the wild-type strain.
The numbers in parentheses are reference numbers.
See Fig. 1.
Data from reference 25.
Value obtained from the EU consortium (Micado database [http://locus.jouy.inra.fr/micado]).
Data from reference 44.
Data from reference 16.
Data from reference 43.
The DNA microarray data are available on the World Wide Web at http://www.genome.ad.jp/kegg/expression.
Transformation and β-galactosidase assay.
Transformation and β-galactosidase activity analyses were carried out as described previously (29). For the lacZ fusions showing low β-galactosidase activities, the integrity of each fusion was checked by nucleotide sequence determination. To determine transformation efficiencies, two or more separate experiments were carried out with DNA from ODF200, and the averages are shown in Table 1.
RESULTS AND DISCUSSION
Basis for interpreting the results of DNA microarray analysis.
DNA microarray analysis of the comK gene should provide valuable information on all aspects of ComK regulation. We expected that the ComK regulon would include the genes or operons that are regulated both directly and indirectly by ComK, the latter being the secondary targets of ComK. A ComK box (17) would be present in the control regions belonging to the first class but not in the control regions belonging to the second class. In addition, a cell culture induced for competence is composed of two cell populations (competent and noncompetent cells), and the competent cells account for a minor fraction. Therefore, if a gene is controlled solely by ComK, it is expected that the ratio of its mRNA level in wild-type cells to its mRNA level in comK-deficient cells will be high. In contrast, if a gene is expressed in growing cells and is controlled by ComK under competence-inducing conditions, the ratio will not be very high. Thus, it is conceivable that in the latter case the level of expression of the ComK target gene in the growing culture will be the same in both ComK+ cells and ComK− cells, and ComK-induced expression will be added only in the ComK+ cells in the competence-inducing period. We, therefore, considered a ratio of more than three to be the criterion for stimulation by ComK.
We carried out microarray analyses three times using RNAs isolated from independent cell cultures of CU741 and the comK-deficient mutant CU714c grown simultaneously in MC medium (see Materials and Methods). The numbers of putative ComK target candidates showing ratios of more than three varied from experiment to experiment (159, 88, and 61 candidates for experiments 1, 2, and 3, respectively). It is known that only a small fraction of a B. subtilis cell population becomes competent (12, 14), and the competent cell fraction is different in different experiments. In the three experiments, the intensity of the comK spot on the microarray was 3.2 to 4.1 times higher in experiment 1 than in experiments 2 and 3 despite the fact that in the three experiments the levels of expression for non-ComK-dependent genes were similar (data not shown). Furthermore, the ratios of fluorescence intensity for known ComK target genes in ComK+ and ComK− cells were much higher in experiment 1 than in experiments 2 and 3 (data not shown). These results suggest that the cells used in experiment 1 contained a higher level of ComK, and therefore, the variation in the number of target candidates may be attributed to different contents of competent cells in the cell population. It is also possible that mRNAs for some ComK-regulated genes were not extracted efficiently in experiments 2 and 3 due to their instability, although the same method was used for each preparation. On the basis of the reproducibility of the results, we selected 32 genes that were detected in all three experiments and 25 genes that were detected in both experiment 1 and either experiment 2 or experiment 3. These genes are identified in Table 1 according to the experiments in which they were found. We found five genes that exhibited more than threefold expression in both experiment 2 and experiment 3 but not in experiment 1. Three of these genes, degU, ywfI, and ycgM, were, however, false positives, since expression of the lacZ fusions of these genes was not dependent on ComK (data not shown). The β-galactosidase activities derived from the lacZ fusions of the remaining two genes, yndF and ytnI, were too low to determine the ComK dependence (data not shown), and therefore, these genes are not shown in Table 1. We also include four genes in Table 1 that were detected only in experiment 1 but showed ComK dependence as determined with the lacZ fusions (see below).
The data in Table 1 indicate the levels of transcription of the genes observed in experiment 1. Some of the genes detected were grouped in operons or putative operons by reference to the JAFAN database (http://bacillus.genome.ad.jp). It should be noted that obviously not all of the ComK-regulated genes were detected by the microarray technique used here, because (i) the RNA was isolated under one set of experimental conditions, (ii) some ComK-regulated genes may not show a threefold change in expression, and (iii) some ComK target genes could have been among the 45 genes not present on the microarray (see Materials and Methods).
Known genes in ComK regulon.
The known ComK-regulated genes include the late competence genes (the comC, comE, comF, and comG operon genes), nucA-nin, recA, addBA, and comK itself (15, 16, 43, 45). All of these ComK-dependent genes except addBA were identified, indicating that gross transcriptional changes caused by comK deficiency could be detected under our assay conditions. One of the ComK-regulated genes, recA, showed the smallest ratio (12.1-fold) within this set of genes. The smaller ratio for recA than for the late competence genes may reflect expression of the recA gene in both the growth and competence development phases. It should be noted that none of the three microarray experiments revealed addBA as a target, although the operon is known to be regulated by ComK and carries the ComK box (15, 17). The DNA for addBA was on the microarray plate, as shown by a relatively strong fluorescence spot on the microarray (data not shown). The reason for the discrepancy, however, is not known.
Examination of ComK dependence by β-galactosidase assay.
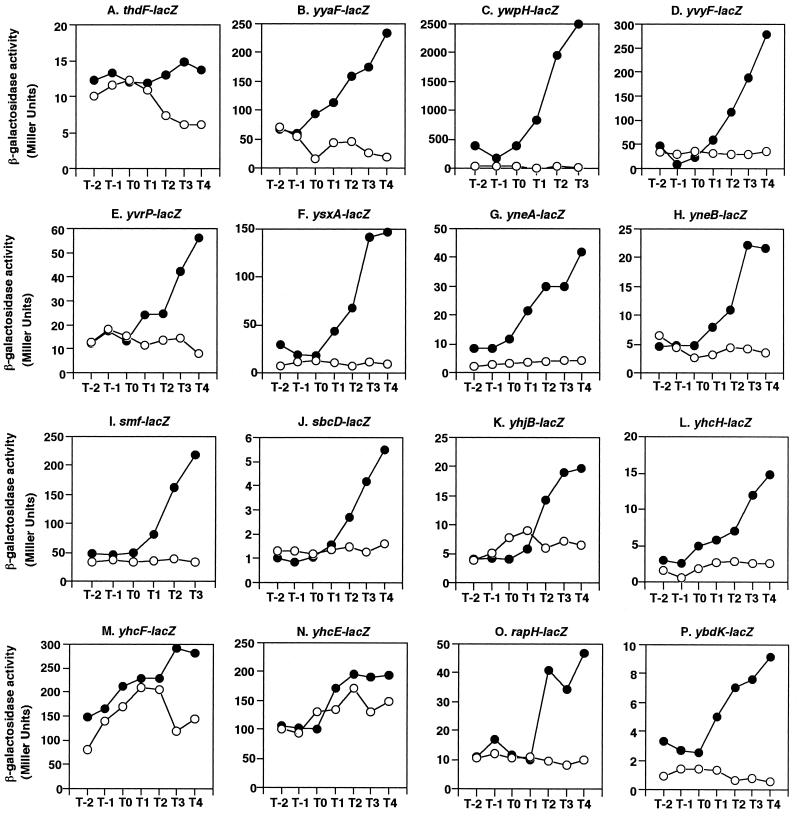
To confirm that the newly identified genes are indeed regulated by ComK, we examined expression of several such genes by using lacZ fusions constructed by and available from the Japan and EU consortia of B. subtilis functional genomics (see Materials and Methods). The expression profiles of the genes are shown in Fig. 1 in the order of appearance in Table 1. We found that expression of most of the lacZ fusions tested started after the end of the exponential growth phase, which is in agreement with the regulatory pattern of ComK. Except for the thdF, yhcF, and yhcE genes (Fig. 1A, M, and N, respectively), the levels of lacZ expression were greatly reduced during the stationary phase in the comK-deficient cells, demonstrating that 13 genes were dependent on ComK for expression. The ComK dependence of the thdF, yhcF, and yhcE genes became apparent only after 2 or 3 h of growth time relative to the end of vegetative growth. It is possible that expression of these genes is also controlled by some other regulator(s) in addition to ComK, which would result in only small differences in the levels of expression between the wild-type and comK-deficient cells. Although ComK dependence was clearly demonstrated for yvrP, yneB, sbcD, and ybdK (Fig. 1E, H, J, and P, respectively), these genes were detected only in experiment 1. This could have been due to one or more of the possible reasons described above. Some of the fusion experiments did not show a difference in β-galactosidase activities, apparently because the activities were too low to be evaluated (Table 1). yjaZ was detected twice as a ComK target candidate in the microarray analysis (Table 1), but there was not a significant difference in the levels of expression of yjaZ-lacZ between the wild-type and comK-deficient cells (Table 1). One interpretation is that expression of yjaZ is autoregulated by YjaZ, since the yjaZ gene is disrupted by insertion of lacZ in the host strain (41). Another possibility is that the structure of the mRNA of yjaZ-lacZ is different from that of yjaZ and the mRNA of yjaZ-lacZ is subject to regulation different from ComK regulation. A definitive conclusion concerning this issue awaits further study.
FIG. 1.
Expression of fusions between ComK target candidates and lacZ in comK disruption mutants. Each strain contains a pMutin-based transcriptional lacZ fusion (41). Cells were grown in MC medium, and β-galactosidase activities were determined as described previously (29). For lacZ fusions of thdF, yhjB, yhcH, yhcF, and yhcE, the assay was performed two or three times, and representative results of these experiments are shown. The numbers on the x axis indicate the growth time (in hours) relative to the end of vegetative growth (T0). Symbols: •, β-galactosidase activity in wild-type cells; ○, β-galactosidase activity in comK mutant cells.
For several genes tested, such as ywpH and yneA, a good correlation was observed between the mRNA levels shown by the signal intensity in the microarray analysis (Table 1) and the levels of β-galactosidase activity from the lacZ fusions (Fig. 1C and G). However, certain lacZ fusions (for example, ywfM, yhjR, and yckE) showed very low β-galactosidase activities (Table 1), in contrast to the significant levels of transcription observed in the microarray analysis. This inconsistency may have been due to (i) the structural difference in mRNAs between the intact genes and the lacZ fusions and (ii) the fact that the signal intensity in our microarray analysis was determined by both the gene size and the copy number of the transcript. Thus, if a gene is large and poorly transcribed, the microarray gives observable signal intensity but the β-galactosidase activity from the lacZ fusion is low. Since the genes studied are not extremely large (JAFAN database), the low β-galactosidase activities could have been due to instability of mRNAs caused by structural differences.
lacZ fusion strains were not available for several of the genes that were tentatively shown to be regulated by ComK (Table 1). We, therefore, examined the pattern of expression of the putative ComK target genes in the cells grown in minimal medium (9) by microarray-based time course analysis and found that expression of all the candidate genes was induced during the stationary phase, although the induction levels were relatively low for ytzD, yitT, yhzC, and yfnG (data not shown). These results are consistent with the notion that the genes are regulated by ComK, but they do not prove it.
Search for ComK boxes in upstream regions of candidate genes.
It has been shown that ComK-regulated genes carry the ComK box (Fig. 2) in their control regions (17). We, therefore, searched for a ComK box on the basis of both the typical nucleotide sequence motifs and the spacing between them and found 21 genes or operons that met the criteria (Table 1 and Fig. 2). Among the 16 genes for which ComK dependence was observed in the lacZ fusion experiments (Fig. 1), possible ComK boxes were found for 10 genes (Fig. 2A), whereas no such sequences were identified for the remaining 4 genes (yvyF, yneA, sbcD, and yhcH). It was previously shown that the yvyF gene is transcribed from the ComK-dependent comF promoter located upstream of this gene (24), and this may explain the ComK dependence of yvyF-lacZ expression (Fig. 1D). The ysxA, yvrP, and smf loci carry a putative ComK box (Fig. 2A), but the sequence deviation from the typical ComK box is somewhat large. Therefore, it is possible that yneA, sbcD, yhcH, ysxA, yvrP, and smf are the secondary targets of ComK. A putative ComK box was found in the upstream region of yjaZ, for which a difference in expression in ComK+ and ComK− cells was detected by the microarray analysis but not by the lacZ fusion assay (see above).
FIG. 2.
Putative ComK boxes found in the upstream regions of the ComK-regulated genes. (A and B) Putative ComK boxes of the genes or operons whose expression was ComK dependent, as shown by both microarray analysis and lacZ fusion experiments and by microarray analysis alone, respectively. (C) Known ComK boxes. Lowercase letters indicate nucleotides that differed from the nucleotides in the ComK consensus sequence. N and dashes indicate nucleotides, and the numbers indicate the spaces between the T and A clusters. The negative numbers on the right indicate the positions of the last nucleotides of the putative ComK boxes relative to the translational start site. The numbers in parentheses are reference numbers.
We also sought ComK boxes in the upstream regions of the genes whose expression was not tested by the lacZ fusion assay, and the results are shown in Fig. 2B. The putative ComK-regulated argBD, carAB, and argF genes constitute an operon with the upstream argCJ genes (33). Although the argCJ genes were detected only once in experiment 1 (data not shown), a putative ComK box was recognized in the upstream region (Fig. 2B).
It should also be noted that the med-comZ operon, which has a canonical ComK box in the regulatory region (29, 30), was detected, although it was detected only once in experiment 1 (8.2- and 4.6-fold for med and comZ, respectively).
Newly identified ComK target genes. (i) Genes involved in transformation.
The smf disruptant exhibited significantly reduced transformation efficiency (Table 1). As determined by the BLAST search described in the JAFAN database, the putative Smf protein has sequence similarity with Haemophilus influenzae DprA, and disruption of the dprA gene results in reduced transformation efficiency (19). It has been suggested that DprA might be part of the cellular apparatus for DNA transport and/or recombination in H. influenzae (19). In Helicobacter pylori, the transformation efficiency is reduced by disruption of dprA (1). Furthermore, H. influenzae DprA is homologous to Streptococcus pneumoniae CilB, and it has been shown that disruption of cilB results in competence deficiency (3). From these observations, it may be reasonable to assume that smf is also involved in transformation in B. subtilis.
Disruption of the rest of the newly identified genes did not result in significant decreases in transformation efficiency (Table 1). These genes may play roles in some processes other than DNA transformation, although we cannot rule out the possibility that multiple disruption of such genes leads to transformation deficiency.
(ii) Gene homologues induced in the competent state in other bacteria.
B. subtilis ssb (Table 1) is homologous to S. pneumoniae cilA, which has been shown to be competence inducible (3, 36). As shown in Fig. 1B, expression of yyaF-lacZ was comK dependent. Since yyaF, rpsF, and ssb were demonstrated to constitute an operon on the basis of Northern experiments (JAFAN database), ssb expression should also be comK dependent. Also, we note that a ysxA homologue of S. pneumoniae is induced during competence development (36). Hence, the two homologues might play similar roles in transformation in the two organisms, although their functions are not known.
(iii) Genes homologous to those acting on DNA structure or cleavage.
Based on sequence similarities, the genes that are thought to affect DNA structure are shown in Table 1; these genes include ssb (encoding single-stranded DNA-binding protein), ywpH (encoding a single-stranded DNA-binding protein homologue), ysxA (encoding a DNA repair protein homologue), yneB (encoding a resolvase homologue), smf (encoding a DNA processing Smf homologue), and sbcD (encoding an exonuclease SbcD homologue, which degrades palindromic nucleotide structure) (5). These proteins might be involved in DNA processing after the DNA translocation or recombination events. In fact, disruption of smf resulted in a decrease in transformation efficiency (see above). ysxA and yneB disruptants, however, showed normal levels of transformation efficiency (95 and 102%, respectively), while ywpH and sbcD disruptants showed slight decreases in transformation (20 and 9%, respectively). The ssb gene is essential (JAFAN database), which precluded us from testing the effect of gene disruption. Recently, NucA has been shown to be required for DNA cleavage during transformation, but nucA deficiency does not affect transformation efficiency (34, 43). Taking these results into account, the slight reduction in transformation efficiency observed for ywpH and sbcD disruptants might indicate that these genes are involved in transformation.
(iv) Genes involved in arginine biosynthesis, transportation, and formation of cell shape.
The yhcHG genes, encoding a putative ABC transporter (35), were found to be regulated by ComK (Table 1 and Fig. 1L), which might suggest that an unknown substrate(s) is transported by YhcHG in competent cells.
Surprisingly, expression of the argGH and argBD-carAB-argF operon genes was greatly reduced in comK cells (Table 1). The comK-deficient cells, however, can grow in minimal medium (data not shown), indicating that these genes are expressed at least in noncompetent cells. Therefore, it is likely that expression of the arginine biosynthetic genes is enhanced in competent cells, although the biological significance of this is not known.
The Maf protein is involved in cell shape determination, and overexpression of maf results in inhibition of septum formation, leading to filamentation of the cells and loss of viability (2). The implications of the effect of ComK on maf are not known.
(v) Regulatory factors.
Expression of eight regulatory genes, including four putative genes (yyaN, sacXY, glcR, sinI, rapH, ybdK, and yhcF), was found to be ComK dependent, which suggests that secondary targets of ComK may be included in the list shown in Table 1. Although sinR (37), the sacXY genes (6), and glcR (38) are known to be involved in certain stationary-phase events, including exoprotease production and sucrose metabolism, it is difficult to identify which genes listed in Table 1 are regulated by these genes. Further microarray analysis should provide valuable information on the regulatory network starting from ComK.
It is interesting that ybdK encoding the sensor kinase of the putative ybdJK two-component system (20, 22) is regulated by ComK (Table 1 and Fig. 1P), which suggests that the YbdJ response regulator might affect competence-specific gene transcription. To examine this possibility further, we performed a microarray analysis of YbdJ-overproducing cells grown in MC medium and found that transcription of several of the comK regulon genes, including comK, was reduced in the range from 0.24- to 0.45-fold compared to transcription in the wild-type cells (data not shown). These results suggest that there is a functional relationship between ComK and the YbdJK two-component system.
We note that the rapH gene encoding a putative aspartyl-phosphate phosphatase for a phosphorylated response regulator (22, 23) is regulated by ComK (Table 1 and Fig. 1O). It is tempting to speculate that the RapH phosphatase might have some functional relationship with the phosphorylated YbdJ response regulator, since rapH and ybdJ are in the same regulon. How these two genes are functionally correlated will be investigated in the future.
Acknowledgments
We thank A. Matsuzawa, T. Ohsawa, A. Negishi, and T. Saito for technical assistance. We greatly appreciate all of the members of the Japan and EU consortia of B. subtilis functional genomics who provided pMutin-based mutants used in this study.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (C) “Genome Biology” and by Grants-in-Aid for Scientific Research (B) and (C) from the Ministry of Education, Science and Sports and Culture.
REFERENCES
- 1.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler, Y. X., Y. Abhayawardhane, and G. C. Stewart. 1993. Amplification of the Bacillus subtilis maf gene results in arrested septum formation. J. Bacteriol. 175:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, E. A., S. Y. Choi, and H. R. Masure. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 27:929-939. [DOI] [PubMed] [Google Scholar]
- 4.Chung, Y. S., and D. Dubnau. 1998. All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis. J. Bacteriol. 180:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connelly, J. C., E. S. de Leau, and D. R. Leach. 1999. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 15:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crutz, A.-E., and M. Steinmetz. 1992. Transcription of the Bacillus subtilis sacX and sacY genes, encoding regulators of sucrose metabolism, is both inducible by sucrose and controlled by the DegS-DegU signalling system. J. Bacteriol. 174:6087-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubnau, D. 1997. Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins. Gene 192:191-198. [DOI] [PubMed] [Google Scholar]
- 8.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Genet. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 9.Dubnau, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 10.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, J., J. Bylund, M. Haines, M. Higgins, and D. Dubnau. 1995. Inactivation of mecA prevents recovery from the competent state and interferes with cell division and the partitioning of nucleoids in Bacillus subtilis. Mol. Microbiol. 18:755-767. [DOI] [PubMed] [Google Scholar]
- 12.Hahn, J., L. Kong, and D. Dubnau. 1994. The regulation of competence transcription factor synthesis constitutes a critical control point in the competence regulation in Bacillus subtilis. J. Bacteriol. 176:5753-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, J., A. Luttinger, and D. Dubnau. 1996. Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol. Microbiol. 21:763-775. [DOI] [PubMed] [Google Scholar]
- 14.Haijema, B. J., J. Hahn, J. Haynes, and D. Dubnau. 2001. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 40:52-64. [DOI] [PubMed] [Google Scholar]
- 15.Haijema, B. J., L. W. Hamoen, J. Kooistra, G. Venema, and D. van Sinderen. 1995. Expression of the ATP-dependent deoxyribonuclease of Bacillus subtilis is under competence-mediated control. Mol. Microbiol. 15:203-211. [DOI] [PubMed] [Google Scholar]
- 16.Haijema, B. J., D. van Sinderen, K. Winterling, J. Kooistra, G. Venema, and L. W. Hamoen. 1996. Regulated expression of the dinR and recA genes during competence development and SOS induction in Bacillus subtilis. Mol. Microbiol. 22:75-85. [DOI] [PubMed] [Google Scholar]
- 17.Hamoen, L. W., A. F. van Werkhoven, J. J. Bijlsma, D. Dubnau, and G. Venema. 1998. The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev. 12:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inamine, G. S., and D. Dubnau. 1995. ComEA, a Bacillus subtilis integral membrane protein required for both DNA binding and transport. J. Bacteriol. 177:3045-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karudapuram, S., X. Zhao, and G. J. Barcak. 1995. DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid DNA transformation. J. Bacteriol. 177:3235-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi, K., M. Ogura, H. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. The comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183:7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst, F., T. Msadek, and G. Rapoport. 1994. Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis, p. 1-20. In P. J. Piggot, C. P. Moran, Jr., and P. Youngman (ed.), Regulation of bacterial differentiation. American Society for Microbiology, Washington, D.C.
- 22.Kunst, F., N. Ogasawara, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 23.Lazazzera, B. A., and A. D. Grossman. 1998. The ins and outs of peptide signaling. Trends Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., and P. Zuber. 1998. A molecular switch controlling competence and motility: competence regulatory factors ComS, MecA, and ComK control σD-dependent gene expression in Bacillus subtilis. J. Bacteriol. 180:4243-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Londono-Vallejo, J. A., and D. Dubnau. 1993. comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases. Mol. Microbiol. 9:119-131. [DOI] [PubMed] [Google Scholar]
- 26.Mohan, S., and D. Dubnau. 1990. Transcriptional regulation of comC: evidence for a competence-specific transcription factor in Bacillus subtilis. J. Bacteriol. 172:4064-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier. 2001. Catabolite repression mediated by CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 28.Ogura, M., and T. Tanaka. 1997. Bacillus subtilis ComK negatively regulates degR gene expression. Mol. Gen. Genet. 254:157-165. [DOI] [PubMed] [Google Scholar]
- 29.Ogura, M., and T. Tanaka. 2000. Bacillus subtilis comZ (yjzA) negatively affects expression of comG but not comK. J. Bacteriol. 182:4992-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogura, M., Y. Ohshiro, S. Hirao, and T. Tanaka. 1997. A new Bacillus subtilis gene, med, encodes a positive regulator of comK. J. Bacteriol. 179:6244-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogura, M., L. Liu, M. Lacelle, M. M. Nakano, and P. Zuber. 1999. Mutational analysis of ComS: evidence for the interaction of ComS and MecA in the regulation of competence development in Bacillus subtilis. Mol. Microbiol. 32:799-812. [DOI] [PubMed] [Google Scholar]
- 32.Ogura, M., H. Yamaguchi, K.-I. Yoshida, Y. Fujita., and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Reilly, M., and K. M. Devine. 1994. Sequence and analysis of the citrulline biosynthetic operon argC-F from Bacillus subtilis. Microbiology 140:1023-1025. [DOI] [PubMed] [Google Scholar]
- 34.Provevedi, R., I. Chen, and D. Dubnau. 2001. NucA is required for DNA cleavage during transformation of Bacillus subtilis. Mol. Microbiol. 40:634-644. [DOI] [PubMed] [Google Scholar]
- 35.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 36.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 37.Strauch, M. A., and J. A. Hoch. 1993. Transition state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol. Microbiol. 3:337-342. [DOI] [PubMed] [Google Scholar]
- 38.Stulk, J., I. Martin-Yerstraete, P. Glaser, and G. Rapoport. 2001. Characterization of glucose-repression-resistant mutants of Bacillus subtilis: identification of the glcR gene. Arch. Microbiol. 175:441-449. [DOI] [PubMed] [Google Scholar]
- 39.Turgay, K., L. W. Hamoen, G. Venema, and D. Dubnau. 1997. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11:119-128. [DOI] [PubMed] [Google Scholar]
- 40.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 42.van Sinderen, D., and G. Venema. 1994. comK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J. Bacteriol. 176:5762-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Sinderen, D., R. Kiewiet, and G. Venema. 1995. Differential expression of two closely related deoxyribonuclease genes, nucA and nucB, in Bacillus subtilis. Mol. Microbiol. 15:213-223. [DOI] [PubMed] [Google Scholar]
- 44.van Sinderen, D., A. ten Berge, B. J. Hayema, L. Hamoen, and G. Venema. 1994. Molecular cloning and sequence of comK, a gene required for genetic competence in Bacillus subtilis. Mol. Microbiol. 11:695-703. [DOI] [PubMed] [Google Scholar]
- 45.van Sinderen, D., A. Luttinger, L. Kong, D. Dubnau, G. Venema, and L. Hamoen. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida, K.-I., K. Kobayashi, Y. Miwa, C.-M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]