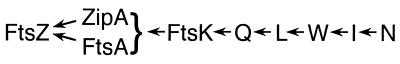Abstract
The septal ring in Escherichia coli consists of at least nine essential gene products whose order of assembly resembles a mostly linear dependency pathway: FtsA and ZipA directly bind FtsZ polymers at the prospective division site, followed by the sequential addition of FtsK, FtsQ, FtsL, FtsW, FtsI, and FtsN. Recruitment of FtsK and all downstream components requires the prior localization of FtsA. Here we show that recruitment of FtsK, FtsQ, FtsL, and FtsN equally requires ZipA. The results imply that association of both FtsA and ZipA with FtsZ polymers is needed for further maturation of the nascent organelle.
Cell division in Escherichia coli is mediated by the septal ring, a membrane-associated structure that assembles at the site of division prior to the onset of septal invagination. At least nine gene products, FtsA, FtsI, FtsK, FtsL, FtsN, FtsQ, FtsW, FtsZ, and ZipA, have been shown to be essential components of this organelle (18, 23). Recent studies determining the ability of these proteins to localize to putative division sites in cells where one of them is either limiting or nonfunctional have suggested a likely order for their assembly into a cytokinetic ring. The picture that has emerged from such work reveals a mostly linear dependency pathway (7). The first visible step involves the movement of FtsZ from the cytoplasm to the inner surface of the inner membrane, where it polymerizes to form a ring-like structure at the prospective division site (4). Both FtsA and ZipA bind FtsZ directly (12, 14, 16, 17, 20, 26) and join the nascent ring in a mutually independent fashion, which is followed by the sequential addition of FtsK, FtsQ, FtsL, FtsW, FtsI, and finally FtsN (1-3, 5, 7, 8, 11, 13, 15, 16, 19, 21, 24, 25, 27, 28). The linear dependency of the pathway is derived from the observation that the recruitment of each component requires the prior localization of all upstream components. However, the mutual independence of FtsA and ZipA localization is a notable exception to this linear sequence and represents a possible branch point in the assembly pathway. It has already been well established that, in addition to FtsZ, FtsA is required for targeting FtsK and all downstream components to the septal ring (1-3, 8, 11, 13, 16, 19, 24, 25, 27, 28), but no such relationship has been established for ZipA. Here we report that green fluorescent protein (GFP) fusions to FtsK, FtsQ, FtsL, and FtsN do not localize to putative division sites in ZipA-depleted filaments. These results imply that recruitment of FtsK, -Q, -L, -W, -I, and -N to the nascent organelle requires the prior formation of an intermediate structure consisting of FtsZ, FtsA, and ZipA.
Localization of FtsK, FtsQ, FtsL, and FtsN
Assembly of FtsK, FtsQ, FtsL, and FtsN into the septal ring was monitored through the use of GFP fusions. Relevant features of these constructs, as well as the strains used in this study, are presented in Table 1. A gfp-tagged derivative of ftsK was obtained by amplifying the 5′ portion of ftsK previously shown to be sufficient to support cell division (10, 25), using primers 5′-GAGCGACATATGAGCCAGGAATACATTGAAGACAAAGAAGTC-3′ and 5′-CGGACTCGAGCGCAGCGTCTGTTTGCCGCCCCATCG-3′. The product was digested with NdeI and XhoI (underlined) and cloned into pET21A (Novagen), yielding pCH203. A gfp tag was then added and the construct was put under the control of the lac promoter in pMLB1113 (9). The resulting plasmid, pCH205, encodes a 57.4-kDa FtsK-Gfp fusion protein, in which Gfpmut2 is fused to the C terminus of FtsK at amino acid 266.
TABLE 1.
Hosts, plasmids, and phages used for localization studies
| Host, plasmid, or phage | Relevant genotype | Source or reference |
|---|---|---|
| Hosts | ||
| CH3 | PB103 recA::Tn10 | 12 |
| CH5 | PB103 zipA::aph recA::Tn10 | 12 |
| Plasmids | ||
| pDB361 | aadA+repA+cI857(ts) PλR::ftsZ | 13 |
| pCH205 | bla+lacIq+ Plac::ftsK(1-266)-gfp | This work |
| Phages | ||
| λCH195 | imm21bla+lacIq+ Plac::gfp-ftsL | This work |
| λCH196 | imm21bla+lacIq+ Plac::gfp-ftsQ | This work |
| λCH201 | imm21bla+lacIq+ Plac::gfp-ftsN | This work |
Gfp-tagged FtsQ, FtsL, and FtsN were expressed from lysogenic λ phages, obtained by first constructing plasmid derivatives in pMLB1113 and then crossing these with phage λNT5 as described previously (9). ftsQ was amplified using primers5′-ACGCGAATTCCATATGTCGCAGGCTGCTC-3′ and 5′-CGCCAAGCTTATTGTTGTTCTGCCTGTGC-3′, andthe product was digested with EcoRI and HindIII (underlined) and ligated to pET21A, yielding pGP1. The NheI-HindIII fragment from pGP1 was cloned into pDR107C (22), creating pCH194, which encodes a 59.9-kDa Gfp-T-FtsQ fusion protein (T represents a T7 tag [Novagen]), in which the Gfpmut2 peptide is fused to the N terminus of FtsQ by the linker peptide ASMTGGQQMGRGSEFH. In pCH196, expression of this fusion protein was placed under the control of the lac promoter by ligating the BglII-HindIII fragment of pCH194 to BamHI- and HindIII-digested pMLB1113.
ftsL was amplified with the primers 5′-CCGAATTCCATATGATCAGCAGAGTGACAG-3′ and 5′-CGTGTCGACTTATTTTTGCACTACGAT-3′. The product was digested with BclI and SalI (underlined) and ligated to BamHI- and SalI-digested pET21C (Novagen), yielding pAB13. The NheI-PstI fragment from pAB13 was cloned into pDR107C, generating pCH193, which encodes a 41.5-kDa Gfp-T-FtsL fusion protein in which the N-terminal methionine of FtsL is replaced with the Gfpmut2 peptide and the linker peptide ASMTGGQQMGR. Expression of this fusion protein was placed under the control of the lac promoter in pCH195 as outlined above.
ftsN was amplified with the primers 5′-CAGCGAATTCCATATGGCACAACGAGATTATG-3′ and 5′-TGAGAAGCTTAACCCCCGGCGGCGAG-3′, and the product was digested with EcoRI and HindIII (underlined) and cloned into pDR107A (22). The resulting plasmid, pCH198, encodes a 64.3-kDa Gfp-T-FtsN fusion protein in which the Gfpmut2 peptide is fused to the N terminus of FtsN by the linker peptide ASMTGGQQMGRGSEFH. As above, the expression of this fusion protein was placed under the control of the lac promoter, yielding pCH201.
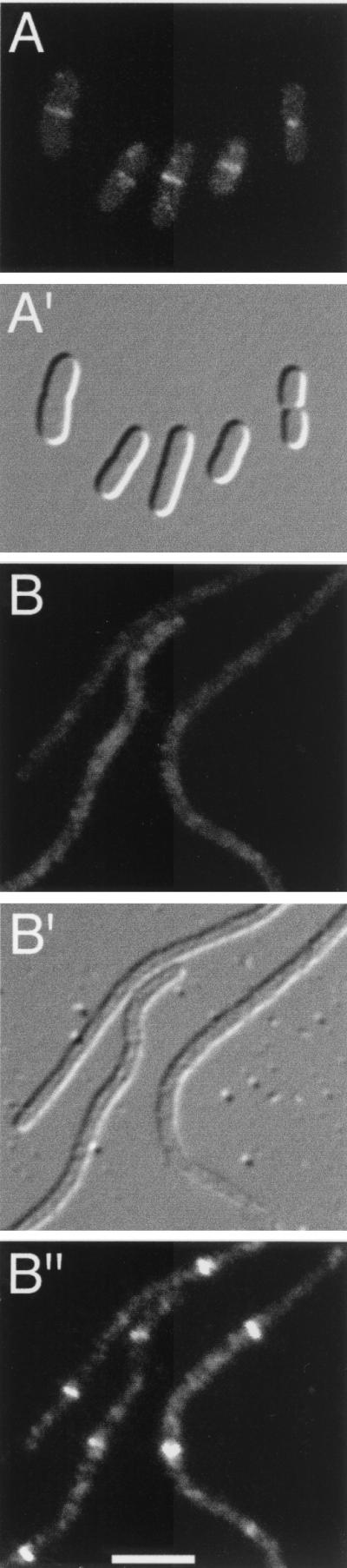
To verify the localization of FtsK(1-266)-Gfp, Gfp-FtsQ, Gfp-FtsL, and Gfp-FtsN, each of the fusions was visualized by fluorescence microscopy in exponentially growing cells of strain CH3/pDB361 (Fig. 1 to 4, A panels). Cells were grown overnight at 30°C in LB-Amp (Luria-Bertani medium containing 50 μg of ampicillin/ml) supplemented with 50 μg of spectinomycin/ml and 0.1% glucose. Cultures were then diluted in fresh LB-Amp plus 5 μM (pCH205), 20 μM (λCH195), 25 μM (λCH196), or 35 μM (λCH201) isopropyl-β-d-thiogalactopyranoside (IPTG) and grown at 30°C to an optical density at 600 nm of 0.35. Cells were chemically fixed, viewed, and imaged as previously described (13). At the low levels of expression used in this study, none of the fusion proteins interfered with the division process, and cells displayed a wild-type phenotype in each case (Fig. 1 to 4, A′ panels). Consistent with earlier observations (2, 6, 8, 11, 25, 28), the majority of the cells expressing FtsK(1-266)-Gfp (66%), Gfp-FtsQ (78%), Gfp-FtsL (58%), or Gfp-FtsN (56%) exhibited a single band of fluorescence at the division site, demonstrating that all four fusions preferentially accumulated at the septal ring (Fig. 1 to 4, A panels). Similar results were obtained when the fusions were expressed in cells of the isogenic ZipA-depleted strain CH5/pDB361 (see below), provided the cells were grown at the permissive temperature (42°C; data not shown.)
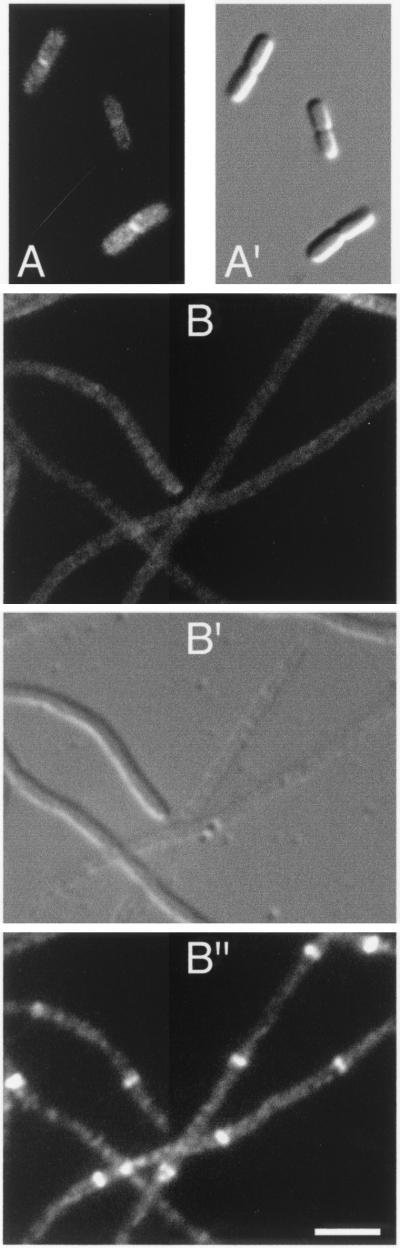
FIG. 1.
Localization of FtsK(1-266)-Gfp. Fluorescence and differential interference contrast micrographs showing the location of FtsK(1-266)-Gfp in normally dividing cells of strain CH3/pDB361/pCH205 (A) and in ZipA-depleted filaments of strain CH5/pDB361/pCH205 (B). (B") Location of native FtsZ in the same filaments shown in panel B. Cells were imaged with Nomarski (A′ and B′) and fluorescence optics using GFP-specific (A and B) or Cy3-specific (B") filter sets. Bar, 2.5 μm.
FIG. 4.
Localization of Gfp-FtsN. Fluorescence (A and B) and differential interference contrast (A′ and B′) micrographs showing the location of Gfp-FtsN in CH3(λCH201)/pDB361 cells (A and A′) and CH5(λCH201)/pDB361 filaments (B and B′). Bar, 2.5 μm.
Localization of FtsK, FtsQ, FtsL, and FtsN in ZipA-depleted filaments
We showed previously that growth of strain CH5/pDB361 (zipA0/λPR::zipA cI857) at 30°C leads to the production of long nonseptate filaments due to the repression of zipA transcription from pDB361 and the consequent depletion of ZipA. Moreover, multiple FtsA-decorated FtsZ rings are still present in these filaments (13).
To assess whether recruitment of FtsK and FtsQ to the septal ring is dependent on ZipA, strain CH5/pDB361 was either transformed with plasmid pCH205 [Plac::ftsK (1-266)-gfp] or lysogenized with phage λCH196 (Plac::gfp-ftsQ). Cells were grown overnight at 42°C in LB-Amp supplemented with 50 μg of spectinomycin/ml and 0.1% glucose. Cultures were diluted in fresh LB-Amp plus 5 μM (pCH205) or 25 μM (λCH196) IPTG and grown at 42°C for 1 h. Cultures were then shifted to 30°C and growth was continued for another 5 to 6 h until cultures reached an optical density of between 0.2 and 0.3. Cells were chemically fixed, immunostained by treatment with affinity-purified anti-FtsZ antibodies and Cy-3-conjugated secondary antibodies, and imaged as described previously (13).
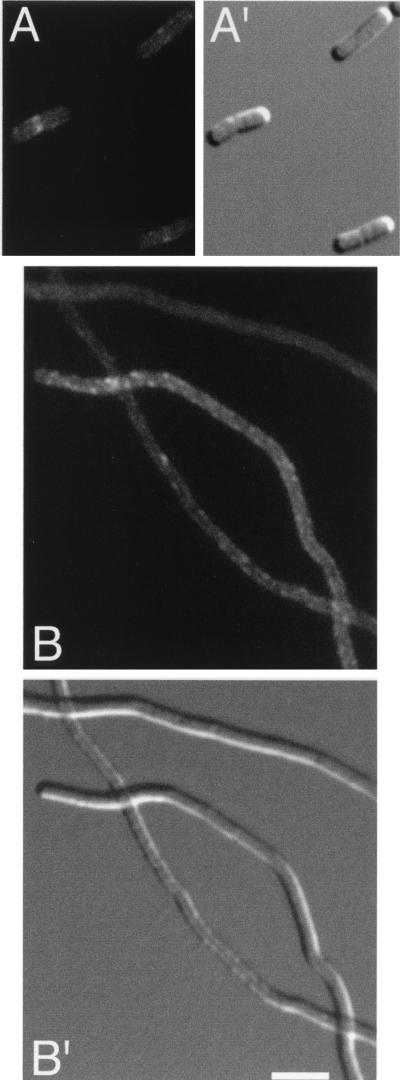
As expected, the ZipA-depleted filaments contained multiple FtsZ rings at regularly spaced intervals along their length (Fig. 1 and 2, B" panels). In contrast, both FtsK(1-266)-Gfp and Gfp-FtsQ completely failed to localize to rings in the vast majority of these filaments (Fig. 1 and 2, B panels). Among hundreds of filaments examined, we observed only a few (<5%) with a single faint accumulation of fluorescence resembling a ring. Instead, a significant portion of FtsK(1-266)-Gfp appeared to be located along the membrane (Fig. 1B), while Gfp-FtsQ showed a mostly diffuse pattern of fluorescence (Fig. 2B). Thus, even though multiple Z rings were present, FtsK(1-266)-Gfp and Gfp-FtsQ failed to associate with these rings due to the depletion of ZipA. We conclude that a minimal amount of ZipA is required for recruitment of FtsK and FtsQ to the septal ring.
FIG. 2.
Localization of Gfp-FtsQ. Fluorescence and differential interference contrast micrographs showing the location of Gfp-FtsQ in CH3(λCH196)/pDB361 cells (A) and in CH5(λCH196)/pDB361 filaments (B). (B") Location of native FtsZ in the same filaments shown in panel B. Cells were imaged with Nomarski (A′ and B′) and fluorescence optics using GFP-specific (A and B) or Cy3-specific (B") filter sets. Bar, 2.5 μm.
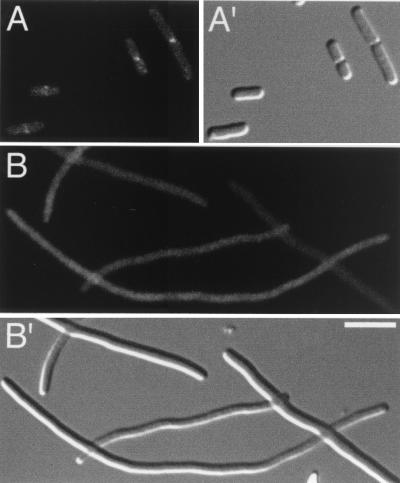
Because FtsK and FtsQ are required for the septal recruitment of FtsL, FtsW, FtsI, and FtsN (2, 7, 11, 19, 27), our results predicted that ZipA would be essential for recruitment of these downstream components as well. To test this prediction, strain CH5/pDB361 was lysogenized with the phages λCH195 (Plac::gfp-ftsL) or λCH201 (Plac::gfp-ftsN) and cells were grown and imaged as above, except that expression of Gfp-FtsL and Gfp-FtsN was induced with 20 and 35 μM IPTG, respectively. As anticipated, neither Gfp-FtsL nor Gfp-FtsN accumulated in rings upon depletion of ZipA (Fig. 3 and 4, B panels). Rather, Gfp-FtsL exhibited a diffuse pattern of fluorescence (Fig. 3B), while Gfp-FtsN appeared distributed along the periphery of the filaments (Fig. 4B).
FIG. 3.
Localization of Gfp-FtsL. Fluorescence (A and B) and differential interference contrast (A′ and B′) micrographs showing the location of Gfp-FtsL in CH3(λCH195)/pDB361 cells (A and A′) and in CH5(λCH195)/pDB361 filaments (B and B′). Bar, 2.5 μm.
We note that at higher levels of expression, all four GFP fusions showed a clear preference for the periphery of cells (data not shown), as might be expected for membrane proteins. Therefore, we suspect that the failure in both this and other (2, 7, 8, 11, 25) studies to detect a more overt membrane localization of FtsQ, FtsL, and (to a lesser extent) FtsK in filaments that lack an upstream septal ring component is due to the level of Fts-specific signal being too low to be clearly distinguishable.
Our observations extend the body of data that addresses the order of assembly of the septal ring components in E. coli. The results, along with those of earlier studies, are summarized in the model shown in Fig. 5. This model is similar to the linear pathway put forth by Beckwith and coworkers (7, 11, 27), but it now incorporates the requirement of ZipA for septal recruitment of FtsK, -Q, -L, -W, -I, and -N. Though we did not directly show here that recruitment of FtsW and FtsI require ZipA, it may be anticipated that this will be the case, as the septal localization of FtsW has been shown to be dependent on the prior localization of FtsQ and FtsL (19), and that of FtsI is dependent on FtsK, -Q, and -L (7, 27).
FIG. 5.
Model showing the dependency pathway for recruitment of division proteins to the developing septal ring organelle. Efficient recruitment of FtsK through FtsN requires the presence of sufficient amounts of functional FtsZ and FtsA, as well as ZipA. The latter three likely form a tripartite complex consisting of FtsZ polymers that are bound to both FtsA and ZipA. Adapted from references 7, 11, and 27.
To date, good evidence for direct physical interactions between the E. coli division proteins has only been obtained for FtsZ and FtsA (14, 17, 26) and for FtsZ and ZipA (12, 14, 16, 20). It seems clear, however, that numerous other interactions among the septal ring components must take place within the organelle. A web of interactions is indicated not only by the recruitment dependency pathway (Fig. 5) but also from the fact that all of the ring's known components remain associated with the invaginating cell wall until septal closure, suggesting that they act in concert with one another throughout the constriction process. An immediate challenge is to precisely define these interactions, as such information should help in meeting the far larger challenge of understanding how the division apparatus, once fully assembled, actually stimulates cell constriction.
Acknowledgments
We thank Gregg Pietz and Anita Boyapati for help with plasmid construction.
This work was supported by NIH grant GM-57059 and by a generous donation from Wyeth-Ayerst Research.
REFERENCES
- 1.Addinall, S. G., E. Bi, and J. Lutkenhaus. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. [DOI] [PubMed] [Google Scholar]
- 3.Addinall, S. G., and J. Lutkenhaus. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178:7167-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 5.Boyle, D. S., M. M. Khattar, S. G. Addinall, J. Lutkenhaus, and W. D. Donachie. 1997. ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 24:1263-1273. [DOI] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., M. E. Aarsman, A. H. Kolk, M. Vicente, and N. Nanninga. 1998. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J. Bacteriol. 180:6107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. C., D. S. Weiss, J. M. Ghigo, and J. Beckwith. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli J. Bacteriol. 181:521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 10.Draper, G. C., N. McLennan, K. Begg, M. Masters, and W. D. Donachie. 1998. Only the N-terminal domain of FtsK functions in cell division. J. Bacteriol. 180:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghigo, J. M., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31:725-737. [DOI] [PubMed] [Google Scholar]
- 12.Hale, C. A., and P. A. J. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 13.Hale, C. A., and P. A. J. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ, and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haney, S. A., E. Glasfeld, C. Hale, D. Keeney, Z. He, and P. A. de Boer. 2001. Genetic analysis of the E. coli FtsZ-ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980-11987. [DOI] [PubMed] [Google Scholar]
- 15.Khattar, M. M., S. G. Addinall, K. H. Stedul, D. S. Boyle, J. Lutkenhaus, and W. D. Donachie. 1997. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J. Bacteriol. 179:784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, Z., A. Mukherjee, and J. Lutkenhaus. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 31:1853-1861. [DOI] [PubMed] [Google Scholar]
- 17.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 19.Mercer, K. L. N., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosyak, L., Y. Zhang, E. Glasfeld, S. Haney, M. Stahl, J. Seehra, and W. S. Somers. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogliano, J., K. Pogliano, D. S. Weiss, R. Losick, and J. Beckwith. 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. USA 94:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raskin, D. M., and P. A. J. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothfield, L., S. Justice, and J. García-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 24.Wang, L., M. K. Khattar, W. D. Donachie, and J. Lutkenhaus. 1998. FtsI and FtsW are localized to the septum in Escherichia coli. J. Bacteriol. 180:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, L., and J. Lutkenhaus. 1998. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol. 29:731-740. [DOI] [PubMed] [Google Scholar]
- 26.Wang, X., J. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, X.-C., A. H. Tran, Q. Sun, and W. Margolin. 1998. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J. Bacteriol. 180:1296-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]