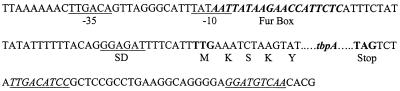Abstract
tbpA, fur, and fldA homologs from two strains (9L and 3384Y) of the sheep pathogen Histophilus ovis were sequenced. The predicted TbpA proteins of these strains are homologs of the Pasteurella multocida TbpA protein and collectively represent the second example of a new subfamily of TonB-dependent receptors. tbpA transcripts were readily detected by reverse transcription (RT)-PCR with RNA isolated from strain 9L grown under iron-restricted conditions in the presence or absence of bovine transferrin (Tf). However, with strain 3384Y and depending on the primer pair, tbpA transcripts were detected by RT-PCR predominantly when the RNA was from cells grown under iron-restricted conditions in the presence of bovine Tf. In both strains, the fldA homolog was found to be immediately upstream of fur and, based on RT-PCR, these genes are transcribed as a single unit; the availability of iron and the presence or absence of bovine Tf in the growth medium had no apparent effect on the relative amounts of the fldA-fur transcripts.
Histophilus ovis, a member of the family Pasteurellaceae, is a sheep pathogen capable of causing a variety of disease syndromes (30), including thrombotic meningoencephalitis (1). The extracellular environment encountered by invading pathogens is extremely iron limiting (10), and in order to survive and cause disease within a host, iron-requiring pathogens must possess mechanisms for the acquisition of iron from host components, such as transferrin (Tf). We demonstrated previously that five strains of H. ovis (9L, 642A, 714, 5688T, and 3384Y) are capable of binding and removing iron from, specifically, ovine, bovine, and goat, but not human or porcine, Tfs (4). Affinity isolation procedures allowed the isolation and identification of a major Tf-binding polypeptide of 78 kDa and a minor one of 66 kDa from all five strains. Such Tf-binding polypeptides could be isolated from total membranes derived from two strains (9L and 642A) grown under iron-restricted conditions alone, whereas Tf-binding polypeptides could be isolated from total membranes derived from the other three strains (714, 5688T, and 3384Y) only if the organisms were grown under iron-restricted conditions in the presence of Tf (4). The majority of the Tf-binding proteins (Tbp proteins) characterized to date are expressed when the respective organisms are grown under conditions of iron restriction (see, e.g., references 20, 21, 23, 25, and 27); such regulation is presumably due to the activities of ferric uptake regulator (Fur) proteins, which, in the presence of iron, would repress the transcription of tbp genes (9). The objectives of the present study were to identify and sequence the tbp genes, including the upstream sequences, from two representative strains of H. ovis to determine their relationship to other characterized tbp genes and to investigate the possibility that the upstream sequences might provide some clue to the mechanism of Tf-dependent regulation of TbpA expression. Furthermore, since it was apparent that iron also plays a role in the expression of Tbp proteins, we sought to determine if H. ovis possesses a fur homolog.
Identification and sequencing of a tbpA homolog
The N-terminal amino acid sequence of the affinity-isolated 78-kDa Tf-binding polypeptide from strain 9L (DSNPATTVPN) was determined previously, but the identities of only the first three to five amino acids were considered to be definitive (see reference 4). Degenerate oligonucleotide primers based on this amino acid sequence and on sequences conserved in a variety of TbpA proteins, and also primers 223 and 224 of Ogunnariwo and Schryvers (22), were used in PCRs with strain 9L DNA, but amplification products of the appropriate size and/or sequence were not obtained. Based on this experience, we found the recent publication by Ogunnariwo and Schryvers (23), describing a novel TbpA protein from Pasteurella multocida, extremely interesting in that they reported having similar difficulties amplifying fragments of P. multocida tbpA by using a rapid-PCR-based approach (22). The authors speculated that the failure to amplify appropriately sized fragments may be due to a lack of signature sequences in the P. multocida tbp genes. Alignment of the deduced amino acid sequences of the TbpA proteins from P. multocida (accession no. AY007725), Pasteurella trehalosi (accession no. AF312919), and Mannheimia (Pasteurella) haemolytica (accession no. U73302) with ClustalW (Biology Workbench; http://workbench.sdsc.edu/) revealed at least three regions with homologous amino acids. The nucleotide sequence of P. multocida tbpA, corresponding to conserved regions AIRGVDK and SKTGYTSKN, was used to design forward (PmF1; 5′ GCTATCCGTGGCGTTGATAAA) and reverse (PmR2; 5′ GTTTTTTGAAGTATAGCCGGTTTTAGA) primers, respectively, and with these primers, DNAs from the five strains of H. ovis and the Expand High Fidelity PCR System (Roche Diagnostics, Laval, Quebec, Canada), an appropriately sized (∼300-bp) fragment was amplified from each strain. These amplification products were ligated into pGEM-T Easy (Promega, Madison, Wis.) and transformed into Escherichia coli DH5α. Plasmids harboring the correct insert, as verified by colony PCR, were isolated from transformants, and such inserts were sequenced with pUC sequencing primers and a BigDye sequencing kit (PE Biosystems, Foster City, Calif.). The ∼300-bp products obtained with the DNAs from all five strains were very similar (results not shown), and a BlastX search revealed that all were homologous to the TbpA of P. multocida. The single-stranded sequence of tbpA from strain 9L was determined by sequencing products generated by inverse PCR (19) and/or by direct genomic sequencing (13). Primers based on this single-stranded sequence were used to amplify tbpA, including upstream and downstream regions, from strains 9L and 3384Y with the Expand High Fidelity PCR System (Roche Diagnostics). The resulting PCR products, purified with the QIAquick PCR Purification Kit (Qiagen, Mississauga, Ontario, Canada), were used directly as templates for the sequencing of both strands with primers based on the single-stranded 9L tbpA sequence.
The deduced amino acid sequences of the mature TbpA proteins from strains 9L (733 amino acids) and 3384Y (734 amino acids) indicate molecular masses of approximately 83 kDa, and the predicted proteins share 97% identity based on BlastP analyses. On the basis of web-based SignalP V2.0 (http://www.cbs.dtu.dk/; 18), the cleavage site, resulting in mature TbpA proteins from both strains, was predicted to be between residues 26 and 27 (ALSLA↓DSNPA). This prediction is in keeping with the results of the N-terminal sequence analysis since the first five amino acids of the affinity-isolated, 78-kDa Tf-binding polypeptide are identical to those following the predicted cleavage site. Furthermore, this confirms that the 78-kDa Tf-binding polypeptide isolated from strain 9L (and, most probably, 3384Y) is TbpA. The first two BlastP results revealed that the H. ovis TbpA proteins share 72 to 73% identity with the TbpA of P. multocida and 31% identity with a hemoglobin receptor of Neisseria meningitidis. As was the case with the TbpA of P. multocida (23), the TbpA proteins of H. ovis share limited identity with classical TbpA proteins of other, comparable, organisms and such findings suggest that the H. ovis TbpA proteins represent, collectively, the second example of a new subfamily (23) of TonB-dependent receptors.
Based on the requirements for the expression of the TbpA proteins of strains 9L and 3384Y, we anticipated that there would be a difference between the promoter regions of the two genes; however, they were found to be identical (Fig. 1). Putative −10 and −35 sequences are identical to consensus σ70 RNA polymerase binding sites (see, e.g., reference 31) and a potential Fur box (13 of 19 bases match the E. coli consensus; 5) overlaps the −10 region. Following the ribosomal binding site is the uncommon start codon TTG, which, in the E. coli genome, constitutes only 1.1% of the start codons (11). Downstream of tbpA are two inverted repeats (Fig. 1) that may be involved in transcriptional termination and on the opposite strand, there is a partial orf encoding the carboxy-terminal end of a diacylglycerol kinase-like protein (see GenBank accession numbers). BlastX searches with the sequence upstream of both tbpA genes, including an additional 650 bases of single-stranded sequence (strain 9L; unpublished data), failed to reveal any orf with significant homology to any protein in the database.
FIG. 1.
Genetic organization of the tbpA genes in strains 9L and 3384Y. Putative −35, −10, and Shine-Dalgarno (SD) sequences are underlined, a potential Fur-binding site is in bold italics, start and stop codons are in bold, and inverted repeats, possibly involved in transcriptional termination, are in underlined italics.
To date, all of the organisms that acquire iron from Tf by a contact-dependent mechanism, with the probable exception of P. multocida (23), employ a two-receptor system composed of the proteins TbpA and TbpB (see, e.g., reference 9). In most organisms, the genes encoding the Tbp proteins are arranged in an operon, with tbpB preceding tbpA, and the promoter controlling the expression of both genes is located upstream of tbpB (see, e.g., references 7, 8, 14, and 24). Although a tbpB homolog did not appear to be present upstream of H. ovis tbpA, the genetic arrangement tbpBA is not conserved in all organisms. For example, tbpA of Moraxella catarrhalis is upstream of tbpB; these genes are separated by an unknown orf, and each of the three genes appears to have its own promoter (17). Notably, recombinant M. catarrhalis TbpB is able to bind Tf after sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transfer to polyvinylidene difluoride (17), a defining characteristic of the TbpB proteins described to date (see, e.g., references 6 and 26,27–28). Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis, however, and transfer to nitrocellulose, the 78- and 66-kDa Tf-binding polypeptides from strains 9L and 3384Y failed to bind bovine Tf (results not shown). Although it is tempting, therefore, to conclude that H. ovis does not possess a TbpB or tbpB homolog, we cannot preclude its existence absolutely; in effect, the significance of the 66-kDa Tf-binding polypeptide remains obscure.
Sequencing of fur and fldA homologs
The availability of iron influences the expression of Tbp proteins in H. ovis (4), suggesting that H. ovis may possess a Fur homolog. Fur, with its corepressor, iron, is able to repress the transcription of iron-regulated genes (12). The conserved amino acid sequences VGLKITEPR and HHDHIICEDC in the Haemophilus influenzae Fur protein (accession no. U32704), as identified by Daniel et al. (3), were used to design degenerate forward (Fur3; 5′ GTIGGIYTIAARATIACIGARCCIMG) and reverse (Fur5; 5′ RCARTCYTCRCARATRATRTGRTCRTGRTG) primers, respectively. With these primers and DNA isolated from strain 9L, PCR yielded a product with an anticipated size of ∼250 bp. This product was cloned and sequenced as described above, and BlastX searches revealed that it encoded a Fur homolog. A strategy similar to that used for the sequencing of tbpA was employed to determine the single-stranded sequence of fur from strain 9L and, subsequently, the complete double-stranded sequence of fur from strains 9L and 3384Y. During initial sequencing, we noted that immediately (16 bp) upstream of fur, there was an orf (fldA) encoding a flavodoxin homolog. It has been suggested that flavodoxin maintains the reduced state of enzymatic Fe-S clusters and is involved in defense against oxidative stress (32). Since the intergenic region lacked an obvious promoter sequence, fldA and the 190 bp upstream of fldA were sequenced to determine if either of these regions contains a promoter. The genetic arrangement, fldA-fur, has been observed in many organisms (see e.g., 32), including P. multocida (15). A well-documented association exists between oxidative stress defenses and the regulation of iron uptake (for a review, see reference 29), suggesting that the genetic organization, fldA-fur, reflects a need for their coordinated expression (32).
RT-PCR of tbpA and fldA-fur transcripts
H. ovis strains 9L and 3384Y were grown in 200 ml of sTYE-H as described previously (4). When the optical density was ∼0.05, 25-ml aliquots were removed quickly to sterile 125-ml screw cap Nalgene flasks, ethylenediamine di(o-hydroxyphenylacetic acid) (to 50 μM for iron-restricted conditions) and bovine Tf (2 mg) were added as appropriate and the flasks were returned to the incubator. Growth was monitored turbidimetrically at 660 nm. The organisms were harvested in exponential phase (optical density, ∼0.3 to 0.75, depending on the strain and growth conditions), washed immediately with phosphate-buffered saline, and resuspended in RNAlater RNA Stabilization Reagent as described by the manufacturer (Qiagen). RNA was isolated with the RNeasy Mini Kit (Qiagen), and contaminating DNA was removed by two consecutive on-column treatments with RNase-free DNase (Qiagen). RNA concentrations were determined spectrophotometrically (260 nm; Lambda 3B UV/VIS spectrophotometer; Perkin-Elmer). RT-PCR was performed with the OneStep RT-PCR Kit (Qiagen) as recommended by the manufacturer, with 1 ng of the appropriate RNA sample (or an aliquot of DNA [positive control]) and primers as indicated in Fig. 2. Negative controls, in which RNA isolated from cells grown under all conditions served as the template for PCRs with the different primer combinations (as described in Fig. 2), did not yield any detectable amplification products (results not shown), confirming that the RNA samples were free of contaminating DNA. Duplicate RT-PCR experiments, with newly isolated RNA, yielded comparable results.
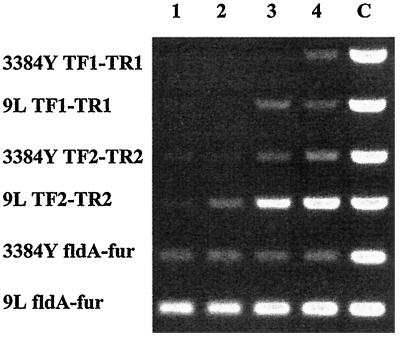
FIG. 2.
RT-PCR with RNA isolated from cells grown under iron-replete conditions (lane 1), under iron-replete conditions with bTf (lane 2), under iron-restricted conditions (lane 3), under iron-restricted conditions with bTf (lane 4), or with total DNA (lane C). The strain from which the RNA (or DNA) was isolated and the primer pairs (described in Table 1) used are indicated on the left.
The genetic organization of fldA-fur suggested that these genes may be cotranscribed, and in keeping with this suggestion, RT-PCR with a forward primer (fldA) inside fldA and a reverse primer (fur) inside fur yielded a product of the appropriate size (Fig. 2). With a forward primer based on a single-stranded DNA sequence (strain 9L; unpublished data) that is farther upstream of fldA than the sequence submitted to the GenBank database, in combination with the reverse fur primer, also resulted in an appropriately sized amplification product (results not shown), suggesting that the promoter controlling the expression of these two genes is located upstream of fldA and beyond the sequence submitted. Although it was not our intention to determine if environmental conditions affect the transcription of fldA-fur, it was noted that the iron content of the medium and the presence or absence of Tf had no effect on the relative amount of the fldA-fur transcript. Since the amount of the fldA-fur transcript, as detected by RT-PCR, remained essentially the same for each strain, regardless of the growth conditions, these transcripts could be used as controls to ensure that equivalent amounts of RNA, from organisms grown under the different conditions, were used in the RT-PCR experiments. While the TF1-TR1 primer pair used in the RT-PCR with RNA isolated from strain 9L, grown under iron-restricted conditions in the presence or absence of bovine Tf, allowed the identification of tbpA transcripts, such transcripts were detected with RNA isolated from strain 3384Y only when the organisms were grown under conditions of iron restriction in the presence of Tf (Fig. 2). Similarly, while TbpA was expressed by cells of strain 9L when grown under conditions of iron restriction in the presence or absence of bovine Tf, TbpA was expressed by cells of strain 3384Y only if the organisms were grown under conditions of iron restriction in the presence of bovine Tf (4). However, when the TF2-TR2 primer pair was used, RT-PCR allowed the identification of tbpA transcripts with RNAs isolated from strains 9L and 3384Y grown under conditions of iron restriction in the presence or absence of bovine Tf. Although RNA isolated from strain 3384Y grown under conditions of iron restriction allowed the identification of tbpA transcripts, cells of 3384Y, which served as the RNA source, exhibited Tf-binding activity in a solid-phase binding assay (as previously described [4]) only if they were grown under iron-restricted conditions in the presence of bovine Tf (results not shown).
The promoter regions of tbpA in strains 9L and 3384Y are essentially identical, suggesting that despite the different genetic backgrounds, initiation of transcription should take place under similar environmental conditions. While the involvement of Fur remains to be confirmed experimentally, the putative Fur boxes in the promoter regions upstream of tbpA and the identification of fur homologs in both strains help to explain the requirement for iron-restricted conditions for the transcription of tbpA. The environmental conditions required for TbpA expression in strain 9L are the same as those that resulted in the detection of increased amounts of tbpA transcripts by RT-PCR; on the other hand, with strain 3384Y, tbpA transcripts were readily detected by RT-PCR even when the growth conditions were such that TbpA expression was not detected (i.e., iron-restricted conditions). It appears, therefore, that the expression of TbpA in strain 9L is regulated at the level of transcription, as is the case with other, comparable, organisms (9). The regulatory mechanisms controlling the expression of 3384Y TbpA, however, appear to be more complex since appreciable tbpA transcript amounts were detected (with primer pair TF2-TR2; Fig. 2) in RNA samples isolated from cells grown under iron-restricted conditions, conditions under which Tf-binding activity could not be detected. This suggests that regulation of TbpA expression in strain 3384Y may occur, to some extent, at the posttranscriptional level. One mechanism for posttranscriptional regulation involves the production of antisense RNA which is complementary to the mRNA from the gene it regulates; antisense RNA may act by inhibiting translation or possibly by destabilizing the mRNA (2, 16). Further studies are needed to fully characterize the mechanism of Tf-dependent regulation of TbpA expression.
Nucleotide sequence accession numbers. The nucleotide sequences of tbpA from strains 9L and 3384Y were submitted to the GenBank database and assigned accession numbers AY040784 and AY040785, respectively. The sequences of fldA-fur from strains 9L and 3384Y were submitted to the GenBank database and assigned accession numbers AF386645 and AF386646, respectively.
TABLE 1.
Primers used in RT-PCRs
| Primer | Description (coordinates) | Sequence (5′ → 3′) |
|---|---|---|
| TF1 | tbpA forward (432-453)a | CCGTTGTGGCGAATGTTGAGCC |
| TR1 | tbpA reverse (1025-1004) | GCGGATTAGGTTTACCGCGATG |
| TF2 | tbpA forward (2065-2086) | AGTCAGCGGTGCTGTTAATGGC |
| TR2 | tbpA reverse (2556-2532) | GCGGCAAAATTACGTTTCGGTGCAG |
| FldA | fldA forward (587-609) | GTTGATGACAATACATTCGTAGG |
| Fur | fur reverse (930-907) | CCTCATCAAATTGGTTAAGTACAC |
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC). A.E. was the grateful recipient of postgraduate scholarships from NSERC and the Fonds pour la Formation de Chercheurs et l'Aide à la Recherche.
We thank Nadia Surdek and Sofia Fuga of the Applied Biotechnology Laboratory, McGill University (Macdonald Campus), for their time and patience.
REFERENCES
- 1.Cassidy, J. P., S. W. J. McDowell, G. A. C. Reilly, W. J. McConnell, F. Forster, and D. Lawler. 1997. Thrombotic meningoencephalitis associated with Histophilus ovis infection in lambs in Europe. Vet. Rec. 140:193-195. [DOI] [PubMed] [Google Scholar]
- 2.Chen, Q., and J. H. Crosa. 1996. Antisense RNA, Fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J. Biol. Chem. 271:18885-18891. [DOI] [PubMed] [Google Scholar]
- 3.Daniel, C., S. Haentjens, M.-C. Bissinger, and R. J. Courcol. 1999. Characterization of the Acinetobacter baumannii Fur regulator: cloning and sequencing of the fur homolog gene. FEMS Microbiol. Lett. 170:199-209. [DOI] [PubMed] [Google Scholar]
- 4.Ekins, A., and D. F. Niven. 2001. Production of transferrin receptors by Histophilus ovis: three of five strains require two signals. Can. J. Microbiol. 47:417-423. [DOI] [PubMed] [Google Scholar]
- 5.Escolar, L., J. Pérez-Martín, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez, G. C., D. L. Caamano, and A. B. Schryvers. 1990. Identification and characterization of a porcine-specific transferrin receptor in Actinobacillus pleuropneumoniae. Mol. Microbiol. 4:1173-1179. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez, G. C., R.-H. Yu, P. R. Rostek, Jr., and A. B. Schryvers. 1995. Sequence, genetic analysis, and expression of Actinobacillus pleuropneumoniae transferrin receptor genes. Microbiology 141:2405-2416. [DOI] [PubMed] [Google Scholar]
- 8.Gray-Owen, S. D., S. Loosmore, and A. B. Schryvers. 1995. Identification and characterization of genes encoding the human transferrin-binding proteins from Haemophilus influenzae. Infect. Immun. 63:1201-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4:185-191. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths, E. 1987. The iron-uptake systems of pathogenic bacteria, p. 69-137. In J. J. Bullen and E. Griffiths (ed.), Iron and infection. Molecular, physiological and clinical aspects. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 11.Hannenhalli, S. S., W. S. Hayes, A. G. Hatzigeorgiou, and J. W. Ficket. 1999. Bacterial start site prediction. Nucleic Acids Res. 27:3577-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 13.Heiner, C. R., K. L. Hunkapiller, S.-M. Chen, J. I. Glass, and E. Y. Chen. 1998. Sequencing multimegabase-template DNA with BigDye terminator chemistry. Genome Res. 8:557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legrain, M., V. Mazarin, S. W. Irwin, B. Bouchon, M.-J. Quetin-Millet, E. Jacobs, and A. B. Schryvers. 1993. Cloning and characterization of Neisseria meningitidis genes encoding the transferrin-binding proteins Tbp1 and Tbp2. Gene 130:73-80. [DOI] [PubMed] [Google Scholar]
- 15.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuno, T., M.-Y. Chou, and M. Inouye. 1984. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. USA 81:1966-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers, L. E., Y.-P. Yang, R.-P. Du, Q. Wang, R. E. Harkness, A. B. Schryvers, M. H. Klein, and S. M. Loosmore. 1998. The transferrin binding protein B of Moraxella catarrhalis elicits bactericidal antibodies and is a potential vaccine antigen. Infect. Immun. 66:4183-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielson, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 19.Ochman, H., A. S. Gerber, and D. L. Hart. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogunnariwo, J. A., C. Cheng, J. Ford, and A. B. Schryvers. 1990. Response of Haemophilus somnus to iron limitation: expression and identification of a bovine-specific transferrin receptor. Microb. Pathog. 9:397-406. [DOI] [PubMed] [Google Scholar]
- 21.Ogunnariwo, J. A., and A. B. Schryvers. 1990. Iron acquisition in Pasteurella haemolytica: expression and identification of a bovine-specific transferrin receptor. Infect. Immun. 58:2091-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogunnariwo, J. A., and A. B. Schryvers. 1996. Rapid identification and cloning of bacterial transferrin and lactoferrin receptor genes. J. Bacteriol. 178:7326-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogunnariwo, J. A., and A. B. Schryvers. 2001. Characterization of a novel transferrin receptor in bovine strains of Pasteurella multocida. J. Bacteriol. 183:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogunnariwo, J. A., T. K. W. Woo, R. Y. C. Lo, G. C. Gonzalez, and A. B. Schryvers. 1997. Characterization of the Pasteurella haemolytica transferrin receptor genes and the recombinant receptor proteins. Microb. Pathog. 23:273-284. [DOI] [PubMed] [Google Scholar]
- 25.Ricard, M. A., F. S. Archibald, and D. F. Niven. 1991. Isolation and identification of a putative porcine transferrin receptor from Actinobacillus pleuropneumoniae biotype 1. J. Gen. Microbiol. 137:2733-2740. [DOI] [PubMed] [Google Scholar]
- 26.Schryvers, A. B. 1989. Identification of the transferrin- and lactoferrin-binding proteins in Haemophilus influenzae. J. Med. Microbiol. 29:121-130. [DOI] [PubMed] [Google Scholar]
- 27.Schryvers, A. B., and L. J. Morris. 1988. Identification and characterization of the transferrin receptor from Neisseria meningitidis. Mol. Microbiol. 2:281-288. [DOI] [PubMed] [Google Scholar]
- 28.Stevenson, P., P. Williams, and E. Griffiths. 1992. Common antigenic domains in transferrin-binding protein 2 of Neisseria meningitidis, Neisseria gonorrhoeae, and Haemophilus influenzae type b. Infect. Immun. 60:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 30.Webb, R. F. 1983. Clinical findings and pathological changes in Histophilus ovis infection in sheep. Res. Vet. Sci. 35:30-34. [PubMed] [Google Scholar]
- 31.Wösten, M. M. S. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 32.Zheng, M., B. Doan, T. D. Schneider, and G. Storz. 1999. OxyR and SoxRS regulation of fur. J. Bacteriol. 181:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]