Abstract
Pseudomonas aeruginosa regulates the production of many exoproteins and secondary metabolites via a hierarchical quorum-sensing cascade through LasR and RhlR and their cognate signal molecules N-(3-oxododecanoyl)-l-homoserine lactone (3O-C12-HSL) and N-(butanoyl)-l-homoserine lactone (C4-HSL). In this study, we found that transcription of the quorum sensing-regulated genes lecA (coding for PA-IL lectin), lasB (coding for elastase), and rpoS appeared to be growth phase dependent and their expression could not be advanced to the logarithmic phase in cells growing in batch culture by the addition of exogenous C4-HSL and 3O-C12-HSL. To identify novel regulators responsible for this growth phase dependency, a P. aeruginosa lecA::lux reporter strain was subjected to random transposon mutagenesis. A number of mutants affected in lecA expression were found that exhibited altered production of multiple quorum sensing-dependent phenotypes. While some mutations were mapped to new loci such as clpA and mvaT and a putative efflux system, a number of mutations were also mapped to known regulators such as lasR, rhlR, and rpoS. MvaT was identified as a novel global regulator of virulence gene expression, as a mutation in mvaT resulted in enhanced lecA expression and pyocyanin production. This mutant also showed altered swarming ability and production of the LasB and LasA proteases, 3O-C12-HSL, and C4-HSL. Furthermore, addition of exogenous 3O-C12-HSL and C4-HSL to the mvaT mutant significantly advanced lecA expression, suggesting that MvaT is involved in the growth phase-dependent regulation of the lecA gene.
Many bacterial species employ complex communication mechanisms linking cell density with gene expression. Diffusible signal molecules termed autoinducers accumulate in the extracellular environment during the growth of a bacterial population, thus reflecting its cell density. Once a critical threshold concentration has been reached, a response is triggered that leads to changes in gene expression and consequently the phenotype of the cells. This type of communication, which regulates many diverse physiological processes, has been termed quorum sensing (13). In gram-negative bacteria, the most intensely studied quorum-sensing systems rely upon the interaction of N-acylhomoserine lactone (AHL) signal molecules, synthesized by LuxI-type AHL synthases, with LuxR-type transcriptional regulator proteins. Together, the LuxR-type protein and its cognate AHL then activate the expression of specific target genes (for reviews, see references 52 and 59).
Pseudomonas aeruginosa, an opportunistic human pathogen, is known to possess at least two AHL-dependent quorum-sensing systems, the las and rhl systems, which are composed of the LuxRI homologues LasRI (14, 32) and RhlRI (23, 30), respectively. LasI directs the synthesis of N-(3-oxododecanoyl)-l-homoserine lactone (3O-C12-HSL) (33), whereas RhlI directs the synthesis of N-(butanoyl)-l-homoserine lactone (C4-HSL) (34, 60). Each system modulates a regulon comprising an overlapping set of genes. However, the las and rhl systems are not independent of each other but form a regulatory hierarchy in which LasR/3O-C12-HSL activates the transcription of rhlR (24, 36). Recently, a third LuxR homologue, termed QscR, has been identified that has been shown to regulate the transcription of lasI (6). Furthermore, an additional signaling molecule has been described that was shown to control the expression of lasB. This molecule was chemically characterized as 2-heptyl-3-hydroxy-4-quinolone, a compound related to the 4-quinolone antibiotics. This was termed the Pseudomonas quinolone signal (PQS), the production of which was shown to be dependent on LasR/3O-C12-HSL (37). In addition to lasB, the PQS was also shown to regulate rhlI expression (25). PQS concentrations were found to be highest in late stationary phase, suggesting that this molecule is not involved in cell density sensing (25). Genes regulated by either LasR/3O-C12-HSL or RhlR/C4-HSL include those coding for elastase (LasB), LasA protease, alkaline protease, exotoxin A, cytotoxic lectins, hydrogen cyanide, and pyocyanin. As well as controlling the expression of many genes for exoproteins and secondary metabolites, the quorum-sensing machinery in P. aeruginosa is known to influence the xcp secretion pathway (4), biofilm maturation (7, 9), catalase gene expression, twitching motility (16), and expression of the stationary-phase sigma factor rpoS (24). Indeed, Whiteley et al. (55) have estimated that 1 to 4% of all P. aeruginosa genes may be controlled, to some extent, by quorum sensing.
Although a few regulatory systems have been described that influence las- or rhl-mediated quorum sensing, no systematic approach has been undertaken to elucidate how the quorum-sensing systems of P. aeruginosa are integrated into the global regulatory network of the cell. A mutant defective in the response regulator gacA was shown to exhibit reduced and delayed formation of C4-HSL and also reduced expression of lasR (40). Furthermore, a CRP homologue termed Vfr was shown to be required for basal-level lasR expression (1). In addition, a regulator termed RsaL has been described that is thought to repress transcription of lasI (8). Whiteley et al. (56) have shown that the stationary-phase sigma factor RpoS negatively regulates C4-HSL production and that, in an rpoS mutant, expression of rhlI, hcnA, and phzA is advanced. Also, a deletion of qscR in P. aeruginosa resulted in premature expression of the quorum sensing-related genes lasI, rhlI, hcnA, and phzA (6). Furthermore, a deletion in the posttranscriptional regulator RsmA led to advanced expression of lasI, rhlI, and hcnA (37a).
We have recently demonstrated that expression of the RhlR/C4-HSL-dependent PA-IL lectin gene lecA cannot not be advanced to the logarithmic growth phase by the addition of either 3O-C12-HSL or C4-HSL and that lecA is expressed in a growth phase-dependent manner (62). This is intriguing because, for instance, in Vibrio fischeri and Erwinia carotovora subsp. carotovora, the quorum sensing-controlled phenotypes of bioluminescence (10, 28) and carbapenem production (58), respectively, could be induced prematurely by the addition of their cognate AHL signal molecules.
In order to define in more detail the observed superregulation of quorum sensing-dependent genes in P. aeruginosa, we isolated transposon mutants that exhibited either increased, decreased, or delayed expression of a quorum sensing-dependent lecA::lux fusion. Further analysis of these mutants revealed that multiple quorum sensing-dependent phenotypes were affected. We describe here the identification of a new regulator involved in the modulation of cell density-dependent gene expression and propose that quorum-sensing regulation of virulence gene expression is linked with the growth phase and metabolic state of the cell.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli JM109 was used for cloning experiments, and E. coli S17-1 λpir was used for conjugation experiments. Bacteria were grown at 37°C in Luria-Bertani (LB) broth or on LB agar plates or Pseudomonas Isolation Agar (Difco). Where indicated, C4-HSL or 3O-C12-HSL (or a combination of the two) was added to the growth medium prior to inoculation at a concentration of either 10 or 100 μM. Standard methods were used for the preparation of competent cells and for plasmid electroporation into E. coli and P. aeruginosa (42, 48). Conjugal transfer was performed as described by Kaniga et al. (18). Where required, tetracycline, chloramphenicol, and ampicillin were added at 40, 34, and 50 μg/ml, respectively, for E. coli. For P. aeruginosa, kanamycin, tetracycline, carbenicillin, and chloramphenicol were added at 50, 40, 300, and 400 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type | Holloway collection |
| PAO1 lecA::lux | lecA::luxCDABE genomic reporter fusion in PAO1 | 62 |
| PAO-P4 | Tn5-B21 lasR mutant derived from PAO1 lecA::lux | This study |
| PAO-P8 | Tn5-B21 mutant derived from PAO1 lecA::lux | This study |
| PAO-P9 | Tn5-B21 rpoS mutant derived from PAO1 lecA::lux | This study |
| PAO-P10 | Tn5-B21 mvaT mutant derived from PAO1 lecA::lux | This study |
| PAO-P19 | Tn5-B21 mutant derived from PAO1 lecA::lux | This study |
| PAO-P34 | Tn5-B21 rhlR mutant derived from PAO1 lecA::lux | This study |
| PAO-P47 | mvaT chromosomal deletion mutant derived from PAO1 | This study |
| PAO-P52 | Tn5-B21 clpA mutant derived from PAO1 lecA::lux | This study |
| PAO-P69 | Tn5-B21 mutant derived from PAO1 lecA::lux | This study |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZΔM15] | 63 |
| S17-1 λpir | thi pro hsdR hsdM+recA RP4-2-Tc::Mu-Km::Tn7 λpir | 46 |
| Plasmids | ||
| pUC18 | Multicopy cloning vector derived from pBR322 carrying polylinker inserted into lacZα (Ampr) | 54 |
| pUCP18 | Same as pUC18 but containing an additional 1.8-kb stabilizing fragment for maintenance in Pseudomonas spp. (Ampr) | 44 |
| pDM4 | Suicide vector carrying the sacBR genes for sucrose sensitivity (Cmr) | 26 |
| pSP73 | Cloning vector | Promega Corp.: Madison, Wis. |
| pSUP102 Tn5-B21 | Transposable promoter probe; B21 in pSUP102 (Cmr Tetr) | 47 |
| pSB1075 | AHL reporter plasmid; P. aeruginosa lasRI and luxCDABE from Photorhabdus luminescens (Ampr) | 61 |
| pSB536 | AHL biosensor; ahyR"::luxCDABE in pAHP13 (Ampr) | 51 |
| pUCmvaT | 1.6-kb BamHI-PstI PAOI chromosomal DNA fragment containing mvaT in pUC18 | This study |
| pUCΔmvaT | Same as pUC::mvaT except contains a 327-bp mvaT deletion in pUC18 | This study |
| pDM4ΔmvaT | pDM4 containing mvaT flanking regions and deletion-containing mvaT gene | This study |
| pUCPmvaT | 1.6-kb PAO1 BamHI-PstI DNA insert containing mvaT gene in pUCP18 | This study |
| pMW47.1 | 2-kb PstI PAO1 DNA insert (rhlRl) in pUCP18 | 23 |
Ampr, Cmr, and Tetr stand for resistance to ampicillin, chloramphenicol, and tetracycline, respectively.
DNA manipulation.
DNA was manipulated by standard methods (42). Restriction enzymes (Promega United Kingdom Ltd.) were used in accordance with the manufacturer's instructions. Agarose gel electrophoresis and Southern blot transfer were done essentially as described by Sambrook et al. (42). DNA probes were labeled with digoxigenin and detected with the Alkaline Phosphatase Direct kit from Amersham. The IsoQuick kit (ORCA Research Inc.) was employed for isolation of chromosomal DNA from P. aeruginosa. For isolation of plasmid DNA from E. coli, the Qiagen Mini and Midi kits (Qiagen Ltd.) were used.
RNA analysis.
Total RNA of P. aeruginosa was isolated as described previously (62). For Northern blot assays, RNA was separated in denaturing formaldehyde gels and transferred to Hybond N+ nylon membranes (Amersham Pharmacia Biotech Ltd.) as described by Sambrook et al. (42). RNA dot blots were prepared with Hybond N+ nylon membranes in accordance with the manufacturer's instructions by using a dot blot apparatus from Bio-Rad. Probes were generated by PCR with chromosomal DNA from P. aeruginosa PAO1 and primers lecA1F (5′-ATATATCGGAGATCAATCATGGCTTGG-3′) and lecA1R (5′-CGTTCAGACCGAAGCGTGTTGAAGC-3′) (62) for lecA, lasB1F (5′-CGATCATGGGTGTTTCGCCG-3′) and lasB1R (5′-GCTCTTGCCGGGACCCTTGA-3′) for lasB, and rposF (5′-CACATCGACTACACGCGCGC-3′) and rposR (5′-GCTCGATGGTCTGGCGGATC-3′) for rpoS. For rhlR, a 0.5-kb rhlR-specific fragment was isolated from pMW47.1 (23) after restriction digestion with BamHI and BglII. Probes were labeled with [α-33P]ATP by using the Random Primers Labeling kit (Life Technologies Inc.). Hybridizations and washings were performed as described by Gerischer and Dürre (15). Transcript sizes were estimated by comparison with the 0.16- to 1.77-kb RNA ladder from Life Technologies Inc.
Transposon mutagenesis and cloning of Tn5-B21 insertion sites.
Equal volumes of overnight cultures of P. aeruginosa PAO1 lecA::lux (62) grown at 42°C and E. coli S17-1 λpir pSUP102 Tn5-B21 (47) grown at 37°C were mixed and incubated for 6 h at 30°C. Cells were then harvested by centrifugation, resuspended in 1 ml of LB broth, and pelleted at 19,000 × g for 5 min before being resuspended in 50 μl of LB broth. A 25-μl volume was then plated on LB agar containing 40 μg of tetracycline per ml and 50 μg of kanamycin per ml and incubated overnight at 37°C. Approximately 6,000 P. aeruginosa Tcr and Kmr colonies containing Tn5-B21 were restreaked onto selective antibiotic LB agar. Colonies were then screened for differences in light production throughout growth with a Luminograph LB 980 photon video camera (EG & G Berthold). A total of 38 mutants demonstrating increased, decreased, or no expression of the reporter fusion were selected and analyzed in greater detail with LUCYI (Anthos), a combined photometer-luminometer. Growth conditions and analysis of bioluminescence with LUCYI were as previously described (62). Tn 5-B21 insertion sites were cloned into pSP73 after digestion of genomic DNAs obtained from the mutants with XhoI. Recombinant clones were analyzed on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates (50 μg/ml) for a color change due to the presence of the transposon-encoded lacZ gene. Alternatively, EcoRI-digested DNA was cloned into the same vector and screened for Tcr, indicating the presence of the Tn5-B21-encoded gene for Tcr. DNA flanking the transposon was sequenced with the primer Tn5.seq (5′-ACGGGAAAGGTTCCGTTCAGGAC-3′). The DNA sequences obtained were analyzed with the National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/).
Construction of a PAO1 mvaT deletion mutant. A PAO1 mvaT chromosomal deletion mutant lacking 327 internal nucleotides was constructed as follows. With PAO1 DNA as the template, the intact mvaT gene (375 bp) plus 549 bp of upstream flanking DNA and 665 bp of downstream flanking DNA was amplified by using primers mvaTUF (containing a BamHI restriction site [underlined]; 5′-AATGGGGATCCTTGCTGGCCATCAGC-3′) and mvaTDR (containing a PstI restriction site [underlined]; 5′-TGCCGCTGCAGCTGAAGCAGATCCAGG-3′). The resulting PCR product was digested with BamHI and PstI and cloned into similarly digested pUC18, resulting in plasmid pUCmvaT (Table 1). To introduce a deletion of the recombinant mvaT gene, primers mvaTUR (5′-ATATTCGTTGATCAGGTGCACGTCAGGTACC-3′) and mvaTDF (5′-GAGAGCTGGGCCACCGTGCACGGCTAAACCA-3′) were used in conjunction with inverse PCR with pUC mvaT DNA as the template. The resulting blunt-ended PCR product, containing a 327-bp deletion in mvaT, was self-ligated, resulting in the plasmid pUCΔmvaT. The PCR product was excised from the vector with BamHI and SphI and cloned into the vector pDM4 (26), digested with BglII and SphI, resulting in the plasmid pDM4ΔmvaT. Allelic exchange using pDM4ΔmvaT contained in E. coli S17-1 λpir with PAO1 resulted in a P. aeruginosa strain (PAO-P47) containing an in-frame deletion of the mvaT gene. This deletion was confirmed by both PCR and Southern blot analysis (data not shown).
DNA sequencing and sequence analysis.
Automated nonradioactive sequencing reactions were carried out with the BigDye terminator cycle sequencing kit in conjunction with a 373A automated sequencer (Perkin-Elmer Applied Biosystems). For radioactive sequencing, the T7 sequencing kit from Amersham Pharmacia Biotech Ltd. was used. Sequence analysis and database searches were performed with the Genetics Computer Group (Madison, Wis.) software package and the National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/). For sequence comparisons, the programs Gap (complete protein sequences) and BestFit (for truncated protein sequences) were used.
Time- and cell density-dependent measurements of bioluminescence.
Bioluminescence was determined as a function of cell density with a combined, automated luminometer-spectrometer (Anthos Labtech LUCYI). Overnight cultures of P. aeruginosa were diluted 1:1,000 in fresh LB medium, and 0.2 ml was inoculated into microtiter plates. The luminescence and turbidity (optical density at 495 nm [OD495]) of the cultures were automatically determined every 30 min. Luminescence is given in relative light units (RLU) per unit of OD495.
Assay for elastolytic activity.
Elastolytic activity of bacterial supernatants was determined with the elastin Congo red (ECR; Sigma) assay (31). A 100-μl aliquot of bacterial supernatant was added to 900 μl of ECR buffer (100 mM Tris, 1 mM CaCl2, pH 7.5) containing 20 mg of ECR and incubated with shaking at 37°C for 3 h. Insoluble ECR was removed by centrifugation, and the absorption of the supernatant was measured at 495 nm. LB medium was used as a negative control.
Assay for pyocyanin production.
Pyocyanin was extracted from culture supernatants and measured by the method of Essar et al. (11). A 3-ml volume of chloroform was added to 5 ml of culture supernatant and mixed. The chloroform layer was transferred to a fresh tube and mixed with 1 ml of 0.2 M HCl. After centrifugation, the top layer (0.2 M HCl) was removed and its absorption at 520 nm was measured.
Staphylolytic activity assay.
LasA protease activity was measured by determining the ability of P. aeruginosa culture supernatants to lyse boiled Staphylococcus aureus cells (20). A 30-ml volume of an overnight culture of S. aureus was boiled for 10 min and then centrifuged for 10 min at 10,000 × g. The resulting pellet was resuspended in 10 mM Na2PO4 (pH 7.5) to an OD600 of approximately 0.8. A 100-μl aliquot of bacterial supernatant was then added to 900 μl of S. aureus suspension, and the OD600 was determined after 2, 6, 10, 14, 18, 22, 26, 30, 45, and 60 min.
AHL detection and analysis.
Aliquots of 900 μl of culture supernatant were taken at specific time intervals throughout growth. Bacterial cells were removed by centrifugation (19,000 × g, 5 min), and the resulting supernatant was filter sterilized through 0.2-μm-pore-size filters (Millipore). For accurate AHL quantification, culture supernatants were first acidified with 1 M HCl (100 μl) and then incubated with agitation at 20°C for 18 h. This was done to ensure that any AHLs hydrolyzed to the ring-open form during growth were recyclized. For detection of 3O-C12-HSL, 1 μl of acidified culture supernatant taken after 8 h of growth was spotted onto normal-phase thin-layer chromatography (TLC) plates (Silica gel 60F254; Merck). For detection of C4-HSL, 5 μl of acidified culture supernatant taken after 8 h growth was spotted onto reverse-phase TLC plates (RP-18 F245; Merck). TLC plates were overlaid with 50 ml of soft top agar containing either 1 ml of an E. coli strain harboring reporter plasmid pSB1075 (to detect 3O-C12-HSL) or pSB536 (to detect C4-HSL). Plates were incubated at 30°C for 18 h. Bioluminescence was detected and quantified with a Luminograph LB 980 photon video camera (EG & G Berthold). Each bacterial strain was tested in quadruplicate, and values were corrected for differences in cell density. Results are presented as percentages of those of the PAO1 lecA::lux parent strain.
SDS-PAGE and immunoblotting.
P. aeruginosa cells were grown in LB broth, and 1-ml samples were taken throughout the growth. All strains demonstrated similar growth patterns. Cells were suspended in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer and lysed by sonication. Samples were then boiled before being loaded onto SDS-15% polyacrylamide gels. Proteins were then electrophoretically transferred onto nitrocellulose membranes (Hybond C) and probed with an anti-PA-IL polyclonal antibody (1:200 dilution). Detection was achieved with a secondary anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (1:3,000 dilution) (Amersham Life Sciences) and developed with the ECL chemiluminescence system (Amersham Life Sciences).
Swarming motility assay.
In order to analyze swarming motility, an assay based on that of Rashid and Kornberg (39) was used. Briefly, a 2-μl aliquot of an overnight culture of P. aeruginosa was inoculated onto the surface of a swarm plate and incubated overnight at 37°C. Swarm plates consisted of 2 g of Bacto Agar (Difco) and 3.2 g of Nutrient Broth No. 2 (Oxoid) in 400 ml of distilled water. After autoclaving, filter-sterilized 10% (wt/vol) d-glucose in distilled water was added to a final concentration of 0.5% (wt/vol). The ability to swarm was assessed by the distance of swarming from the central inoculation site.
Detection of siderophores.
To detect siderophore production, 10 μl of an overnight culture was inoculated onto Chrome azurol S plates (45) and incubated for 18 h at 37°C. Siderophore production was detected by the appearance of orange halos around bacterial growth due to the release of free iron from an iron-dye complex within the plates.
Synthesis of N-acylhomoserine lactones.
C4-HSL and 3O-C12-HSL were synthesized as previously described by Chhabra et al. (5). AHLs were dissolved in acetonitrile before addition to culture media at the indicated concentrations.
RESULTS
Transcription of rhlR, rpoS, lecA, and lasB cannot be advanced by exogenous AHLs.
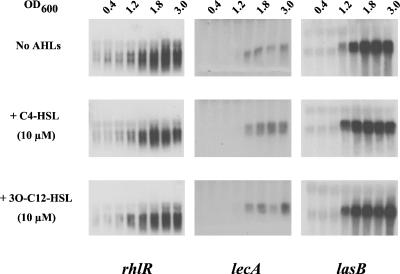
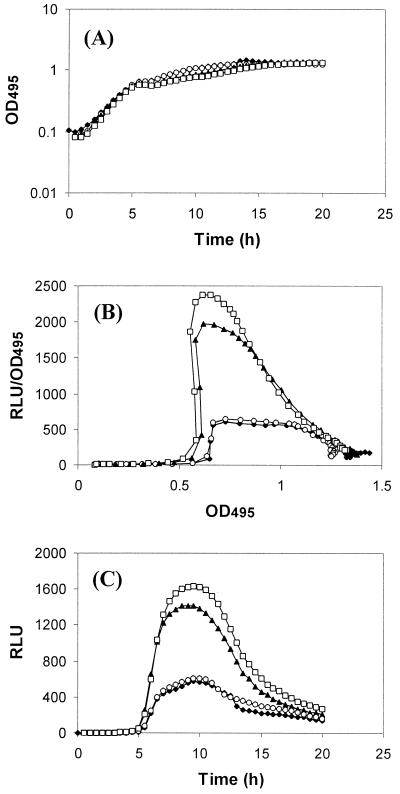
The RhlR/C4-HSL-dependent lecA::lux reporter fusion, which is expressed in a growth phase-dependent manner, could not be advanced by exogenous addition of either 3O-C12-HSL or C4-HSL (62). To test whether this is also the case for other quorum sensing-dependent genes or even components of the quorum-sensing cascade itself, the transcript levels of rhlR, rpoS, lecA, and lasB were analyzed in the presence of 10 μM C4-HSL or 3O-C12-HSL by Northern blotting (Fig. 1) and dot blot analysis (data not shown). Addition of these AHLs did not significantly advance the transcription of rhlR, lecA, lasB (Fig. 1), or rpoS (data not shown), indicating that a high autoinducer concentration, on its own, is insufficient to advance gene expression. The possibility remained that a high concentration of both the long- and short-chain AHLs is required for premature induction of these genes. This hypothesis was tested with the chromosomal lecA::lux fusion (62) and RNA dot blot analysis. Growth of P. aeruginosa in the presence of C4-HSL or a combination of C4-HSL and 3O-C12-HSL at 100 μM did not advance (or only very slightly advanced) expression of the lecA::lux fusion (Fig. 2) and lasB transcription (data not shown). Similar results were also obtained at lower concentrations (data not shown). However, in combination, both AHLs significantly elevated the expression of both genes. Thus, the quorum-sensing cascade and onset of quorum sensing-dependent gene expression appear to be tightly regulated.
FIG. 1.
Northern blot analysis of rhlR, lecA, and lasB in the presence and absence of exogenous AHLs. P. aeruginosa cells were grown in LB broth in the absence or presence of 10 μM C4-HSL or 3O-C12-HSL. RNA was isolated from cells harvested throughout growth and separated by horizontal electrophoresis as detailed in Materials and Methods. The lecA and lasB structural genes were amplified with primers lecA1F and lecA1R and primers lasB1F and lasB1R, respectively. The rhlR gene was derived from pMW47.1 (23). The resulting products were labeled with [α-33P]ATP and used as probes.
FIG. 2.
Expression of lecA::lux in P. aeruginosa. PAO1 lecA::lux was grown in LB medium in the absence of exogenously added AHL (⧫) or in the presence of C4-HSL (▴), 3O-C12-HSL (○), or a combination of the two (□) at 100 μM. RLU and OD495 were determined as described in Materials and Methods. The results represent a single experiment, although the experiment was repeated three times with similar results. Panels: A, growth of PAO1 lecA::lux in the presence or absence of AHLs; B, expression of lecA as a function of light output per cell; C, light output (lecA expression) of the culture with time.
Isolation of mutants displaying altered lecA::lux expression.
To identify new regulators that modulate quorum sensing-dependent gene expression, we subjected PAO1 lecA::lux to transposon mutagenesis. A collection of ∼6,000 mutants was generated by random insertion of the transposon Tn5-B21 (47). The resulting mutants were screened for altered bioluminescence, due to changes in lecA::lux expression, with a photon imaging camera. A total of 38 mutants were isolated that demonstrated increased, decreased, or delayed expression of the reporter fusion. To quantify the expression of the reporter fusion throughout growth, each of these mutants was grown in LB broth and analyzed in a combined photometer-luminometer (Anthos Labtech LUCYI). A number of mutants showing significant alterations in the expression of the reporter fusion were selected for further analysis (Table 2). Southern blot analysis showed that all of the mutants contained only a single copy of the transposon and that it had not been inserted into the lecA::lux reporter fusion (data not shown). Of the mutants selected, four showed little (less than 15% compared to the parent strain) or no expression of the fusion. In contrast, two mutants demonstrated a significant up-regulation (greater than 190% compared to the parent strain) of bioluminescence and, hence, lecA expression (Table 2). The transposon-flanking regions were cloned and sequenced, and the genes identified were compared with the genome databases (Table 2). The genes found to be inactivated included the known quorum-sensing regulators lasR (mutant PAO-P4) and rhlR (PAO-P34), as well as the gene for the stationary-phase sigma factor RpoS (mutant PAO-P9), but also a set of new loci that had not been described in the context of cell-cell communication or virulence gene expression.
TABLE 2.
Identification of genes in mutants carrying a Tn5-B21 insertiona
| Mutant | Cumulative index of lecA::lux expression (% of PAO1 lecA::lux expression) | Gene no. in Pseudomonas genome sequence | Tn5-B21 insertion site on PAO1 chromosome | Gene name or product | Reference |
|---|---|---|---|---|---|
| PAO-P4 | 4 | PA1430 | 1558663 | lasR | 14 |
| PAO-P8* | 38 | PA4341 | 4870418 | Putative regulator | |
| PAO-P9 | 4 | PA3622 | 4058513 | rpoS | 53 |
| PAO-P10 | 223 | PA4315 | 4844118 | mvaT | 41 |
| PAO-P19 | 47 | PA5265 | 5927833 | Hypothetical protein | |
| PAO-P34 | 4 | PA3477 | 3890556 | rhlR | 29 |
| PAO-P52 | 191 | PA2620 | 2963477 | clpA | 17 |
| PAO-P69 | 10 | PA4207 | 4707888 | Probable RND-like efflux transporter |
For each mutant, the assigned gene number in the P. aeruginosa complete genome sequence (49) is provided. Insertion sites were identified by comparing the DNA sequence flanking Tn5-B21 with the complete P. aeruginosa genome sequence. In PAO-P8*, Tn5-B21 was inserted just upstream of gene PA4341. The lecA::lux expression value for each mutant is shown as percentages of that of the parent PAOI lecA::lux strain. Each value was calculated by dividing each 30-min reading of RLU per unit of OD495 from the LUCYI experiment by that for the PAOI lecA::lux parent strain.
In mutant PAO-P10, Tn5-B21 had been inserted into a gene termed mvaT (49), as the putative gene product shows 82% similarity to the P16 subunit encoded by mvaT in the soil bacterium Pseudomonas mevalonii (41). In P. mevalonii, MvaT is a heteromeric transcriptional regulator of the mvaAB operon (41). Disruption of this gene in P. aeruginosa significantly increased (223%) expression of the lecA::lux reporter fusion (Table 2).
PAO-P8 showed a Tn5-B21 insertion site upstream of a gene (PA4341) with similarity to the IclR family of transcriptional regulators. Members of the IclR family have been shown to act as both repressors and activators of genes. In PAO-P8, expression of the lecA::lux reporter was greatly reduced (to 38% of that of PAO1 lecA::lux).
PAO-P52 showed an insertion within the putative clpA gene (PA2620). ClpA in E. coli can interact with ClpP to form a protease involved in the degradation of misfolded proteins (17), but it also acts as a chaperone.
In PAO-P69, the transposon had disrupted open reading frame (ORF) PA4207, coding for a putative efflux pump component. Expression of the lecA::lux fusion in this mutant was significantly down-regulated (10% of that in the parent strain).
The Tn5-B21 insertion site in the mutant PAO-P19 was shown to be in an ORF termed PA5265 (49). This ORF had no similarity to any known genes, but a mutation did affect lecA::lux expression, showing 47% of the expression in PAO1 lecA::lux.
Multiple quorum sensing-regulated phenotypes are altered in Tn5-B21 mutants.
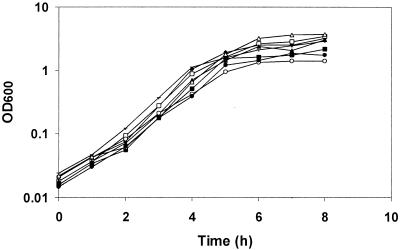
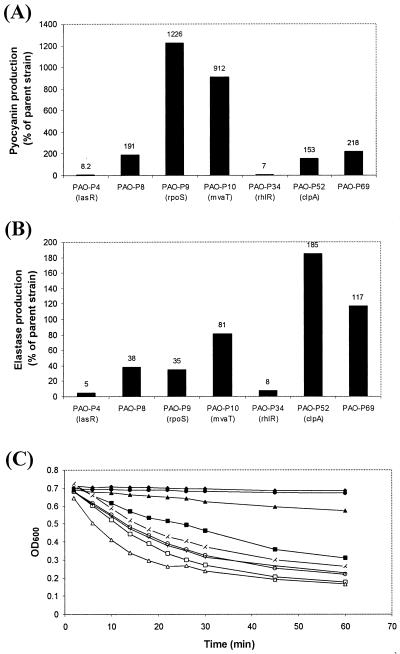
To determine whether any of the genes listed in Table 2 control lectin expression directly via quorum sensing rather than as a consequence of impaired growth, we first examined the growth of each Tn5-B21 mutant in LB. The mutants examined possessed similar rates of growth in LB (Fig. 3). This suggests that any phenotypic differences observed between each mutant and the parent strain could not be attributed to differences in growth. The production of pyocyanin, elastase, LasA protease, and siderophore, as well as swarming motility, was analyzed in a selection of the Tn5-B21 mutants. Production of the phenazine pigment pyocyanin 8 h postinoculation was shown to be altered in all of the mutants tested compared with that in PAO1 and PAO1 lecA::lux (Fig. 4A). As expected, very little pyocyanin was detected in PAO-P34 (rhlR mutant), a finding in keeping with that of previous workers (3). Both PAO-P9 (rpoS mutant) and PAO-P10 (mvaT mutant) showed very high levels of pyocyanin production (1,226 and 912% of the parent strain level, respectively). Interestingly, levels of pyocyanin in the lasR mutant (PAO-P4) increased to levels greater than those in PAO1 lecA::lux, but only after 14 h of growth. For all of the other mutants, the comparative pyocyanin levels, calculated as a percentage of the PAO1 lecA::lux reporter, after 14 h of growth were the same as those found after 8 h.
FIG. 3.
Growth curves of P. aeruginosa lecA::lux Tn5-B21 mutants. Strains were grown at 37°C for 8 h in 100 ml of LB medium with shaking at 200 rpm, and the OD600 was determined. Growth of PAO1 lecA::lux (⧫), PAO-P4 (□), PAO-P8 (▴), PAO-P9 (○), PAO-P10 (▪), PAO-P34 (▵), PAO-P52 (•), and PAO-P69 (+) is shown.
FIG. 4.
Influence of Tn5-B21 mutations on pyocyanin production (A), elastolytic activity (B), and LasA protease activity (C). Samples for pyocyanin analysis were taken after 8 h of growth, after which all strains were in stationary phase. Pyocyanin levels are expressed as percentages of that of the parent strain, PAO1 lecA::lux. For elastolytic and LasA protease activities, culture supernatants were analyzed after 13 h of growth. Elastase activity is shown as a percentage of that of the parent strain. LasA protease activity was determined by measuring the changes in OD600 caused by the lysis of S. aureus cells after 2, 6, 10, 14, 18, 22, 26, 30, 45, and 60 min. LB culture medium was used as a control. All of the experiments shown represent a single experiment, although each experiment was repeated at least three times with similar results. The results shown are for the control (⧫), PAO1 lecA::lux (□), PAO-P4 (▴), PAO-P8 (○), PAO-P9 (▪), PAO-P10 (X), PAO-P34 (•), PAO-P52 (▵), and PAO-P69 (+).
Elastase, a zinc metalloprotease encoded by lasB (2), is capable of degrading and inactivating a wide range of biological and immunological tissues. As with pyocyanin, elastase production was affected in all of the mutants tested. Figure 4B shows the results of ECR assays (31) of culture supernatants sampled after 13 h of growth. As expected, both the lasR (PAO-P4) and rhlR (PAO-P34) mutants produced very little elastase. PAO-P52 (clpA mutant) showed high levels of elastase production compared to the parent strain (185%), and in contrast, both PAO-P9 (rpoS mutant) and PAO-P10 (mvaT mutant) showed reduced elastase production compared with the parent strain (35 and 81%, respectively).
Analysis of LasA protease activity with a staphylolytic assay (20) showed LasA to be affected in all of the transposon mutants tested (Fig. 4C), with PAO-P4 (lasR mutant) and PAO-P34 (rhlR mutant) showing little or no activity. PAO-P9 (rpoS mutant) and PAO-P10 (mvaT mutant) both demonstrated reduced levels compared with those of PAO1 lecA::lux. Interestingly, PAO-P52 (clpA mutant) showed greater LasA activity than the PAO1 lecA::lux parent strain.
A selection of mutants was also analyzed for siderophore production with the Chrome azurol S assay. Both PAO-P4 (lasR mutant) and PAO-P10 (mvaT mutant) showed levels of siderophore activity similar to those of PAO1 lecA::lux, but activity was reduced in both PAO-P8 and PAO-P9 (rpoS mutant) as determined by halo size (data not shown). PAO-P34 (rhlR mutant) exhibited virtually no siderophore activity (data not shown).
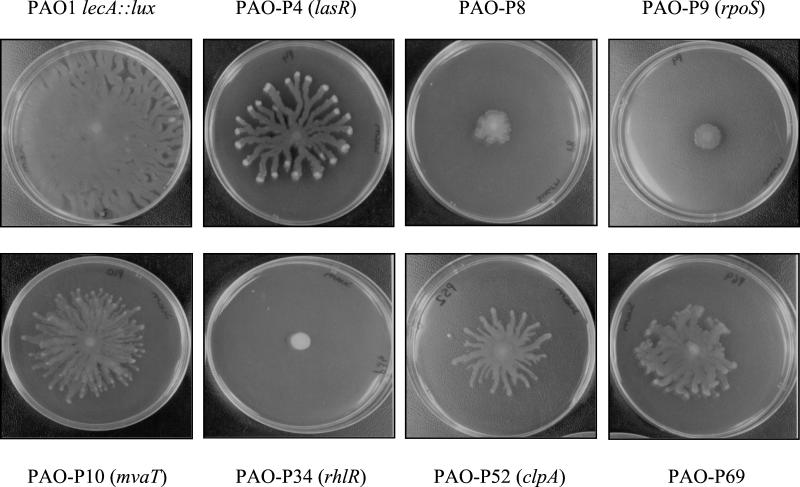
Finally, swarming motility was tested in a selection of mutants (Fig. 5). Compared with PAO1 lecA::lux, strains PAO-P4 (lasR mutant), PAO-P10 (mvaT mutant), PAO-P52 (clpA mutant), and PAO-P69 exhibited reduced swarming behavior while PAO-P8 and PAO-P9 (rpoS mutant) showed very little swarming. The rhlR mutant (PAO-P34) did not swarm, a finding in keeping with the observations of Kohler et al. (21).
FIG. 5.
Swarming motility assessed with swarm plates. The ability to swarm was determined by measuring growth from the central inoculation site as previously described (39).
AHL analysis.
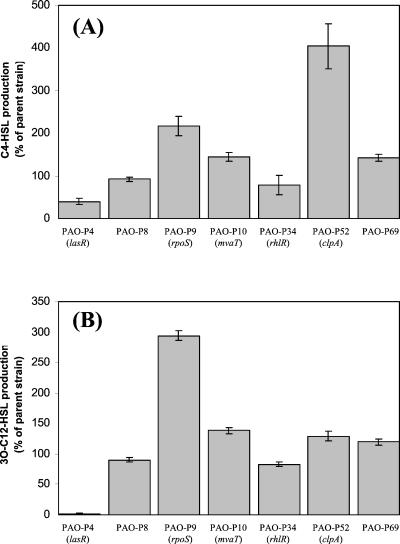
To determine whether the observed pleiotropic effects in the Tn5-B21 mutants were due to alteration of the quorum-sensing cascade, we analyzed the production of both C4-HSL and 3O-C12-HSL in these mutants. Samples of P. aeruginosa culture (900 μl) were taken 8 h postinoculation and acidified with 100 μl of 1 M HCl. The detection and quantification of AHLs are described in Materials and Methods. Figure 6 shows that PAO-P9 (rpoS mutant), PAO-P10 (mvaT mutant), PAO-P52 (clpA mutant), and PAO-P69 all produced high levels of both C4-HSL (Fig. 6A) and 3O-C12-HSL (Fig. 6B) compared with PAO1 lecA::lux, suggesting that these genes influence the quorum-sensing circuitry to some extent.
FIG. 6.
Influence of Tn5-B21 mutations on C4-HSL (A) and 3O-C12-HSL (B) production. Bacterial culture supernatants were taken after 8 h of growth and acidified with 1 M HCl. Culture supernatants were spotted onto either reverse-phase TLC plates and overlaid with soft top agar containing an E. coli bioluminescent reporter strain to detect C4-HSL or onto normal-phase TLC plates and overlaid with an E. coli bioluminescent reporter strain to detect 3O-C12-HSL. C4-HSL and 3O-C12-HSL levels were determined by quantifying the bioluminescence observed with the reporter strains. Values were adjusted for differences in OD600, and the level observed for the parent strain (PAO1 lecA::lux) was defined as 100%.
Addition of C4-HSL and 3O-C12-HSL to mvaT mutant PAO-P10 advances lecA::lux expression.
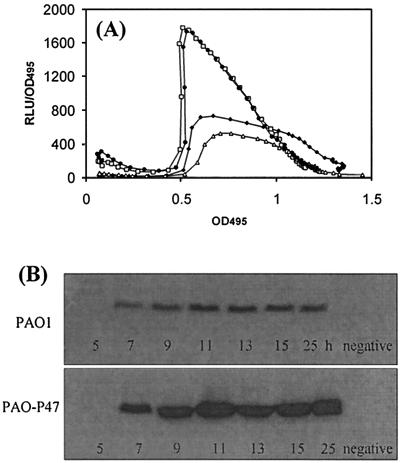
Since mvaT appeared to encode an important novel regulator of the expression of a number of quorum sensing-regulated phenotypes, we sought to determine whether this gene plays a role in controlling the growth phase-dependent expression of lecA. Firstly, experiments were performed to confirm that the observed effects on lecA expression in PAO-P10 were indeed due to inactivation of mvaT and not a secondary mutation on the chromosome. It was possible to reduce expression of the lecA::lux fusion in PAO-P10 to levels lower than those in the parent strain by expressing intact mvaT on pUCP18 (Fig. 7A). In addition, pyocyanin production was also significantly decreased in this trans-complemented mutant (data not shown). Furthermore, a defined mvaT deletion mutant (PAO-P47) form of PAO1 was constructed (see Materials and Methods) that showed significantly enhanced PA-IL lectin (Fig. 7B) and pyocyanin (data not shown) levels compared with the PAO1 wild type. In addition, elastase and LasA protease levels were decreased in this mutant compared with those in strain PAO1 (data not shown).
FIG. 7.
(A) Complementation of PAO-P10 with plasmid-based mvaT. The results shown are for PAO1 lecA::lux (⧫), PAO-P10 (□), PAO-P10 pUCP18 (•), and PAO-P10 pUCP mvaT (▵). (B) PA-IL lectin production in PAO1 and an mvaT chromosomal deletion mutant (PAO-P47) after 5, 7, 9, 11, 13, 15, and 25 h of growth. Shown is a Western blot of a P. aeruginosa cellular lysate after SDS-PAGE. The blot was developed with PA-IL antisera as described in Materials and Methods. PAO-P34 (rhlR mutant) was used as a PA-IL-negative control.
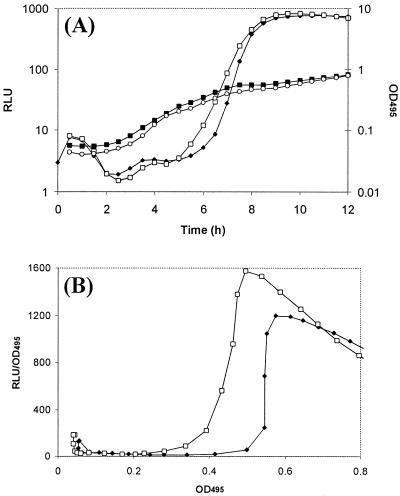
To investigate whether expression of lecA::lux could be advanced in the absence of a functional mvaT gene, PAO-P10 was grown in the presence of either C4-HSL, 3O-C12-HSL, or a combination of the two at a 10 μM concentration. In contrast to that in the PAO1 lecA::lux parental strain (Fig. 2), expression of lecA::lux was advanced in PAO-P10 with respect to time (Fig. 8A) and cell density (Fig. 8B) when both AHLs were added in combination to the medium.
FIG. 8.
Advancement of lecA expression in PAO-P10 (mvaT mutant). (A) Growth (OD495) of PAO-P10 in the absence (▪) or presence (○) of both C4-HSL and 3O-C12-HSL at 10 μM. Also shown is absolute lecA expression (RLU) with time in the absence (⧫) or presence (□) of both C4-HSL and 3O-C12-HSL at 10 μM. (B) Representation of relative lecA expression with cell density in the absence (⧫) or presence (□) of a combination of C4-HSL and 3O-C12-HSL at 10 μM. The results represent a single experiment, although the experiment was repeated three times with similar results.
DISCUSSION
In P. aeruginosa, many virulence genes have been reported to be regulated by quorum sensing. By definition, quorum sensing is a mechanism by which bacterial cells regulate specific target genes in response to a critical concentration of signal molecules, which is, in turn, a measurement of the cell density of the population (13). Previous work has shown that deletion of qscR (6), rpoS (56), and rsmA (37a) in P. aeruginosa results in advanced expression of quorum sensing-regulated genes. In addition, work by Whiteley et al. (55) with a lasI rhlI double mutant has shown that expression of a number of genes can be induced prematurely in the presence of AHLs. This is, in fact, the case for bioluminescence and carbapenem production in wild-type strains of V. fischeri (10, 28) and E. carotovora subsp. carotovora (58), respectively. In contrast, the present report and also a previous publication (62) demonstrate that at least some quorum sensing-regulated virulence genes in P. aeruginosa are “superregulated” in a growth phase-dependent manner. For instance, under the conditions employed in this study, induction of lecA and lasB occurred during the transition to stationary phase and transcription of these genes could not be advanced by addition of exogenous C4-HSL and 3O-C12-HSL, indicating that a critical autoinducer concentration is required but is not sufficient for their expression. Similarly, expression of rhlR, the gene encoding one of the LuxR homologues that mediate quorum sensing in P. aeruginosa, could not be advanced under these conditions. Thus, it appears that growth phase-dependent superregulation of quorum sensing is conducted on at least two levels, (i) control of the quorum-sensing cascade itself and (ii) target gene expression. To identify new genes involved in quorum sensing or growth phase-dependent regulation of the lecA gene, we used the bioluminescent lecA::lux reporter strain (62) as a target for transposon mutagenesis. This approach resulted in the isolation of mutants that were all affected in multiple quorum-sensing phenotypes. Furthermore, the isolation of lasR and rhlR mutants demonstrated that this approach could also be used for the identification of regulators involved in control of the quorum-sensing hierarchy itself. Among the mutants isolated, only PAO-P10 showed a greater-than-twofold increase in lecA::lux expression. Furthermore, in contrast to the parental strain, addition of exogenous C4-HSL and 3O-C12-HSL in combination advanced lecA::lux expression in PAO-P10 significantly. This suggested that the growth phase-dependent control mechanisms observed with the parent strain were no longer fully functional in this mutant. The locus inactivated in PAO-P10 was highly similar to a gene encoding the P16 subunit of MvaT, a novel heteromeric transcriptional regulator in P. mevalonii (41). Hence, the putative mvaT homologue of P. aeruginosa merited a more detailed analysis. In P. mevalonii, MvaT positively regulates the mvaAB operon encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase and 3-hydroxy-3-methylglutaryl coenzyme A lyase, enzymes that catalyze the initial reactions of mevalonate catabolism in this organism. In order to determine whether MvaT regulates a similar operon in P. aeruginosa, we searched for homologues of mvaA and mvaB with the complete Pseudomonas genome sequence. A putative homologue of mvaB was identified that showed 74% similarity to mvaB from P. mevalonii. However, no homologue to mvaA was found. Analysis of the mvaT gene region in P. aeruginosa with the genome sequence revealed only two genes with an assigned function. Downstream of mvaT and convergently transcribed is the sbcB gene coding for the protein exo-DNase I (also known as exonuclease I). This protein showed 66% similarity to exonuclease I in E. coli, where it degrades single-stranded DNA from the 3′ end and is involved in DNA replication, recombination, modification, and repair (38). Upstream of mvaT and divergently transcribed is the purU1 gene, which encodes a putative formyltetrahydrofolate deformylase (27). This protein showed 67% similarity to the formyltetrahydrofolate deformylase of Aquifex aeolicus. It seems unlikely that polar effects of the transposon insertion were responsible for the observed phenotypic changes, as mvaT does not form an operon with these genes. Proteins similar to MvaT may represent a new family of transcriptional regulators that are unique to pseudomonads, as database analysis revealed putative homologues in P. putida, P. syringae, and P. fluorescens and also a second homologue (PA2667) in P. aeruginosa but not in other bacterial species. Interestingly, analysis of the gene region encoding one MvaT homologue in P. putida KT2440 revealed that this gene was also flanked by the sbcB and purU1 genes in a manner identical to that seen in P. aeruginosa.
In P. aeruginosa PAO-P10, inactivation of mvaT not only resulted in increased expression of the lecA::lux reporter fusion but also caused ninefold overproduction of pyocyanin. As expected, these effects could be reversed by providing an intact mvaT gene in trans. Furthermore, increased production of the PA-IL lectin and pyocyanin was confirmed with an in-frame deletion of mvaT in P. aeruginosa PAO1. AHL analysis of PAO-P10 demonstrated that this mutant produced increased levels of both C4-HSL and 3O-C12-HSL, suggesting that MvaT may act as a repressor of quorum sensing in P. aeruginosa. The increase in AHLs could provide an explanation of why lecA expression and pyocyanin production are dramatically increased in PAO-P10. However, it is important to note that although AHL levels were increased, other quorum sensing-dependent phenotypes were down-regulated in this mutant. For instance, PAO-P10 showed reduced production of elastase and LasA protease and decreased swarming motility. Thus, in P. aeruginosa, MvaT appears to be a global regulator that modulates the expression of several virulence genes and is part of a growth phase-dependent control system.
Addition of both C4-HSL and 3O-C12-HSL is required, in the absence of MvaT, to bring lecA::lux expression forward (Fig. 8). Since the parent strain did not show advanced expression with regard to time and cell density in the presence of both AHLs (Fig. 2), it can be argued that a critical AHL concentration is required, but is not sufficient, for the expression of some quorum sensing-regulated genes. We hypothesize that early expression of these genes may be prevented by additional control elements that ensure that they are only switched on at a certain stage of growth. MvaT may represent one such control element. Interestingly, Whiteley et al. (55) also identified a number of genes (so-called class II and IV genes) whose expression was enhanced but not advanced by the addition of AHLs. Some of these genes may also be superregulated in a growth phase-dependent manner. Furthermore, the increase in lecA expression in the presence of both AHLs, taken together with data provided by Whiteley et al. (55), supports the idea that posttranslational regulation of the rhl system, due to the competition of C4-HSL and 3O-C12-HSL for RhlR binding (36), does not occur in P. aeruginosa but that both autoinducers can act in a synergistic manner.
It is not unreasonable to assume that growth phase-dependent control of quorum sensing-dependent genes is multifactorial. One of the control elements involved appears to be RpoS. Transcription of rpoS was shown to be induced upon entry into stationary phase (56), although transcripts are already detectable during mid-exponential phase (K. Winzer and P. Williams, unpublished). Mutations in either the las or rhl quorum-sensing system or addition of exogenous AHLs did not advance or inhibit induction of the rpoS gene (56). How rpoS is regulated in P. aeruginosa has not been studied in detail, but Kojic and Venturi (22) have shown that expression is largely dependent on PsrA, a regulator of the TetR family. Intriguingly, regulators belonging to this family are activated upon interaction with a diffusible molecule. Our results demonstrate that the stationary-phase sigma factor is required not only for lecA expression but also for the production of elastase and LasA protease, as well as swarming. This requirement of RpoS for swarming has not been described before. The requirement for RpoS could also explain the late expression of class II and IV genes described by Whiteley et al. (55). In a previous study by Suh et al. (50), only a small decrease in elastase and LasA activities was reported for an rpoS mutant whereas our mutant, PAO-P9, showed lower activity levels. Still, in both studies, pyocyanin production by rpoS mutants was shown to be significantly increased, a phenotype also reported by Whiteley et al. (56). It has been shown that rhlI expression is increased in an rpoS mutant (56). In this study, we showed that, accordingly, our rpoS mutant (PAO-P9) produced greater amounts of C4-HSL than did the parental strain. In addition, PAO-P9 also produced more 3O-C12-HSL, a finding not previously described. Whether the extremely high pyocyanin levels seen in this strain were due to high levels of both AHLs is unknown, although it is interesting that other mutants (PAO-P10, PAO-P52, and PAO-P69), up-regulated in pyocyanin and downregulated in other phenotypes, also produced higher levels of both AHLs compared with the parent strain.
Pearson et al. (35) have shown that 3O-C12-HSL, but not C4-HSL, accumulates in P. aeruginosa cells, with the intracellular concentration being three times higher than the external level. The mexA-mexB-oprM-encoded efflux pump is involved in the active efflux of 3O-C12-HSL, as the intracellular level in a mexA-mexB-oprM deletion mutant was eightfold higher than the external concentration (35). In our mutant, PAO-P69, a gene encoding a component of a putative active efflux system had been inactivated. This mutant was significantly different from the parent strain with regard to phenotypes regulated by quorum sensing. Intriguingly, this gene (PA4207) was identified by Whiteley et al. (55) as a class IV (late response) gene (J. Pearson, personal communication) that only responds to the addition of both C4-HSL and 3O-C12-HSL in a lasI-rhlI double mutant. This suggests that this gene is controlled by quorum sensing and is also involved in the regulation of a number of quorum sensing-dependent phenotypes. The expression of quorum sensing-regulated genes in the mexA-mexB-oprM mutant has not been described (35), but according to the present model (24, 36), earlier and stronger induction of LasR/3O-C12-HSL-dependent genes should be expected due to the high intracellular 3O-C12-HSL concentration. Earlier data were in agreement with this model, since Evans et al. (12) had demonstrated that hyperexpression of mexA-mexB-oprM resulted in a significant reduction of extracellular elastase and pyocyanin. However, mutant PAO-P69 showed, with the exception of pyocyanin production, the opposite of what was expected. Elastase levels were shown to be similar to those of the parent strain, while LasA protease activity was reduced. The quorum-sensing system of P. aeruginosa is still not well understood and may involve complex cross-regulation between the las and the rhl systems, as well as the third LuxR homologue, QscR (6).
In E. coli, the ATPase ClpA is a chaperone and also provides substrate specificity to the ClpP protease (19, 57). How inactivation of clpA influences quorum sensing-regulated phenotypes in P. aeruginosa still needs to be established. Inactivation of clpA could reduce the concentration of properly folded regulatory proteins. Alternatively, ClpA, together with ClpP, could have a regulatory function like that of ClpX and ClpP in E. coli, which degrade RpoS during the exponential phase (43). It will be interesting to unravel the link between these proteases and virulence gene expression in P. aeruginosa.
In summary, we have shown that in P. aeruginosa, growth phase-dependent control of gene expression can override activation by quorum sensing. We have isolated and characterized a number of mutants, all of which are affected in multiple quorum-sensing phenotypes. A new global regulator, MvaT, was identified that is involved in the growth phase-dependent control of the lecA gene. However, more work is required to unravel the complicated regulatory network that links cell density-dependent gene expression with the growth phase and metabolic state of the cell.
Acknowledgments
This work was supported in part by grant BIO4-CT96-0119 from the European Union (IVth Framework Biotechnology Programme) and by a grant and studentship from the Biotechnology and Biological Sciences Research Council, United Kingdom (to P.W.).
We thank Andrea Hardman for useful comments and suggestions on the preparation of the manuscript.
Footnotes
For a commentary on this article, see page 2569 in this issue
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. H. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bever, R. A., and B. H. Iglewski. 1988. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J. Bacteriol. 170:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapon-Herve, V., M. Akrim, A. Latifi, P. Williams, A. Lazdunski, and M. Bally. 1997. Regulation of the xcp secretion pathway by multiple quorum sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169-1178. [DOI] [PubMed] [Google Scholar]
- 5.Chhabra, S. R., P. Stead, N. J. Bainton, G. P. C. Salmond, G. S. A. B. Stewart, P. Williams, and B. W. Bycroft. 1993. Autoregulation of carbapenem biosynthesis in Erwinia carotovora by analogues of N-(3-oxohexanoyl)-l-homoserine lactone. J. Antibiot. 46:441-449. [DOI] [PubMed] [Google Scholar]
- 6.Chugani, S. A., M. Whiteley, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 8.DeKievit, T. R., P. C. Seed, L. Passador, J. Nezezon, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeKievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberhard, A., A. L., Burlingame, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 11.Essar, D. W., L. Eberly, A. Hadero, and I. Crawford. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, K., L. Passador, R. Srikumar, E. Tsang, J. Nezezon, and K. Poole. 1998. Influence of the MexA-MexB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 180:5443-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerischer, U., and P. Dürre. 1992. mRNA analysis of the adc gene region of Clostridium acetobutylicum. J. Bacteriol. 174:426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glessner, A., R. S. Smith, B. H. Iglewski, and J. B. Robinson. 1999. Roles of the Pseudomonas aeruginosa las and rhl quorum sensing systems in control of twitching motility. J. Bacteriol. 181:1623-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman, S., C. Squires, E. Pichersky, M. Carrington, M. Hobbs, J. S. Mattick, B. Dalrymple, H. Kuramitshu, T. Shiroza, T. Foster, W. P. Clark, B. Ross, C. L. Squires, and M. R. Maurizi. 1990. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc. Natl. Acad. Sci. USA 87:3513-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide host range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 19.Katayama, Y., A. Kasahara, H. Kuraishi, and F. Amano, F. 1990. Regulation of activity of an ATP-dependent protease, Clp, by the amount of a subunit, ClpA, in the growth of Escherichia coli cells. J. Biochem. (Tokyo) 108:37-41. [DOI] [PubMed] [Google Scholar]
- 20.Kessler, E., M. Safrin, J. C. Olson, and D. E. Ohman. 1993. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 268:7503-7508. [PubMed] [Google Scholar]
- 21.Köhler, T., L. Kocjancic Curty, F. Barja, C. Van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kojic, M., and V. Venturi. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. A. B. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 24.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR to expression of the stationary phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 25.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum-sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, P. L., G. M. McCorkle, and H. Zalkin. 1993. purU, a source of formate for PurT-dependent phosphoribosyl-N-formylglycinamide synthesis. J. Bacteriol. 175:7066-7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nealson, K. H., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial bioluminescent system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochsner, U. A. A. K. Koch, A. Fiechter, and J. Reiser. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in P. aeruginosa. Proc. Natl. Acad. Sci. USA 92:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohman, D. E., S. J. Cryz, and B. H. Iglewski. 1980. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of the Pseudomonas aeruginosa virulence genes requires cell-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 33.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of P. aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by P. aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. G. Holden, M. Cámara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips, G. J., and S. R. Kushner. 1987. Determination of the nucleotide sequence for the exonuclease I structural gene (sbcB) of Escherichia coli K12. J. Biol. Chem. 262:455-459. [PubMed] [Google Scholar]
- 39.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 41.Rosenthal, R. S., and V. W. Rodwell. 1998. Purification and characterization of the heteromeric transcriptional activator MvaT of the Pseudomonas mevalonii mvaAB operon. Protein Sci. 7:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweizer, H. P. 1991. Improved broad host range lac-based plasmid vectors for the isolation and characterization of protein fusions in Pseudomonas aeruginosa. Gene 103:87-92. [DOI] [PubMed] [Google Scholar]
- 45.Schwyn, B., and J. B. Nylands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 46.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 47.Simon, R., J. Quandt, and W. Klipp. 1989. New derivatives of transposon Tn 5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene 80:161-169. [DOI] [PubMed] [Google Scholar]
- 48.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:105-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 50.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. H. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swift, S., A. V. Karlyshev, E. L. Durant, M. K. Winson, P. Williams, S. Macintyre, and G. S. A. B. Stewart. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologues AhyRI and AsaRI and their cognate signal molecules. J. Bacteriol. 179:5271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swift, S., J. A. Downie, N. A. Whitehead, A. M. L. Barnard, G. P. C. Salmond, and P. Williams. 2001. Quorum sensing as a population-density-dependent determinant of bacterial physiology. Adv. Microb. Physiol. 45:199-270. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka, K., and H. Takahashi. 1994. Cloning, analysis and expression of an rpoS homologue gene from Pseudomonas aeruginosa PAO1. Gene 150:81-85. [DOI] [PubMed] [Google Scholar]
- 54.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derivied system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 55.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wickner, S., S. Gottesman, D. Skowyra, J. Hoskins, K. McKenney, and M. R. Maurizi. 1994. A molecular chaperone, ClpA, functions like DNAK and DNAJ. Proc. Natl. Acad. Sci. USA 91:12218-12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams, P., N. J. Bainton, S. Swift, S. R. Chhabra, M. K. Winson, G. S. A. B. Stewart, G. P. C. Salmond, and B. W. Bycroft. 1992. Small molecule-mediated density-dependent control of gene expression in prokaryotes: bioluminescence and the biosynthesis of carbapenem antibiotics. FEMS Microbiol. Lett. 100:161-168. [DOI] [PubMed] [Google Scholar]
- 59.Williams, P., M. Camara, A. Hardman, S. Swift, D. Milton, V. J. Hope, K. Winzer, B. Middleton, D. I. Pritchard, and B. W. Bycroft. 2000. Quorum sensing and the population-dependent control of virulence. Philos. Trans. R. Soc. Lond. B 355:667-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winson, M. K., M. Camara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. C. Salmond, B. W. Bycroft, A. Lazdunski, G. S. A. B. Stewart, and P. Williams. 1995. Multiple N-acylhomoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in P. aeruginosa. Proc. Natl. Acad. Sci. USA 92:9427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. A. B. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum-sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 62.Winzer, K., C. Falconer, N. C. Garber, S. P. Diggle, M. Camara, and P. Williams. 2000. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 182:6401-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]