Abstract
Antisense RNA-mediated transcriptional attenuation is a regulatory mechanism operating in the replication control of two groups of plasmids in gram-positive bacteria, the pT181 group and the inc18 family, represented by pIP501. In contrast, this control mechanism has so far not been identified in gram-negative bacteria or their plasmids. In this work we asked whether such a mechanism can be supported by Escherichia coli. The core replication control regions of plasmids pT181 and pIP501 were transferred into this heterologous host. In vivo lacZ reporter gene assays showed that the antisense RNAs of these plasmids can inhibit lacZ expression and that most of this effect can be accounted for by reduced mRNA readthrough. Northern analyses confirmed that the ratio of attenuated to readthrough target RNA was increased in the presence of the cognate antisense RNA, as expected for this mechanism. Similarly, both antisense RNAs induced premature termination of their cognate target RNAs in an E. coli in vitro transcription system, whereas the noncognate antisense RNAs had no effect. Thus, this report shows that antisense RNA-mediated transcriptional attenuation is supported by at least one gram-negative host, although the data indicate that inhibitory efficiencies are lower than those for, e.g., Bacillus subtilis. Possible explanations for the apparent absence of this control mode in plasmids of gram-negative bacteria are discussed.
Regulation of gene expression in bacteria is carried out by a number of different mechanisms. Transcriptional control works primarily through repressor or activator proteins as in regulation of metabolic processes (25). Posttranscriptional control is exerted at the mRNA level, either by proteins or RNAs. Many accessory genetic elements use antisense RNAs for regulation. Antisense RNAs act on their complementary target RNAs to block translation, induce mRNA decay, inhibit primer maturation, or induce premature termination of target RNA transcription. All these antisense RNA control mechanisms can be found in bacterial plasmids, phages, and transposons (14, 28, 30). Interestingly, antisense RNA-mediated transcriptional attenuation is known to be the principal mechanism by which many plasmids of gram-positive hosts control their copy numbers (13, 19, 23), whereas so far no plasmid in gram-negative hosts is known to use this control mode. In antisense RNA-mediated transcriptional attenuation, the fate of the target (sense) RNA is affected by the antisense RNA. Upon the binding of antisense RNA, the nascent transcript folds into one of two mutually exclusive conformations, so that a Rho-independent terminator forms, entailing premature termination. If the antisense RNA fails to bind during a critical time interval of transcription, the nascent RNA refolds into an alternative structure, which prevents termination and promotes readthrough. Novick and coworkers (23) were first to establish this mechanism for regulation of replication of staphylococcal plasmid pT181. Subsequently, streptococcal plasmids pIP501 and pAMβ1, belonging to the inc18 family of broad-host-range plasmids (11), were shown to be regulated by the same mechanism (13, 19). Details of this type of control in pIP501 and pT181 were later characterized in vitro (7, 10).
Attenuation can be mediated by factors other than antisense RNA, but the shared property of all systems is that of inducible, alternative RNA conformations that promote either readthrough or termination: a nascent mRNA carries sequences with the potential to fold into alternative, mutually exclusive, secondary structure elements. In the classical example of attenuation in the Escherichia coli trp operon, worked out by the pioneering studies of Yanofsky, ribosomal stalling at tandem trp codons in the leader of the mRNA affects the choice between readthrough and termination (34). Attenuation can also be mediated by RNA binding proteins, as in the E. coli bgl operon and the Bacillus subtilis sac, pyr, and trp operons, and by the binding of cognate uncharged tRNA to leader transcripts of aminoacyl-tRNA synthetase mRNAs in gram-positive bacteria (16). Interestingly, an in vitro study showed that oligodeoxyribonucleotides can induce termination by forming a short helix with a region of a nascent transcript immediately upstream of a run of uridines by mimicking the structural features of a terminator (35). The list of chromosomal genes and operons controlled by attenuation mechanisms thus contains representatives of both gram-positive and gram-negative bacteria. It may therefore appear surprising that all of the cases of antisense RNA-mediated attenuation known to date occur in gram-positive hosts. This could imply that such systems are yet to be discovered in gram-negative bacteria. Alternatively, this particular mechanism might not be supported due to properties of, e.g., the transcriptional machinery of E. coli. A further possibility is that this type of regulation confers properties to a plasmid copy number control system which are not compatible with maintenance in gram-negative hosts.
To approach these questions, we asked here whether control regions derived from plasmids pT181 and pIP501, when present in E. coli cells, can support cognate antisense RNA-dependent transcriptional attenuation.
MATERIALS AND METHODS
Enzymes and chemicals.
Chemicals used were of the highest purity available. Taq DNA polymerase was purchased from Perkin-Elmer. T7 RNA polymerase was obtained from New England Biolabs, and the E. coli RNA polymerase holoenzyme was bought from LaRoche. Sequencing reactions were performed with a Sequenase kit from Amersham Pharmacia Biotech.
Bacterial strains.
E. coli strains TG1 and DH5α (26) were used for cloning and subsequent analyses. Isogenic E. coli strains IBPC 5321 (rnc+) and IBPC 5321 rnc105 (3) were used for Northern blot analyses.
Determination of β-galactosidase activity.
Overnight cultures of plasmid-carrying E. coli strains were grown in L broth containing appropriate antibiotics, diluted to an optical density at 550 nm (OD550) of 0.05, and grown to an OD550 of 1 before harvesting. Specific β-galactosidase activity was determined as described previously (4).
Construction of plasmids for in vivo RNA analysis.
All plasmids are listed in Table 1. Three plasmids containing the control regions of pIP501 and pT181 were constructed. These regions were truncated within the rep genes, and either a −35 or −10 mutation was introduced into the antisense RNA gene promoters to yield pGTR6 (pIP501 segment) and pGTR181/35 and pGTR181/10 (pT181 segments). All three plasmids were pGK14 replicons (17) (Table 1). Into each plasmid, a DNA fragment containing an rrnB terminator sequence was inserted. This fragment was obtained by PCR from plasmid pMG25 (M. Mikkelsen and K. Gerdes, unpublished data) with primers SB136 (5′ AGA TCT GAA TTC GTC GAC GAA TGC TTA ATG AAT TAC AA) and SB137 (5′ GGA TCC GAA TTC TCT AGA GCG GCG GAT TTG T). The resulting 225-bp fragment was cleaved within the primer sequences by EcoRI, followed by insertion into the unique EcoRI site of plasmid pGK14 (Table 1) (8), yielding plasmid pGTR. The orientation of the insert was confirmed by sequencing.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pUC19 | E. coli cloning vector; Apr, MCS | 26 |
| pGK14 | pGK13 (17) derivative lacking the Cmr gene; Emr | R. Breitling, unpublished |
| pRS10 | pBR322 replicon containing a translational repR-lacZ fusion; Ampr | 5 |
| pPR6 | pIP501 derivative containing a point mutation in the −35 box of promoter pIII | 6 |
| pMG25 | ColE1 derivative containing the lacI gene, an IPTG-inducible promoter, and the rrnB terminator; Ampr | M. Mikkelsen and K. Gerdes, unpublished |
| pGW380L | pSC101 derivative containing the promoterless lacZ gene with SDa sequence; Spcr | M. Lindell, unpublished |
| pGTC4 | pGK14-based vector containing the copR gene, pII/IR2, pIII, the attenuator, and 100 bp downstream | 9 |
| pUCpIII | pUC19 derivative containing the rnaIII gene with its own promoter | This study |
| pMGI | pMG25 derivative containing the promoterless pT181 rnaI gene | This study |
| pGTR | pGK14 derivative containing the rrnB terminator | This study |
| pGTR6 | pGTR derivative containing a pPR6-derived fragment upstream of the rrnB terminator | This study |
| pGTR181/35 | Like pGTR6, but with a fragment of pT181 carrying a point mutation in the −35 box of the RNAI promoter | This study |
| pGTR181/10 | Like pGTR6, but with a fragment of pT181 lacking the −10 box of the RNAI promoter | This study |
| pGW501 | pGW380L derivative containing a pPR6 sequence | This study |
| pGW181 | pGW380L derivative with a pGT181/35 sequence | This study |
| pGCZ6 | pGK14 derivative containing a pPR6 sequence fused in frame with the pRS10-derived lacZ gene | This study |
| pGT181/35 | pGK14 derivative with pGTR181/35 derived sequence | This study |
| pGKZ181 | pGT181/35 derivative carrying an in frame fusion with the pPRS10-derived lacZ gene | This study |
SD, Shine-Dalgarno sequence; MCS, multiple cloning site.
To construct pGTR6, a PCR product was generated from plasmid pPR6 (6), which contains a mutation in the −35 box of the pIP501 antisense promoter, with primers B 846 (5′ GAA TTC GGA TCC AAC AGA ACC AGA ACC AG) and B845-31 (5′ GAA TTC GTC GAC CGT CAT GAA GCA CAG TTT C). This DNA fragment was cleaved with BamHI and SalI and inserted into vector pGTR.
To obtain plasmid pGTR181/35, a mutation was introduced into the −35 box of the pI (antisense) promoter of pT181 in the following way. Two PCR steps were performed with oligodeoxyribonucleotide SB146 (5′ ATA TTT AAA TAT ACA TAA AGA TAT ATA TTT GGG) in combination with SB54 (10) and oligodeoxyribonucleotide SB147 (5′ CCC AAA TAT ATA TCT TTA TGT ATA TTT AAA TAT) in combination with SB91 (10) on a pT181 DNA template. The purified PCR fragments were combined, denatured, annealed, and used as templates for a third PCR step with outer primers SB91 and SB54 to generate a fragment carrying the mutated control region, bordered again by BamHI and SalI sites. Introduction of this segment into pGTR resulted in plasmid pGTR181/35. The same approach was used for the construction of pGTR181/10, which had a deletion of the −10 box of the pI promoter, except that primers SB146 and SB147 were replaced by SB144 (5′ TAT AAT CTT GTA TAT TTA GAA CGA TAT TTA AAT ATA CAT) and SB145 (5′ ATG TAT ATT TAA ATA TCG TTC TAA ATA TAC AAG ATT ATA). The sequences of the pT181 region inserts in pGTR181/35 and pGTR181/10 contain pIP501 sense promoter pII, which is active in E. coli, immediately followed by the sequence encoding the pT181 repC mRNA.
The two antisense RNA donor plasmids were constructed as follows. The EcoRI fragment of pGKIII, containing the entire antisense RNA gene of pIP501 with its own promoter (8), was inserted into the EcoRI site of pUC19 to yield plasmid pUCpIII. PCR performed on pT181 DNA with primers SB135 (5′ GGA TCC GAA TTC ATA CAA GAT TAT AAA AAC AAC T) and SB143 (5′ GAA TTC GGA TCC AAA ATA AAA AGG AGT CGC TCA) gave a DNA fragment carrying the gene encoding the antisense RNA of pT181 lacking its native promoter. After digestion with EcoRI and BamHI, this fragment was inserted into the EcoRI/BamHI-linearized pMG25 vector. The resulting plasmid, pMGI, allows for inducible expression of RNAI. The sequences of all PCR-derived fragments were determined to ensure that no additional mutations had been introduced.
Construction of translational lacZ fusion plasmids.
The repR[pIP501]-lacZ fusion plasmid pGCZ6 was constructed in several steps. A segment of the pIP501 control region containing a pIII −35 mutation, bordered by EcoRI and BamHI sites, was cleaved out of pPR6 and inserted into pBR322. Into the unique HindIII site of the pIP501 rep gene, a lacZ segment (lacking its promoter), derived from pRS10 (5) by HindIII cleavage, was inserted, generating an in-frame repR-lacZ fusion. The copR repressor gene (BamHI-KpnI fragment of pGTC4) (8) was now inserted upstream of the repR-lacZ segment. Finally, a BamHI-EcoRI fragment was transferred from the pBR322-based plasmid to intermediate-copy-number vector pGK14, cleaved with the same enzymes. The resulting plasmid was named pGCZ6.
The repC[pT181]-lacZ fusion plasmid pGKZ181 was constructed as follows. A segment of the pT181 control region containing a pI −35 mutation, bordered by BamHI and HindIII sites, was generated by PCR from pGTR181/35 (see above) DNA with primers SB91 and SB140 (5′ AGA TCT AAG CTT CGT CCA ACC GGC TAT TAG A). This fragment was inserted into pGK14, yielding pGT181/35. As above for pGCZ6, the HindIII lacZ fragment of plasmid pRS10 was used to create an in-frame fusion between the repC sequence and lacZ by insertion into the unique HindIII site. The resulting plasmid was named pGKZ181.
Construction of transcriptional lacZ fusion plasmids.
To construct transcriptional lacZ fusion plasmids, control regions of pIP501 or pT181 (with antisense RNA promoter down-mutations) were placed in front of a promoterless lacZ gene. The rep sequences carried premature termination codons, and intact ribosome binding sites were present in front of lacZ. A 500-bp PCR fragment was generated from pPR6 template DNA with SB162 (5′ GGA TCC GAA TTC AAC AGA ACC AGA ACC AGA AAC) and SB163 (5′ GAA TTC GGA TCC TTA GCC GTC ATG AAG CAC AGT TTC). After cleavage with EcoRI and BamHI, this fragment was inserted into EcoRI- and BamHI-cleaved plasmid pGW380L (M. Lindell, unpublished data), resulting in plasmid pGW501. The same approach was used to construct plasmid pGW181, but here pGTR181/35 was used as the template and SB164 (5′ GGA TCC GAA TTC TTG CTT ATT TTT TTA AAA ACG GAT ATA CTA) and SB165 (5′ GAA TTC GGA TCC TTA CAA CCG GCT ATT AGA GTA) served as primers. In both cases, TAA stop codons were introduced by the downstream primers to ensure translational termination of the pIP501 and pT181 rep coding sequences, upstream of the lacZ Shine-Dalgarno sequence.
DNA templates for in vitro transcription.
All transcription templates were generated by PCR from plasmid DNA. For PCR fragments encoding repC mRNA (pT181), SB59 was used in combination with SB61, and for PCR fragments encoding RNAI, SB57 was used in combination with SB58 (10). PCR fragments encoding RNAII and RNAIII of pIP501 were obtained with SB3 and SB4 or SB1 and SB2, respectively, as described previously (7). The PCRs with all the above primer sets produced fragments carrying a T7 RNA polymerase promoter in front of the sequences to be transcribed. For optimal transcription, two additional G residues were encoded at the transcription start sites. The 500-bp pIP501 DNA fragment and the 420-bp pIPT2 DNA fragment used as templates in the single-round transcription assay for pIP501- and pT181-specific sequences, respectively, were generated by PCR as described previously (7, 10).
In vitro transcription.
Labeled RNAI, RNAII, RNAIII, and repC mRNA, used as probes for Northern blotting, were synthesized in vitro by T7 RNA polymerase from PCR-generated DNA templates in the presence of [α-32P]UTP as described before (7). Unlabeled RNAI and RNAIII were synthesized in the presence of all four unlabeled nucleoside triphosphates (NTPs) and were gel purified before use.
Single-round transcription assays.
Single-round transcription assays were performed with a 500- (pIP501) or a 420-bp (pT181) DNA fragment (see above). The transcription buffer contained 40 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 150 mM KCl, and 0.01% Triton X-100. In a preincubation step, initiation complexes were allowed to form by the inclusion of 100 μM dinucleotide ApA (Sigma), 10 μCi of [α-32P]ATP, an NTP mixture containing 1 μM ATP and 20 μM CTP, template DNA (∼10−9 M), and 0.5 to 1 U of E. coli RNA polymerase. After 5 min at 30°C, solution 2 was added to start elongation of the preformed, short RNA chains. Solution 2 contained, in transcription buffer, rifampin (200 μg/ml) and all four NTPs to give final concentrations of 100 μM each. This allowed for elongation of initiated transcripts and prevented new initiations. Aliquots withdrawn at appropriate times were extracted with phenol, precipitated, dissolved in formamide loading dye, boiled, and separated on 6% sequencing gels. Gels were dried, autoradiographed, and quantitated on a Fuji PhosphorImager.
The protocol of the attenuation experiments was as follows. Tubes containing unlabeled RNAs appropriately diluted in transcription buffer were prewarmed at 30°C, aliquots of the preincubation mixture were added, and elongation of rep RNA transcription was started by addition of solution 2. For gel analysis, the reactions were stopped after 10 min.
Isolation of total RNA for Northern blot analyses.
Total RNA for Northern analysis was prepared from exponentially growing cultures as follows. Overnight cultures of E. coli strains carrying plasmids were diluted to an OD550 of 0.03 and grown in L broth with the appropriate antibiotics to an OD550 of 1. When RNAI was transcribed from the lac promoter, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 2 mM at an OD550 of 0.1 to 0.2 and growth was continued to an OD550 of 1. Then, 1.5 ml of each culture was immediately frozen in liquid nitrogen. When RNA half-lives were analyzed, rifampin was added (final concentration: 100 μg/ml) at an OD550 of 1 and aliquots were stopped by being frozen in liquid nitrogen after certain time intervals. Frozen samples were stored at −70°C for later preparation of total RNA. Centrifuged cells were suspended in 100 μl of lysis buffer 1 (100 mM NaCl, 50 mM EDTA [pH 8.0], 10% sucrose). Immediately, 300 μl of lysis buffer 2 (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 mM sodium acetate [NaOAc]) was added, and one hot phenol extraction (65°C) and two cold phenol-chloroform extractions were performed. Supernatants were precipitated with 1/10 volume of 3 M NaOAc, pH 7.0, and two volumes of ethanol. After centrifugation, the RNA was dissolved in 13 μl of water and stored at −20°C.
Northern blot analysis.
Northern blot analysis was performed as described previously (8). Reprobing the membrane to correct for loading errors was performed with [γ-32P]ATP-labeled oligodeoxyribonucleotide SB24 (5′ TAC GGC GTT TCA CTT CTG AGT TTG GG), which is complementary to E. coli 5S rRNA; both prehybridization and hybridization were performed at 42°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-3× Denhardt solution-0.5% sodium dodecyl sulfate (probe: 3 × 105 cpm of SB24/ml). Filters were washed and quantitated in a Fuji PhosphorImager with PCBAS software. For determination of 5′ or 3′ truncation of the attenuated transcripts in Fig. 2A (see text for details), two oligodeoxyribonucleotides were used: SB156 (5′ ATT CAG TTC GTT GTT TCG TT), complementary to the 5′ end of the repR mRNA, and SB157 (5′ AAA GGC AAT CAA CTA AGT TT), complementary to the 3′ end of the attenuated message. The same hybridization conditions as those for SB24 were applied.
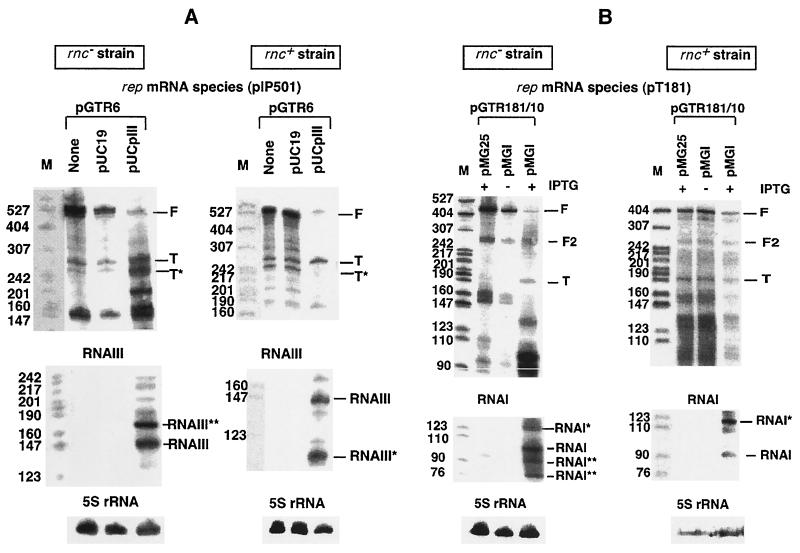
FIG. 2.
In vivo attenuation patterns in E. coli. An analysis of RNAs encoded by the control region of pIP501 (A) and a corresponding analysis of pT181 (B) are shown. The top portions of both panels show autoradiograms obtained after hybridization with labeled cognate antisense RNAs as probes (RNAIII for pIP501 and RNAI for pT181). The middle portions are autoradiograms from reprobing the same membranes with labeled sense RNAs. The bottom portions are loading controls (hybridized with a probe against 5S rRNA). (A) pGTR6 carries an antisense RNA promoter down-mutation. Lanes none, no antisense RNA donor plasmid present; lanes pUC19, empty vector; lanes pUCpIII, pUC19 derivative providing RNAIII in trans. F and T, positions of full-length (terminated at the rrnB terminator) and prematurely terminated sense RNA (terminated at the attenuator), respectively. T*, processed form of T (see Results). (B) Both pGTR181/10 and pGTR181/35 carry antisense RNA promoter down-mutations. Lanes pMG25, insertless vector; lanes pMGI, pMG25 derivative providing RNAI in trans. Cultures were treated with IPTG for induction of antisense RNA transcription (+) or were not treated with IPTG (−) (see Materials and Methods). RNAI*, RNAI**, RNAIII*, and RNAIII** are discussed in the text. The band corresponding to F2 has its 3′ end at the 5′ side of the rrnB sequence. M, pBR322 MspI marker.
RESULTS
Antisense RNA inhibits translational and transcriptional rep-lacZ fusion gene expression.
To assess transcriptional attenuation in E. coli, we made use of the control regions of plasmids pT181 and pIP501. Initial experiments tested whether promoters of the sense and antisense RNA genes were recognized by the heterologous RNA polymerase. For pIP501, both sense and antisense promoters were active. Reverse transcription mapping identified the same 5′-terminal nucleotide for the antisense RNA, RNAIII, previously found in B. subtilis (not shown). The antisense RNA of pT181, RNAI, was synthesized in E. coli, whereas sense RNA transcription was not observed (data not shown). Therefore, the pT181 sense RNA gene promoter was replaced by that of pIP501. Determination of half-lives of RNAIII and RNAI yielded values comparable to those measured for B. subtilis (30 and 5 min, respectively) (data not shown).
Translational and transcriptional fusions of rep (sense) sequences of pIP501 and pT181 to the lacZ gene were constructed to test the inhibitory effect of the corresponding antisense RNAs. In the translational fusions (plasmids pGCZ6 [segment from pIP501] and pGKZ181 [segment from pT181]) (Fig. 1; Table 1), the rep coding region was fused in frame to lacZ; in the transcriptional fusions (pGW501 [pIP501], pGW181 [pT181]) the truncated rep gene preceded the promoterless lacZ sequence, which carried its own translation initiation region. To maintain the characteristic level of pIP501 sense RNA transcription, plasmid pGCZ6 also carried the copR repressor gene. The promoters of the antisense RNA genes in these plasmids had been mutated to decrease the synthesis of RNAIII and RNAI to very low levels. These fusions were present on intermediate-copy-number plasmids. Antisense RNA was supplied from ColE1-based vectors carrying the rnaIII (pUCpIII) or rnaI (pMGI) genes. Synthesis of RNAI from the latter plasmid was driven by the IPTG-inducible lac promoter. Insertless vectors were used as controls.
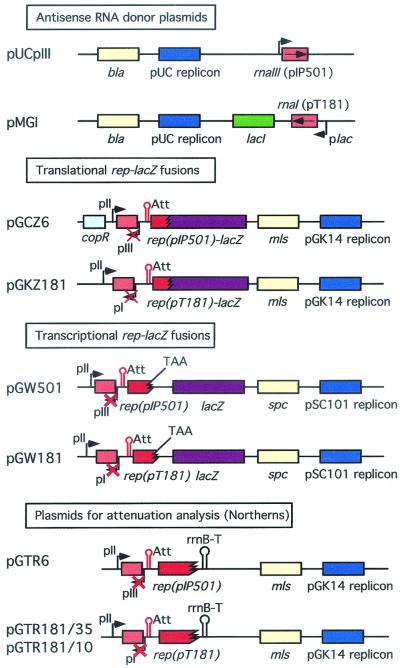
FIG. 1.
Schematic linear maps of the plasmids used in this study. The linear plasmids and genes and sites contained within them are not drawn to scale. Genes and sites of importance are highlighted as follows: replicon type, blue; selectable marker genes, yellow; rep genes or gene fragments, red; antisense RNA gene, light red; lacZ reporter gene, purple. Zigzag lines, in-frame (translational) fusions between rep and lacZ. Premature stop codons (TAA) separate the rep and lac reading frames, with the latter containing translational start signals. Some promoters are indicated by arrows. Inactivated promoters are crossed over. Approximate positions of attenuators (Att) and rrnB terminators (rrnB-T) are shown. For more details, see the text.
E. coli host cells carrying fusion plasmids and antisense RNA donor plasmids or insertless vectors were assayed for Rep-LacZ (translational fusions) or LacZ (transcriptional fusions) activity in vivo. RNAIII supplied from donor plasmid pUCpΙΙΙ decreased (pIP501)Rep-LacZ synthesis (from pGCZ6) in logarithmically grown cultures by a factor of ∼3.0 compared to values obtained in the presence of the control plasmid, pUC19 (Table 2). In vivo (pT181)Rep-Lac fusion protein synthesis (from pGKZ181) was decreased by RNAI in trans from donor plasmid pMGI by ∼2.6-fold compared to the control. Heterologous antisense RNA supplied in trans had no effect (data not shown).
TABLE 2.
β-Galactosidase activities of strains carrying translational or transcriptional sense RNA-lacZ fusions in the presence and absence of antisense RNAa
| Antisense donor | Translational fusions
|
Transcriptional fusions
|
||
|---|---|---|---|---|
| Fusion plasmid | β-Gal sp act | Fusion plasmid | β-Gal sp act | |
| None | pGCZ6 | 1.0 | pGW501 | 1.0 |
| pUC19 | pGCZ6 | 1.0 ± 0.15 | pGW501 | 0.9 ± 0.1 |
| pUCpIII | pGCZ6 | 0.33 ± 0.05 | pGW501 | 0.33 ± 0.08 |
| None | pGKZ181 | 1.0 | pGW181 | 1.0 |
| pMG25b | pGKZ181 | 1.0 ± 0.1 | pGW181 | 1.0 ± 0.17 |
| pMGIb | pGKZ181 | 0.38 ± 0.13 | pGW181 | 0.5 ± 0.05 |
Relative β-galactosidase (β-Gal) activities ± standard deviations from three independent experiments are given. Specific activities from strains containing fusion plasmids but no antisense RNA donor plasmids were set to unity. The β-galactosidase activities were in the range of 30 Miller units for pGCZ6 and pGKZ181 and in the range of 100 Miller units for pGW501 and pGW181. Three independent activity measurements were carried out.
Assayed after induction of the lac promoter by IPTG.
An attenuation mechanism would produce effects of the same magnitude when transcriptional fusions are analyzed. The rep-lacZ transcriptional fusions carried by pGW501 (pIP501 rep) and pGW181 (pT181 rep) were analyzed as described above by providing cognate antisense RNA from compatible donor plasmids. The results are shown in Table 2. For cloning reasons, the copR gene was in this case not included in the pIP501 fusion construct. Logarithmically grown cultures of cells containing pGW501 and pUCpIII showed an ∼3.5-fold decrease in LacZ activity compared to the control. For pT181, the induction of RNAI from pMGI resulted in a twofold decrease in LacZ synthesis, whereas no effect was obtained in the absence of IPTG induction.
These results suggest that (i) the cognate antisense RNAs decrease expression of their corresponding target genes, (ii) the similar magnitudes of the effect in translational and transcriptional fusions are consistent with attenuation, and (iii) both systems appear to operate at lower efficiency in E. coli than in, e.g., B. subtilis.
Northern analysis demonstrates antisense RNA-mediated transcriptional attenuation in vivo.
A hallmark of the mechanism in question is that the ratio of readthrough transcripts to terminated transcripts should be affected by the cognate antisense RNA, i.e., this ratio must decrease when antisense RNA is present and binds to the target RNA. An in vivo analysis for E. coli of the ratio between attenuated transcript RNA and readthrough rep-lacZ RNA was not feasible due to the vastly different lengths of the two RNA species. Therefore, the target gene segments (carrying either of two antisense RNA gene promoter down-mutations) were inserted in front of the rrnB terminator, and the resulting plasmids were denoted pGTR6, pGTR181/35, and pGTR181/10 (Fig. 1; Table 1). Two of these (pGTR6 and pGTR181/35) carried −35 box mutations, and the other (pGTR181/10) carried a deletion of the −10 promoter box. These mutations were included to abolish synthesis of the antisense RNAs. In all three plasmids, the distance between the attenuator and the rrnB terminator was ∼100 bp. Consequently, bands corresponding to attenuated and readthrough transcripts could easily be visualized and evaluated by Northern analysis.
Two-plasmid strains in which one plasmid carries the target segment and the other provides the antisense RNA were constructed. In controls, the antisense RNA donor plasmid was replaced by the insertless vector. To evaluate a possible contribution of RNase III to the expected attenuation pattern, all analyses were performed in parallel with isogenic wild-type (RNase III-proficient) strains and strains with mutant rnc105 (inactive RNase III). Strains were grown logarithmically and harvested, and total RNA was isolated and subjected to Northern blotting. For pT181, RNAI expression was induced by IPTG, followed by further cultivation for four or five generations before harvesting. Figure 2A shows the attenuation patterns obtained for the pIP501-specific RNAs, and Fig. 2B shows those for pT181.
Figure 2A, middle, shows that, in the absence of a donor plasmid or in the presence of the pUC19 plasmid, very little or no RNAIII was present, indicating that the antisense RNA promoter in plasmid pGTR6 (−35 mutation) indeed was impaired. When cells carried, in addition to pGTR6, pUCpIII, two prominent bands were visible in both wild-type and rnc105 strain backgrounds. For the wild type, the upper one represents full-length RNAIII and the lower one (Fig. 2A, RNAIII*) represents a relatively stable degradation intermediate. For the rnc105 strain, the full-length RNAIII and a longer species of ∼170 nucleotides (nt) (RNAIII**), most likely resulting from inefficient termination, are visible. Figure 2A, top, shows the effect of RNAIII in trans on attenuation. In the absence of antisense RNA or in the presence of only minute amounts of RNAIII, 25 to 30% intrinsic termination of the target RNA (cf. the intensities of T and T* to that of F in lanes none and pUC19) was observed, similar to values obtained previously for B. subtilis (8). The reason why two attenuator bands were found is not clear. Reprobing the membrane with specific deoxyribooligonucleotides complementary to the extreme 5′-end (SB156; see Materials and Methods) and 3′-end (SB157) regions of the attenuated RNA showed that the RNA in the faster-migrating band was shortened at its 5′ end (data not shown). When an excess of RNAIII was supplied in trans from plasmid pUCpIII, about 65% of all transcripts were attenuated, i.e., RNAIII induced premature termination of rep mRNA transcription in E. coli cells, consistent with an attenuator mechanism. A comparison of the RNA patterns in wild-type and rnc105 strains indicates similar ratios between F and T bands, i.e., attenuation functions comparably in both backgrounds. However, upon addition of antisense RNA, the total amount of F and T is drastically reduced in the wild-type strain but not in the rnc105 strain. The RNA species of ∼150 nt in the rnc105 strain background could represent the signal of a sense/antisense RNA duplex that fails to be degraded in the absence of functional RNase III.
Similar results were obtained with the control region of pT181 (Fig. 2B). In line with results from previous studies with B. subtilis (10), the rep mRNA of pT181 shows very little premature termination in the absence of its cognate antisense RNA when tested in the E. coli rnc105 strain (2 to 3%) (Fig. 2B, lanes pMG25) and about 30% premature termination in the isogenic wild-type strain. Small amounts of RNAI, provided from the leaky lac promoter of pMGI, only marginally increased attenuation (Fig. 2B, lanes pMGI without IPTG). Upon induction of RNAI synthesis by IPTG, large amounts of RNAI accumulated (Fig. 2B, bottom, lanes pMGI with IPTG) and termination of target RNA was increased to ∼30% in the rnc105 strain (Fig. 2B, lanes pMGI with IPTG) and ∼48% in the wild-type strain. The analysis of pT181 transcription in E. coli resulted in a more complex pattern than the one for pIP501 (see above). For example, as can be seen in Fig. 2B, bottom, full-length RNAI was a minor fraction of the sum of all RNAI-specific transcripts. In the wild-type strain, considerable readthrough of the RNAI terminator is indicated by a band of ∼125 nt (RNAI*). For the rnc105 strain, two degradation products (RNAI**; 85 and 70 nt) and a readthrough product of RNAI (RNAI*; 125 nt) were observed. Whether or not all these RNA species are inhibitory is at present not known. Similarly, several rep mRNA-specific bands were present (Fig. 2B, top). In addition, Fig. 2B, top, indicates that the RNAI-dependent increase in the ratio of T/F is accompanied by a decrease in the total amount of target RNA F and T species in both wild-type and rnc105 strains. Again, this decrease is apparently related to RNase III-dependent degradation of target RNA subsequent to interaction with RNAI: in the rnc105 strain, but not in the wild-type strain, a strong band of approximately 95 nt is induced by RNAI and may represent a duplex which persists in the absence of RNase III.
In vitro transcription with E. coli RNA polymerase confirms the in vivo data.
The Northern analyses in Fig. 2 suggested that both antisense RNAs can induce premature termination of their cognate target RNAs. However, the pattern in Fig. 2 could alternatively be explained by antisense RNA-induced differential changes in the decay rates of the full-length and attenuated RNAs. Therefore, we resorted to analyses of antisense RNA-mediated attenuation in an in vitro system, which is not complicated by the presence of RNase III or other RNases.
In an assay system previously developed for pIP501 RNAIII/II (7), B. subtilis RNA polymerase was replaced by the E. coli enzyme. This system has the following characteristics. Sense RNA transcription starts simultaneously, and restarts are prevented by addition of rifampin, so that synchronous single rounds of transcription can be monitored over time. Antisense RNA is not transcribed in this assay, and hence the effect of defined concentrations of added antisense RNA on the termination frequency can be assessed. Since the fate of the nascent transcripts is determined by the action of given concentrations of antisense RNA during a defined time window, end point analyses can yield accurate values for inhibition rate constants (7).
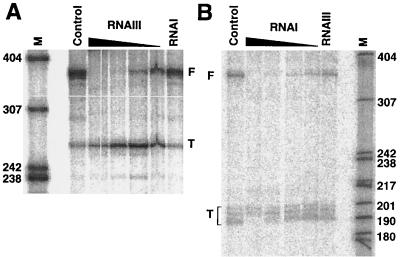
A 500-bp PCR-generated fragment was used as a template for pIP501 sequences, and a 420-bp fragment was used as a template for pT181 sequences (see Materials and Methods) (10). The control region of pT181 carried pIP501 repR promoter pII in place of the repC promoter (inactive in E. coli). In a preincubation step, rep RNA transcription was primed by inclusion of dinucleotide ApA, [α-32P]ATP, CTP, and E. coli RNA polymerase. This confines initiation complex formation to the rep promoter, since ApA is unable to function as a start dinucleotide at the antisense RNA promoter (12, 23). The omission of two NTPs stalls initiation complexes after incorporation of only a few nucleotides. Synchronous transcript elongation was started by addition of all four unlabeled NTPs. First, to determine whether the time windows for antisense RNA binding for B. subtilis and E. coli RNA polymerases differ, aliquots were withdrawn at different times for gel-electrophoretic analysis. Pilot experiments showed that transcription rates on these fragments, using E. coli RNA polymerase, were comparable to those obtained previously with B. subtilis RNA polymerase (7, 10). Results of in vitro attenuation assays are shown in Fig. 3. For pIP501, two prominent bands of ∼365 nt (full-length runoff transcript [F]) and ∼260 nt (terminated RNA [T]) accumulated (Fig. 3A, lane control) as seen previously (7). For pT181 (Fig. 3B), the full-length runoff transcript (F) of ∼360 nt was of the expected size, whereas the prematurely terminated RNA occurred as three bands in the size range of 190 to 201 nt (expected size ≈ 190 nt). Thus, in the absence of added antisense RNA, both target RNAs showed, in vitro, an intrinsic propensity to terminate. Upon addition of increasing amounts of unlabeled, in vitro-synthesized, and gel-purified cognate antisense RNA, the intensities of the F bands of both the pIP501 and pT181 target RNAs were decreased (or even became undetectable at high antisense RNA concentrations). In contrast, even high concentrations of noncognate antisense RNA failed to induce termination (Fig. 3A, lane RNAI, and B, lane RNAIII), indicating that the antisense RNA-mediated effect was dependent on proper antisense and target RNA complementarity. The approximate concentration of RNAIII required to obtain close to complete premature termination was 2 × 10−7 to 5 × 10−7 M, in good agreement with the results obtained with B. subtilis RNA polymerase (7) on the same pIP501 transcription template. Thus, the decrease of target RNA readthrough caused by the cognate antisense RNAs is consistent with an antisense RNA-driven attenuation mechanism.
FIG. 3.
Antisense RNA-dependent attenuation in a single-round E. coli transcription system. The protocol for the single-round transcription assay is described in Materials and Methods. Autoradiograms for pIP501 (A) and pT181 (B) are shown. In all assays, transcription was terminated at 10 min. Where indicated, increasing concentrations of unlabeled homologous antisense RNA were included in the attenuation assay at concentrations between 4 × 10−8 and 8 × 10−7 M. Lanes RNAI (A) and RNAIII (B), incubations in which heterologous (unlabeled) antisense RNA at ∼8 × 10−7 M was included; lanes control, incubations in the absence of antisense RNA; lanes M, size markers. F and T, positions of full-length (runoff at the end of the template fragment) and attenuated sense RNAs, respectively.
DISCUSSION
In this work, we have shown that antisense RNA-mediated transcriptional attenuation, a regulatory mechanism previously described only for gram-positive bacteria, also functions in gram-negative E. coli. The control regions of plasmids pIP501 and pT181 were used in the heterologous host to determine whether this mode of control can operate. The results from translational and transcriptional rep gene fusions to lacZ (Table 2) indicated that E. coli can support this mechanism. Similar inhibition values obtained with translational and transcriptional fusion plasmids already suggested that the major effect of the antisense RNA was to block readthrough of the attenuator. The Northern analyses corroborated this finding; antisense RNAs caused an increase in the ratio of the band intensity of attenuated sense RNA to that of readthrough sense RNA (Fig. 2). Finally, a qualitatively similar pattern was obtained when an in vitro attenuation assay was performed with E. coli RNA polymerase. Cognate, but not noncognate, antisense RNAs decreased readthrough in this minimal system (Fig. 3). Thus, the requirements for this mechanism to work are inherent in the ability to form the alternative RNA structures on which the regulatory decision rests and are not dependent on the source of the host RNA polymerase.
Since thorough studies of the pIP501 and pT181 systems have been performed previously (7, 8, 10, 23), one can compare the results obtained for E. coli with those for B. subtilis. A striking observation is the relatively low efficiency of regulation in E. coli. Inhibition of rep-lacZ (translational) and of lacZ (transcriptional) activities by cognate antisense RNA in vivo ranged from two- to fivefold. This level of repression is only moderate, considering that large amounts of antisense RNA were supplied from high-copy-number plasmids in trans and that the fusions were carried on intermediate-copy-number plasmids (Fig. 1; Table 2). In contrast, fusion plasmid analyses with Staphylococcus aureus indicated that β-lactamase activity was reduced at least sevenfold by RNAI in trans, both as a transcriptional fusion (pT181: repC-bla) (23) and as a translational fusion (31). In the latter case, the antisense RNA gene was present at 50 to 60 copies/cell and the repC-bla fusion was carried by a plasmid at ∼15 copies/cell. It is difficult to compare the inhibition values reported in the above experiments to the ones presented here. The replicons carrying fusion genes varied and replicated at different copy numbers. Intracellular concentrations of the antisense RNAs differed, due to gene dosage effects and possibly to promoter strengths, in the two heterologous bacterial species. In addition, the RNA-metabolizing enzymatic activities present in B. subtilis and E. coli are clearly different. This point is illustrated by the pattern of rep(pIP501)-specific bands in E. coli (Fig. 2A), which is distinct from that seen in B. subtilis (8). A further complication arises when one attempts to estimate the intracellular concentrations of active RNAI and RNAIII. RNAI, in particular, is present as species of different sizes (Fig. 2B). Thus, the question of whether shorter (processed) or longer (readthrough product) variants of RNAI are active as inhibitors cannot be answered, making comparative estimates of specific inhibitory efficiencies uncertain. For pIP501, RNAIII* (Fig. 2A) is known to be essentially inactive, since a plasmid construct from which this species was produced as the major RNA species in vivo proved inefficient as an antisense RNA donor (data not shown).
A lower efficiency of attenuation is also suggested by a comparison of the pIP-specific RNA patterns between E. coli and B. subtilis. Figure 2 shows that, even at high concentrations of antisense RNA provided in trans, a fraction of readthrough sense transcripts were still detected. In contrast, a similar analysis for B. subtilis showed that RNAII readthrough products were below the detection limit when high concentrations of RNAIII were present (8). Similar conclusions can be drawn from experiments on pT181 in S. aureus (23) and on pAMβ1 in B. subtilis (19). Thus, it appears clear that the overall efficiency of antisense RNA control in gram-positive bacteria such as B. subtilis and S. aureus is higher than that observed in E. coli.
An unexpected observation concerns the effects of the antisense RNAs on the RNA patterns in vivo and in vitro (Fig. 2 and 3). In B. subtilis cells and in in vitro systems containing B. subtilis RNA polymerase, antisense RNA of pIP501 or pT181 affected the ratio of readthrough/attenuated target RNA. However, Fig. 2 shows that this is accompanied by a decrease in the total amount of sense RNA in an E. coli wild-type strain. The comparison between the sense RNA patterns in wild-type and rnc105 strains (Fig. 2) indicates that this decrease is at least in art due to duplex-dependent degradation of sense RNA by RNase III of E. coli (3). However, a similar reduction in signal was seen in vitro (Fig. 3). Recent studies indicated that the RNA polymerases of E. coli and B. subtilis show greatly different properties in vitro with respect to promoter recognition, sensitivity to pause signals, propensity for abortive elongation, and effects of downstream DNA sequence (2). Whether these differences can account for the effects observed here remains to be elucidated.
In conclusion, the work presented here indicates that the intracellular environment of at least one gram-negative bacterium, E. coli, can support an antisense RNA-mediated attenuation mechanism. This raises a question: given the ubiquitous use of similar control pathways in a wide range of microorganisms, why has this particular mode of regulation never been found in gram-negative bacteria? This analysis cannot provide an unequivocal answer. However, the possibility that properties of the transcriptional machinery (e.g., altered pausing intervals, involvement of accessory factors influencing folding of target RNA during transcription, etc.) make this mechanism nonfunctional in E. coli is tentatively ruled out by the data presented. Both RNAI and RNAIII induce premature termination of nascent sense RNAs in vitro with E. coli RNA polymerase as the only enzyme present. A more striking difference between results in both hosts concerns the RNA species generated from sense and antisense DNA templates. The Northern analysis indicates a complex pattern of both antisense and sense RNA species (Fig. 2), which suggests that some nucleolytic activities present in E. coli may be absent in B. subtilis (for a review, see reference 21). Since RNA processing and decay are expected to affect the concentrations of the interacting RNAs and probably the distributions of their inactive and active processing products, the efficiency of regulation could be different.
An intriguing hypothesis for the absence of such systems can be put forward when one considers the genetic elements in which they reside: antisense RNA-mediated attenuation controls the replication frequencies of bacterial plasmids. Many plasmids of gram-negative hosts use antisense RNA for control as well, but the mechanisms involve either translational inhibition or inhibition of primer maturation (28, 30). Plasmids such as R1 and ColE1, as representatives for these two mechanisms, are characterized by tight control curves, resulting in narrow copy number distributions in bacterial populations (15, 22, 24, 29). In E. coli, broad copy number distributions, and hence mechanisms that result in them, are counterselected, since at the lower end of the distribution plasmids can be lost at cell division and at the upper end cells die (runaway replication) (27). In gram-positive bacteria, runaway replication of plasmids has so far not been observed (6, 18). Plasmids that lack functional antisense RNA altogether experience dramatic copy number increases but fail to show a runaway phenotype (5, 23). Thus, it is tempting to speculate that the apparent absence of plasmids with attenuation control systems in gram-negative hosts may reflect the consequences of broad copy number distributions. In transcriptional attenuation, any mRNA that escapes induced termination will, in a burst, be translated into many Rep molecules. In contrast, when an antisense RNA acts by inhibiting translation of a rep mRNA, as in R1 (20), any degree of inhibition can in principle be obtained and control curves are smooth. A mechanism that results in bursts of Rep translation, and consequently in sudden increases in replication potential, is expected to lead to greater copy number fluctuations. Thus, bacterial species in which overreplication entails cell death, such as E. coli, may have counterselected the attenuation mechanism that is widespread in plasmids of gram-positive bacteria. Its lower efficiency in E. coli, suggested by our present data, may be one reason for the disfavor of this mechanism. The crucial test for this conjecture is a test of a pT181 or pIP501 control region incorporated into the basic replicon of a plasmid normally maintained in E. coli. We have so far been unable to construct such a viable plasmid.
Finally, it is entirely possible that the apparent absence of antisense RNA-driven attenuation mechanisms in gram-negative bacteria merely reflects our limited knowledge about the prevalence of regulatory RNAs in bacteria. In total, very few chromosomally encoded regulatory RNAs, including antisense RNAs, were known until recently (for a review, see reference 32). Two recent reports showed that the number of small, putatively regulatory RNAs in E. coli is much greater than expected (1, 33). Thus, with the advent of similar searches of gram-positive bacteria, it can be extrapolated that many new RNAs may be found. It will be of great interest to ask whether any of them may be able to regulate target genes by an attenuation mechanism.
Acknowledgments
We are grateful to Eckehard Birch-Hirschfeld, Institute for Virology, Friedrich-Schiller-University Jena, for synthesis of the oligodeoxyribonucleotides.
This work was supported by grant Br1552/2-3 from the Deutsche Forschungsgemeinschaft to S.B.
REFERENCES
- 1.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. H. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 2.Artsimovitch, I., V. Svetlov, L. Anthony, R. R. Burgess, and R. Landick. 2000. RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J. Bacteriol. 182:6027-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomberg, P., E. G. H. Wagner, and K. Nordström. 1990. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 9:2331-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brantl, S. 1994. The copR gene product of plasmid pIP501 acts as a transcriptional repressor at the essential repR promoter. Mol. Microbiol. 14:473-483. [DOI] [PubMed] [Google Scholar]
- 5.Brantl, S., and D. Behnke. 1992. The amount of RepR protein determines the copy number of plasmid pIP501 in Bacillus subtilis. J. Bacteriol. 174:5475-5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brantl, S., and D. Behnke. 1992. Copy number control of the streptococcal plasmid pIP501 occurs at three levels. Nucleic Acids Res. 20:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brantl, S., and E. G. H. Wagner. 1994. Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J. 13:3599-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brantl, S., and E. G. H. Wagner. 1996. An unusually long-lived antisense RNA in plasmid copy number control: in vivo RNAs encoded by the streptococcal plasmid pIP501. J. Mol. Biol. 255:275-288. [DOI] [PubMed] [Google Scholar]
- 9.Brantl, S., and E. G. H. Wagner. 1997. Dual function of the copR gene product of plasmid pIP501. J. Bacteriol. 179:7016-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brantl, S., and E. G. H. Wagner. 2000. Antisense RNA-mediated transcriptional attenuation: an in vitro study of plasmid pT181. Mol. Microbiol. 35:1469-1482. [DOI] [PubMed] [Google Scholar]
- 11.Brantl, S., D. Behnke, and J. C. Alonso. 1990. Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAM beta 1 and pSM19035. Nucleic Acids Res. 18:4783-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brantl, S., B. Nuez, and D. Behnke. 1992. In vitro and in vivo analysis of transcription within the replication region of plasmid pIP501. Mol. Gen. Genet. 234:105-112. [DOI] [PubMed] [Google Scholar]
- 13.Brantl, S., E. Birch-Hirschfeld, and D. Behnke. 1993. RepR protein expression on plasmid pIP501 is controlled by an antisense RNA-mediated transcription attenuation mechanism. J. Bacteriol. 175:4052-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eguchi, Y., T. Itoh, and J. Tomizawa. 1991. Antisense RNA. Annu. Rev. Biochem. 60:631-652. [DOI] [PubMed] [Google Scholar]
- 15.Ehrenberg, M. 1996. Hypothesis: hypersensitive plasmid copy number control for ColE1. Biophys. J. 70:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henkin, T. M. 1996. Control of transcription termination in prokaryotes. Annu. Rev. Genet. 30:35-57. [DOI] [PubMed] [Google Scholar]
- 17.Kok, J., J. M. van der Vossen, and G. Venema. 1984. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 48:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, C. C., and R. P. Novick. 1985. Plasmid pT181 replication is regulated by two countertranscripts. Proc. Natl. Acad. Sci. USA 82:638-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Chatelier, E., S. D. Ehrlich, and L. Janniere. 1996. Countertranscript-driven attenuation system of the pAM beta 1 repE gene. Mol. Microbiol. 20:1099-1112. [DOI] [PubMed] [Google Scholar]
- 20.Malmgren, C., H. M. Engdahl, P. Romby, and E. G. H. Wagner. 1996. An antisense/target RNA duplex or a strong intramolecular RNA structure 5′ of a translation initiation signal blocks ribosome binding: the case of plasmid R1. RNA 2:1022-1032. [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson, A. W. 1999. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23:371-390. [DOI] [PubMed] [Google Scholar]
- 22.Nordström, K., and E. G. H. Wagner. 1994. Kinetic aspects of control of plasmid replication by antisense RNA. Trends Biochem. Sci. 19:294-300. [DOI] [PubMed] [Google Scholar]
- 23.Novick, R. P., S. Iordanescu, S. J. Projan, J. Kornblum, and I. Edelman. 1989. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell 59:395-404. [DOI] [PubMed] [Google Scholar]
- 24.Paulsson, J., and M. Ehrenberg. 2000. Molecular clocks reduce plasmid loss rates: the R1 case. J. Mol. Biol. 297:179-192. [DOI] [PubMed] [Google Scholar]
- 25.Roy, S., S. Garges, and S. Adhya. 1998. Activation and repression of transcription by differential contact: two sides of a coin. J. Biol. Chem. 273:14059-14062. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Uhlin, B. E., and K. Nordström. 1978. A runaway-replication mutant of plasmid R1drd-19: temperature-dependent loss of copy number control. Mol. Gen. Genet. 165:167-179. [DOI] [PubMed] [Google Scholar]
- 28.Wagner, E. G. H., and R. W. Simons. 1994. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 48:713-742. [DOI] [PubMed] [Google Scholar]
- 29.Wagner, E. G. H., and S. Brantl. 1998. Kissing and RNA stability in antisense control of plasmid replication. Trends Biochem. Sci. 23:451-454. [DOI] [PubMed] [Google Scholar]
- 30.Wagner, E. G. H., S. Altuvia, and P. Romby. Antisense RNAs in bacteria and their genetic elements. Adv. Genet., in press. [DOI] [PubMed]
- 31.Wang, P. Z., V. B. Henriquez, S. J. Projan, S. Iordanescu, and R. P. Novick. 1991. The effect of plasmid copy number mutations on pT181 replication initiator protein expression. Plasmid 25:198-207. [DOI] [PubMed] [Google Scholar]
- 32.Wassarman, K. M., A. Zhang, and G. Storz. 1999. Small RNAs in Escherichia coli. Trends Microbiol. 7:37-45. [DOI] [PubMed] [Google Scholar]
- 33.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanofsky, C. 2000. Transcription attenuation: once viewed as a novel regulatory strategy. J. Bacteriol. 182:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611-615. [DOI] [PubMed] [Google Scholar]