Abstract
In a previous study, a quorum-sensing signaling system essential for genetic competence in Streptococcus mutans was identified, characterized, and found to function optimally in biofilms (Li et al., J. Bacteriol. 183:897-908, 2001). Here, we demonstrate that this system also plays a role in the ability of S. mutans to initiate biofilm formation. To test this hypothesis, S. mutans wild-type strain NG8 and its knockout mutants defective in comC, comD, comE, and comX, as well as a comCDE deletion mutant, were assayed for their ability to initiate biofilm formation. The spatial distribution and architecture of the biofilms were examined by scanning electron microscopy and confocal scanning laser microscopy. The results showed that inactivation of any of the individual genes under study resulted in the formation of an abnormal biofilm. The comC mutant, unable to produce or secrete a competence-stimulating peptide (CSP), formed biofilms with altered architecture, whereas the comD and comE mutants, which were defective in sensing and responding to the CSP, formed biofilms with reduced biomass. Exogenous addition of the CSP and complementation with a plasmid containing the wild-type comC gene into the cultures restored the wild-type biofilm architecture of comC mutants but showed no effect on the comD, comE, or comX mutant biofilms. The fact that biofilms formed by comC mutants differed from the comD, comE, and comX mutant biofilms suggested that multiple signal transduction pathways were affected by CSP. Addition of synthetic CSP into the culture medium or introduction of the wild-type comC gene on a shuttle vector into the comCDE deletion mutant partially restored the wild-type biofilm architecture and further supported this idea. We conclude that the quorum-sensing signaling system essential for genetic competence in S. mutans is important for the formation of biofilms by this gram-positive organism.
Bacteria are known to regulate diverse physiological processes through a mechanism called quorum sensing (22). Quorum sensing is a bacterial intercellular communication mechanism for controlling gene expression in response to population density (12, 41). Through quorum sensing, bacteria respond to their population density and regulate gene expression and cellular differentiation to optimize their physiology for a particular environmental stimulus. The ability of bacterial cells to communicate and behave collectively as a group probably provides significant benefits in the colonization of hosts, defense against competitors, adaptation to varying physical conditions, cellular differentiation, and species evolution (39).
In gram-negative bacteria, quorum-sensing systems generally consist of two components, a small, soluble signal molecule and a transcriptional regulatory protein or R protein (41). A large number of gram-negative quorum-sensing systems studied so far utilize N-acyl homoserine lactones as signal molecules (35). When a critical concentration is reached in a growing culture, these molecules can diffuse into the cells, binding to R protein and, in turn, activating transcription of target genes responsible for the cell density-dependent phenotypes (12). Studies of Pseudomonas aeruginosa have shown that cell-to-cell signaling through quorum sensing plays an important role in biofilm differentiation of this organism. A mutant (lasI) defective in the production of N-acyl homoserine lactones had a dramatic effect on the maturation of P. aeruginosa biofilms, resulting in a biofilm that lacked the three-dimensional architecture observed in the parent strain (11). In addition, detection of these quorum-sensing signals in the sputum of a cystic fibrosis patient has been suggested to be an indicator of infection by P. aeruginosa biofilms (40).
In gram-positive bacteria, quorum-sensing systems generally consist of three components, a signal peptide and a two-component regulatory system (TCS) that has a membrane-bound histidine kinase (HK) sensor and an intracellular response regulator (RR) (14, 22). Quorum sensing in gram-positive bacteria has been found to regulate a number of physiological activities, including competence development in Streptococcus pneumoniae (25) and Streptococcus mutans (30), sporulation in Bacillus subtilis (16), antibiotic biosynthesis in Lactococcus lactis (17), and induction of virulence factors in Staphylococcus aureus (21). Although these systems have been well characterized for several gram-positive organisms, their involvement with signaling in biofilm initiation and formation has not yet been defined. The first evidence to suggest that these systems operate in gram-positive biofilms came from a recent study of Streptococcus gordonii Tn916 mutants that were defective in biofilm formation (32). One of these mutants was found to have a transposon insertion in the comD gene, encoding the HK sensor protein of the TCS required for genetic competence. This evidence suggested that biofilm formation by S. gordonii might involve cell-cell communication through quorum sensing.
S. mutans is a bacterium that has evolved to depend on a biofilm “lifestyle” for survival and persistence in its natural ecosystem, dental plaque. It is also considered to be one of the principal etiological agents of dental caries (4, 18). The tooth surface is an indispensable natural habitat for S. mutans, since this organism cannot be detected in the mouth until eruption of the teeth and disappears soon after loss of the teeth (5). S. mutans is capable of forming biofilms through a number of well-studied mechanisms, including expression of the surface adhesin SpaP (27) and the ability to synthesize insoluble, extracellular polysaccharides that enhance its accumulation on the tooth (18). It is, however, unclear whether biofilm formation by S. mutans involves coordinated activity via cell-cell communication.
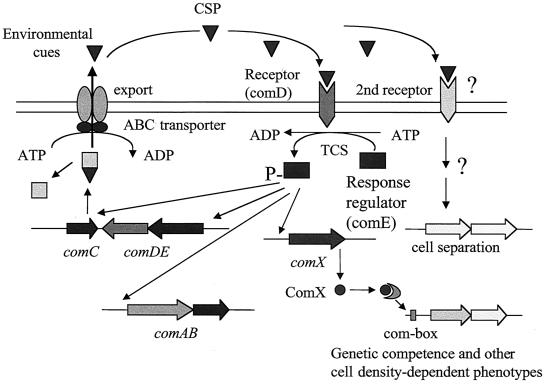
A quorum-sensing system essential for genetic competence in S. mutans was recently identified. This cell-cell signaling system involves at least five gene products encoded by cslAB(comAB) (36) and comCDE (30) (Fig. 1). The comC, comD, and comE genes respectively encode a competence-stimulating peptide (CSP) precursor, its HK sensor protein, and a cognate RR. comC and comDE lie adjacent on the chromosome and, together with their gene products, constitute a peptide (CSP)-signaling system including a generating pathway and a responding pathway, respectively. The other two genes, cslA and cslB, are located in a separate region of the chromosome and encode a CSP-specific secretion apparatus consisting of an ATP-binding cassette (ABC) transporter (ComA) and its accessory protein (ComB), which are presumably involved in the processing and export of the CSP (36). This quorum-sensing system functions optimally when the cells are living in actively growing biofilms (30), suggesting that the cell-cell signaling system might play a role in the formation of S. mutans biofilms. Another gene suspected of being involved in signal transduction via CSP is comX, a homolog of an alternate sigma factor found in S. pneumoniae that directs transcription of RNA polymerase to a number of competence-related genes as part of the CSP-induced signal cascade (25). To test this hypothesis, we examined the ability of S. mutans mutants defective in various components of the quorum-sensing and signal transduction system to form biofilms.
FIG. 1.
Hypothetical two-receptor model of S. mutans cell-cell signaling in genetic competence and biofilm formation. The CSP is involved in genetic competence (30), acid tolerance (29), and cell separation. cslAB (comAB) encodes an ABC-type export permease that cleaves the product of comC to generate a peptide pheromone, CSP. When CSP reaches a critical density, it is detected by neighboring cells via a sensor kinase, ComD (encoded by comD), which then phosphorylates an RR, ComE (comE), initiating transcription of an alternate sigma factor (comX) that initiates transcription of the genes required for DNA uptake and recombination. This model is based on the known functions of gene homologs in S. pneumoniae combined with the experimental evidence described herein.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
S. mutans wild-type strain NG8; its knockout mutants defective in comC, comD, comE, and comX; and a comCDE deletion mutant were assayed for their ability to initiate biofilm formation. All the strains, along with their relevant characteristics and sources, are listed in Table 1. The wild-type strains were subcultured routinely on Todd-Hewitt yeast extract (THYE) agar plates (BBL; Becton Dickinson, Cockeysville, Md.), whereas the mutants were maintained on THYE agar containing 10 μg of erythromycin/ml. THYE liquid medium was routinely used to grow the strains for most experiments unless otherwise specified. To grow biofilms, a semidefined minimal (SDM) medium was prepared by a modification of a method described previously (32). The medium contained 58 mM K2HPO4, 15 mM KH2PO4, 10 mM (NH4)2SO4, 35 mM NaCl, and 2 mM MgSO2·7H2O and was supplemented with filter-sterilized vitamins (0.04 mM nicotinic acid, 0.1 mM pyridoxine HCl, 0.01 mM pantothenic acid, 1 μM riboflavin, 0.3 μM thiamine HCl, 0.05 μM d-biotin), amino acids (4 mM l-glutamic acid, 1 mM l-arginine HCl, 1.3 mM l-cysteine HCl, 0.1 mM l-tryptophan), 0.2% (wt/vol) Casamino Acids, and 20 mM glucose. Biofilms of all strains were developed on polystyrene microtiter plates in SDM medium at 37°C with 5% CO2 for 16 h before quantification and further assessment.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| S. mutans | ||
| NG8 | Wild type, Ems Kms | A. S. Bleiweis, University of Florida |
| BM71 | Wild type, Ems Kms | G. H. Bowden, University of Manitoba |
| SMCC1 | NG8::pComC-KO ComC− Emr Kms | 30 |
| SMCD1 | NG8::pComD-KO ComD− Emr Kms | 30 |
| SMCE1 | NG8::pComD-KO ComE− Emr Kms | 30 |
| SMComX1 | NG8::pComX-KO ComX1− Emr Kms | This study |
| SMCDE-L4 | NG8 ΔcomCDE::PcEm Emr Kms | This study |
| SMComX1 | NG8 ΔcomX1::PcEm Emr Kms | This study |
| SMCDE-L4-pCOMC | SMCDE-L4::pCOMC Spr Emr | This study |
| SMCC1-pCOMC | SMCDE-L4::pCOMC Spr Emr | This study |
| Plasmids | ||
| pDL289 | E. coli-Streptoccus shuttle vector, Kmr | 3 |
| pDL277 | E. coli-Streptococcus shuttle vector, Spr | 24 |
| pCOMC | PDL277 harboring a wild-type comC gene | This study |
Construction of the comCDE and comX deletion mutants.
To determine if exogenous CSP affected biofilm formation by an S. mutans mutant defective in the quorum-sensing system, we constructed a mutant with a complete deletion of the comC, comD, and comE loci in wild-type strain NG8. The mutant was constructed using an insertion-deletion strategy. The primers used to construct and confirm gene deletion are listed in Table 2. Briefly, a 992-bp fragment 5′ from the comC start codon (ComCDE-up) was amplified from S. mutans NG8 genomic DNA by using primers ComCDE-P1 and ComCDE-P2 (containing an AscI site at its 5′ end). Another fragment, designated ComCDE-dw and containing 1,407 bp 3′ from comCDE, was amplified using primers ComCDE-P3 (with an FseI site at the 5′ end) and ComCDE-P4. An erythromycin resistance marker, PcEm (860 bp), from a synthetic Emr cassette (7) was amplified by using the Erm cst-P1 and Erm cst-P2 primers with AscI and FseI sites engineered into their 5′ ends, respectively. These amplicons were subjected to restriction enzyme digestion and subsequent ligation to produce a ComCSE-up::PcEm::ComCDE-dw fragment. The ligated product was used directly for transformation of S. mutans wild-type strain NG8 with the aid of synthetic CSP. Following double-crossover homologous recombination, the internal region of comCDE was completely replaced by the erythromycin cassette (PcEm). Template genomic DNA was prepared from transformants selected on plates containing THYE agar plus erythromycin (10 μg/ml) as described previously (10). Confirmation of the Emr marker's presence at the desired locus was performed by PCR. Primers P1 and P4 were used in combination with those designed for the Emr cassette to amplify fragments with the correctly predicted sizes from the mutants. Wild-type NG8 chromosomal DNA was used as a negative control. The mutant was then assayed for genetic transformation to confirm its competence-negative phenotype by the methods previously described (30). The comCDE mutant had a 100-fold impairment in natural transformation efficiency (data not shown) typical of competence-defective mutants (30). Addition of the synthetic CSP into the culture of the comCDE mutant could not restore the wild-type competence phenotype, which further confirmed the competence-defective phenotype of SMCDE-L4.
TABLE 2.
Primers used to construct the comCDE and comX deletion mutants by PCR restriction-ligation mutagenesis
| Primer | Nucleotide sequence (5′ → 3′)a | Amplicon (bp) |
|---|---|---|
| ComCDE-P1 | CAAAAGGAAAAAAACGGCAG | 992 |
| ComCDE-P2 | GGCGCGCCCGGAAAAATGTTGATAGGCTTC | |
| ComCDE-P3 | GGCCGGCCCGCAATGGTGGTTTCAAGACG | 1,407 |
| ComCDE-P4 | CGGGTTTCAGGAACAGAAGC | |
| Erm cst-P1 | GGCGCGCCCCGGGCCCAAAATTTGTTTGAT | 860 |
| Erm cst-P2 | GCTGGCCGGCCAGTCGGCAGCGACTCATAGAAT | |
| ComX-P4 | GGTTCTACAATTTCACCTTTACCTG | 685 |
| ComX-P3C | GCTGGCCGGCCCACTTTTTGGGAAGGCAAAG685 | |
| ComX1-P2B | GGCGCGCCTTGTTGCCAATCTTCACGAG | 876 |
| ComX1-P1 | AACACAGCAGTTAAGCCCTAGC |
The following restriction enzyme sites are indicated in bold: AscI (GGCGCGCC) and FseI (GGCCGGCC).
A mutant with an inactivated comX gene was constructed by a strategy similar to the one described above. To construct the strain SMComX1 (ΔcomX1), a 5′ 876-bp fragment proximal to comX1 (ComX1-up) which contained the promoter region and the ATG translational start codon was amplified from S. mutans NG8 chromosomal DNA using primers ComX1-P1 and ComX1-P2B (with an AscI site at the 5′ end). ComX1-dw (685 bp), which contained 68 bp of the 3′ end of comX1 plus 617 bp of the downstream region, was amplified by using primers ComX1-P3C (with an FseI site at the 5′ end) and ComX1-P4. An Emr marker from a synthetic Emr cassette, PcEm (860 bp) (7), was amplified by using the Erm cst-P1 and Erm cst-P2 primers with AscI and FseI sites engineered into their 5′ ends, respectively. These amplicons were subjected to restriction enzyme digestion and subsequent ligation to produce a ComX1-up::PcEm::ComX1-dw fragment. Ligated product was transformed into S. mutans wild-type NG8 using synthetic S. mutans CSP (30) to facilitate double-crossover homologous recombination, where the comX internal region was replaced with PcEm. Transformed colonies were selected on THYE agar-erythromycin (10 μg/ml) plates, and their correct amplicon sizes were confirmed by PCR.
Biofilm formation and quantification.
Biofilm initiation and formation by all strains was assayed and quantified by a modification of methods described previously (9, 32). To facilitate quantification and microscopy, both 96-well and 24-well polystyrene microtiter plates were used to develop biofilms. The growth of biofilms was initiated by inoculating 5 μl of pregrown cell suspension into 200 μl of SDM medium in the individual wells of a 96-well microtiter plate or by inoculating 25 μl of cell suspension into 2 ml of SDM medium in 24-well plates. The microtiter plates were then incubated at 37°C with 5% CO2 for 16 h without agitation. After the 16-h incubation, liquid medium was removed and wells were rinsed once with sterile distilled water (dH2O). The plates (96 wells) were then air dried and stained with 0.1% safranin for 10 min. After being stained, the plates were rinsed with dH2O to remove excess dye and then air dried for 3 h. Biofilms were quantified by measuring the absorbance of stained biofilms at 490 nm with an enzyme-linked immunosorbent assay microplate reader (model 3550; Bio-Rad Laboratories, Richmond, Calif.). Each assay was performed in triplicate, and wells without biofilms were used as blank controls after safranin staining. Biofilms formed in 24-well plates were photographed without staining immediately after the removal of planktonic cells. Our experiments confirmed that the modified minimal medium favored sessile biofilm growth of S. mutans and that it facilitated analysis of the biofilm-producing phenotype of mutant strains. During growth in an aerobic atmosphere with 5% CO2, S. mutans showed little difference in biofilm formation from growth under anaerobic conditions. This result differed from that of a previous study in which S. gordonii formed heavier biofilms when it was grown anaerobically (32). Therefore, we routinely grew biofilms of all strains in air with 5% CO2 for 16 h before further analysis.
Adherence assay.
All the strains were assayed for initial adherence to a mucin-coated polystyrene surface to determine if inactivation of individual genes involved in quorum sensing affected bacterial attachment. The surfaces of the polystyrene microtiter plates were first conditioned with 2 ml of 1% hog gastric mucin in an adherence buffer (10 mM KPO4, 50 mM KCl, 1 mM CaCl2, 0.1 mM MgCl2 [pH 7.0]) (26). The plates were incubated at room temperature for 2 h with gentle shaking and air dried for 2 h after removal of excess mucin solution. Resting cells were prepared by centrifugation of an overnight culture that was washed twice and resuspended in the adherence buffer at a density of 108 cells/ml (optical density at 600 nm [OD600], 0.6). Adherence to the mucin-coated surface was initiated by the addition of 2 ml of an already prepared resting cell suspension, and the plate was then incubated at 37°C with gentle shaking for 2 h. After incubation, the liquid cell suspension was removed and adherent cells were dissociated into 2 ml of the buffer by gentle sonication. Both adherent and nonadherent cells were assayed by viable-cell counting to determine percentages of adherent cells.
Growth kinetics.
All the strains were grown in THYE medium to assay their growth kinetics by using a Bioscreen C Labsystems (Helsinki, Finland) Microbiology Reader with multiwell disposable microtiter plates. The Bioscreen reader was equipped with Biolink software that allowed automatic recording and conversion of optical density readings into growth curves. An aliquot (4 μl) of cell suspension was inoculated into each well containing 400 μl of fresh medium. All cell suspensions were adjusted to the same OD600 before inoculation. The microtiter plates were then placed in the Bioscreen reader, which was set up to read optical density automatically every 15 min with shaking. Each sample was assayed in triplicate, and wells without inocula were used as blank controls during the 16-h incubation. S. mutans CSP was added at a final concentration of 500 ng/ml. As a negative control, S. pneumoniae CSP-1 (19) was added at a concentration of 500 ng/ml in parallel growth experiments.
Complementation of the comC mutant.
A 21-amino-acid synthetic peptide (CSP) based on the deduced amino acid sequence encoded by the comC gene (30) was used to determine if the CSP could restore defective phenotypes of the mutants, including genetic competence and the initiation of biofilm formation. During the experiments, the CSP was freshly dissolved in sterile dH2O to a final concentration of 1 mg/ml before use. The CSP solution was then added to the cultures at a saturating concentration 2 h after inoculation at a final concentration of 500 ng/ml as previously described (30).
To ensure that the comC mutant biofilm phenotype resulted from the absence of active CSP and to negate the possibility of an effect from the N-terminal cleavage product, we constructed an S. mutans strain that harbored a shuttle vector, pDL277, carrying a wild-type copy of comC to assay for restoration of the wild-type biofilm phenotype. To construct the comC-complemented strain, we used PCR to amplify a 651-bp fragment containing the entire comC gene and its promoter region from S. mutans NG8 genomic DNA using primers ComC-F5 and ComC-B5. The amplicon was then ligated into the multiple cloning site of plasmid pDrive (Qiagen, Mississauga, Ontario, Canada) and transformed into an Escherichia coli host (XL-10 Gold; Stratagene, La Jolla, Calif.). Transformants selected from Luria-Bertani agar plates containing ampicillin (100 μg/ml) and 2% X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were confirmed further by restriction analysis. The recombinant plasmid DNA was digested with SacI and BamHI to generate a 650-bp SacI/BamHI fragment harboring the comC region. This fragment was then ligated into the same restriction sites of plasmid pDL277. The resulting plasmid, pCOMC, was first transformed into the E. coli XL-10 host for propagation. pCOMC was then transformed into the comC mutant (SMCC-1) by a CSP-mediated transformation technique (30). Transformants were selected on THYE plates containing spectinomycin (1,200 μg/ml). The resulting complemented strain, designated SMCC1-pCOMC, was assayed for competence and its ability to form biofilms.
Microscopy.
To examine the spatial distribution and architecture of biofilms by scanning electron microscopy (SEM), biofilms formed on the surface of a polystyrene microtiter plate were washed once with 10 mM phosphate-buffered saline, fixed by the addition of 2 ml of 3.7% formaldehyde in 10 mM phosphate-buffered saline, and incubated at room temperature for 1 day. The samples were then dehydrated through a series of ethanol rinses (30, 50, 70, 95, and 100%) and dried at critical point with liquid CO2. The bottom surface of the well was cut off, mounted, and sputter coated with gold. The samples were then examined by SEM (model S-2500; Hitachi Instruments, San Jose, Calif.).
Confocal scanning laser microscopy (CSLM) was performed to characterize a larger field view of real-time biofilms on the surfaces of polystyrene microtiter plates. Biofilms were grown for 16 h in SDM medium and stained with LIVE/DEAD BacLight bacterial viability stain (Molecular Probes, Eugene, Oreg.) at a ratio of 1:10,000 of SYTO 9 to propidium iodide in dH2O, ensuring that biofilms were completely covered with the stain. The biofilms were examined with a 40× water immersion objective lens lowered directly into the biofilm-coated polystyrene plates with careful avoidance of biofilm disruption. Samples were examined by CSLM, and images were collected and analyzed with the packaged software (microscope model LSM 510; Carl Zeiss Canada, Toronto, Canada).
RESULTS
Mutants unable to produce CSP are defective in forming wild-type biofilms.
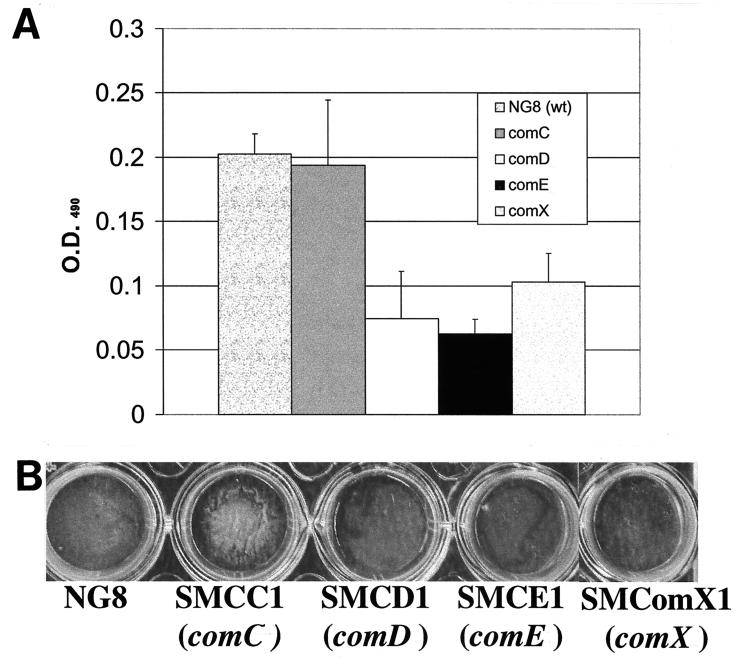
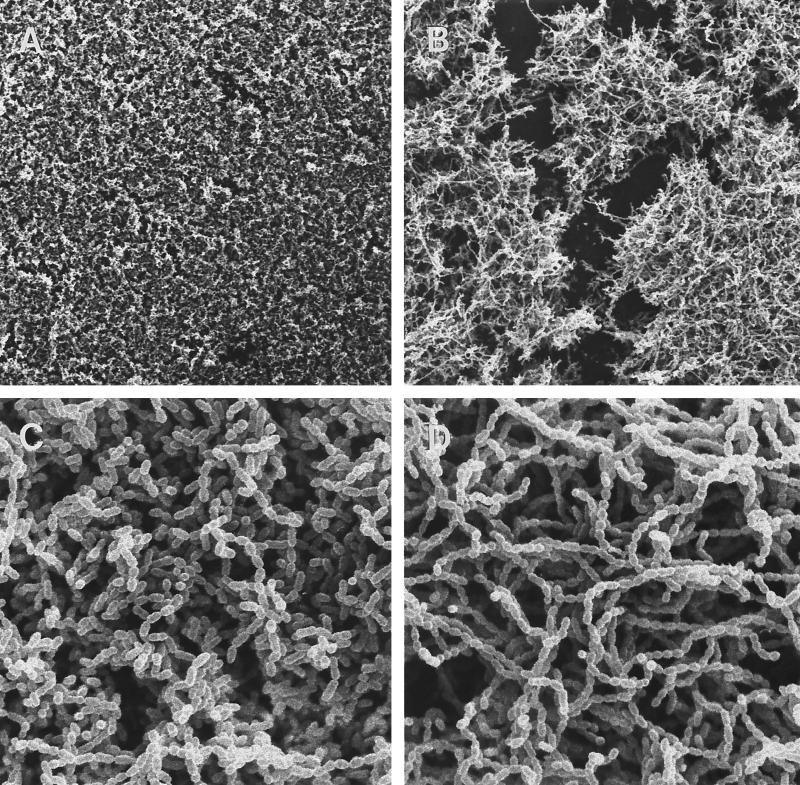
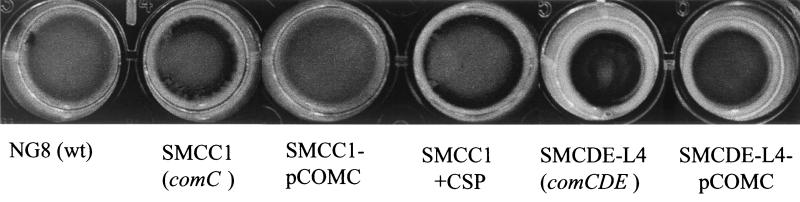
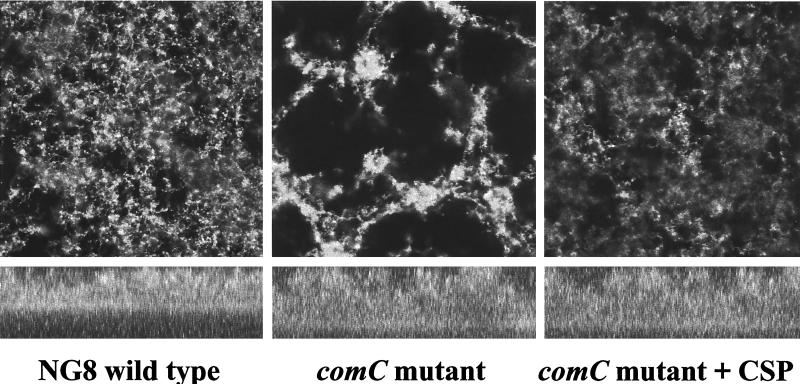
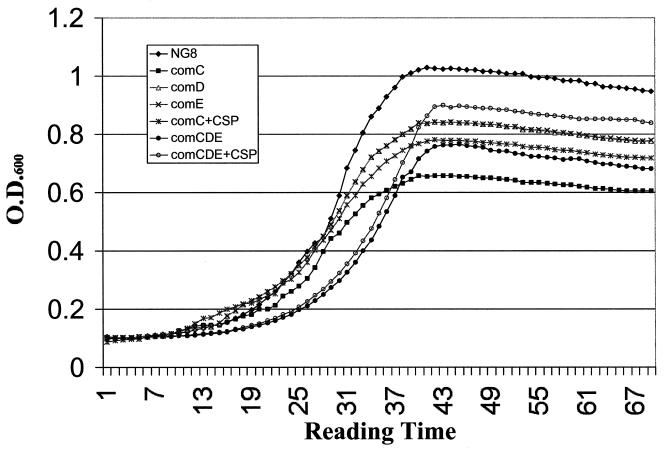
comC, the gene encoding the CSP precursor, and cslAB (comAB), the operon encoding the CSP-specific secretion system, are responsible for the synthesis and export of active CSP (30, 36). Disruption of these genes greatly diminished genetic transformation (30, 36). comC mutants were also defective in forming biofilms typical of the wild type. As shown in Fig. 2, the comC mutant had a noticeable difference in biofilm architecture on the polystyrene surface compared to that of the wild-type strain NG8, although the total biomass of the mutant biofilm was very similar to that of the wild-type strain. The biofilm formed by this mutant appeared to be clumped together with web-like microcolonies. SEM showed that cells in the mutant biofilms tended to form large aggregates or clumps with large spaces between them (Fig. 3B). Under higher magnification (Fig. 3D), the variation in biofilm structure appeared to have likely resulted from the formation of extremely long chains of cells by the mutants. Interestingly, addition of the synthetic peptide (CSP) into the growing culture of the comC mutant almost completely restored the wild-type biofilm architecture (Fig. 4 and 5), suggesting a link between quorum sensing and cell separation. Likewise in the CSLM image (Fig. 5), the NG8 wild type showed typical semiconfluent cells clustered in microcolony formations throughout the biofilm incubated for 16 h (xy plane). The comC mutant biofilms exhibited a consistent pattern of detachment of adherent cells from the growing microcolonies, resulting in gaps in biofilm confluence. Addition of CSP partially restored the wild-type phenotype of clustered semiconfluent microcolonies. Representative images of the biofilm depths (xz plane) of the wild-type strain, comC mutant, and comC mutant with CSP added did not show significant depth variation in the samples. In the liquid cultures, the comC mutant also formed extremely long chains, which may have allowed the cells to form large aggregates in the fluid phase. This may account for the decrease in optical density in the fluid culture compared to that of the wild-type strain (Fig. 6). Addition of the synthetic CSP into the liquid culture of the comC mutant partially restored the optical density reading. Mutants with inactivated comD, comE, or comX formed biofilms with a reduced biomass. Complementation of the comC mutant with pCOMC appeared to completely restore the wild-type phenotype (Fig. 4).
FIG. 2.
Biofilm formation and quantification of the comC, comD, and comE mutants on polystyrene surfaces. Mutants from three independent cultures were stained with safranin, and their OD490s were measured. (A) Mean OD490 values ± standard deviations; (B) photograph of typical biofilms grown on polystyrene microtiter plates. wt, wild type.
FIG. 3.
Scanning electron micrographs of S. mutans biofilms formed on polystyrene surface. (A and C) Wild-type biofilms (NG8); (B and D) comC mutant biofilms. Magnifications, ×480 (A and B) and ×4,800 (C and D).
FIG. 4.
Biofilm formation by the comC and comCDE mutants grown on polystyrene microtiter plates. The mutants are shown with and without plasmid pCOMC, which harbors the wild-type (wt) S. mutans comC gene.
FIG. 5.
CSLM image of biofilm formation by the comC and comCDE mutants with and without CSP. Top panels represent the xy plane, while bottom panels represent the xz plane. Magnification, ×300.
FIG. 6.
Growth curves of S. mutans NG8 and the mutants as generated by a Bioscreen Microbiology Reader.
A previous study demonstrated that the comD and comE genes, respectively, encoded an HK sensor protein and its cognate RR of the two-component signal transduction system responsible for sensing and responding to the CSP (30). This TCS was believed to play a key role in sensing and transducing the input signal to activate expression of the genes responsible for cell density-dependent induction of genetic transformation. A mutation in either comD or comE resulted in a significant impairment in genetic competence that was not restored upon addition of synthetic CSP. In the present study, inactivation of either the comD or comE gene resulted in the formation of a biofilm with a reduced biomass, as measured by the optical density readings after safranin staining of the biofilms (Fig. 2). Visual observation of the microtiter wells revealed that the biofilms formed by the comD and comE mutants were much thinner than the wild-type biofilms (Fig. 2). The reduction of biofilm mass in the comD and comE mutants might result partially from the decrease in their growth yields, since both mutants in fluid cultures had approximately 30% lower cell densities than the wild-type strain (Fig. 6). However, the comD and comE mutants had a nearly 70% decrease in biomass, relative to that of the parent strain, when they were grown in biofilms. Interestingly, the growth curves of these two mutants (in liquid culture) entirely overlapped, suggesting that inactivation of either of the genes encoding the TCS resulted in the same degree of effect on their growth and, consequently, the cell density. Addition of the chemically synthesized CSP into the culture of a comD or comE mutant could not restore the biofilm mass, reinforcing the notion that comDE encoded a TCS with a central role in sensing and transducing the native CSP signal. In addition, the comD mutant had a decreased ability to adhere to a mucin-coated polystyrene surface: 11.4% ± 0.5% of wild-type NG8 cells versus 7.7% ± 0.4% of comD mutant cells adhered to the surface. The difference was statistically significant (P < 0.05). This difference in ability to adhere to the substratum may have contributed to the decrease in biofilm mass.
S. mutans comX appears to be related to the S. pneumoniae comX gene in that inactivation leads to a competence-defective phenotype (data not shown). The S. mutans comX mutant formed biofilms that were indistinguishable from those of the comD and comE mutants (Fig. 2).
Two biofilm-defective phenotypes suggest the existence of a second CSP receptor.
Our results clearly showed that individual inactivation of the comC, comD, comE, or comX gene resulted in the formation of two types of abnormal biofilms, one with a defect in the formation of the wild-type architecture or a thin biofilm with a reduced biomass. These results suggested that there might be a second receptor in S. mutans which sensed the same CSP but activated a different pathway, possibly related to cell septation or separation. To test this hypothesis, we made a comCDE deletion mutant, a strain unable to produce and respond to the CSP through the identified pathway. The comCDE deletion mutant was defective in genetic competence, resulting in a significant impairment in natural transformation whether or not synthetic CSP was added (data not shown). The comCDE deletion mutant formed a biofilm with a reduced biomass which was similar to the comD, comE, and comX mutant biofilms, but its architecture appeared more like that of the comC mutant biofilm (Fig. 4). Addition of synthetic CSP to the culture of the comCDE deletion mutant partially restored the wild-type biofilm architecture, also suggesting the existence of a second receptor that interacted with CSP. Growth kinetics of the comCDE mutant in liquid cultures showed that it had a significantly reduced growth rate (doubling time, 1.92 h−1) and yield compared to those of the wild-type strain (1.2 h−1) (P < 0.01) (Fig. 6). As expected, partial complementation of the comCDE mutant, strain SMCDE-L4, with pCOMC generated a biofilm phenotype similar to that of the comD and comE mutants (Fig. 2 and 4). Addition of the synthetic CSP into the SMCDE-L4 culture resulted in a biofilm phenotype similar to that of SMCC1-pCOMC (data not shown); SMCDE-L4 also had an increased growth yield but showed no change in growth rate (Fig. 6). It was likely that the increased growth yield resulted from decreased formation of aggregates in the liquid culture after the addition of the CSP. Aggregate formation by the parent and comD and comE mutant cultures appeared to have a minimal effect on growth rate measurements.
DISCUSSION
S. mutans uses a typical gram-positive CSP secretion and detection system for sensing cell density and becoming genetically competent (30). It was recently shown that the CSP-mediated regulon mediates other functions, including the activation of a cellular response that increases resistance to acid (30). The present work describes the effects of mutations in several com loci with regard to biofilm formation. Our results have clearly shown that inactivation of any one of the genes encoding the components of the quorum-sensing system results in the formation of an abnormal biofilm. In particular, the comC mutant (unable to produce or secrete the signal peptide [CSP]) formed biofilms lacking the wild-type architecture whereas the comD and comE mutants defective in sensing and responding to the signal peptide formed biofilms with a reduced biomass. We also examined a mutant defective in cslAB (comAB) and found that it had the same phenotype as the comC mutant (data not shown). Previous work by other labs has demonstrated that a defect in streptococcal TCSs may generate the phenotype of a decreased biofilm biomass (1, 32). The present study demonstrates conclusively that the comCDE system is directly connected to the ability of S. mutans to form biofilms.
Molecular genetic dissection of biofilm development has revealed that the formation of biofilms is a complex, dynamic process that involves multiple, convergent signaling pathways (34). A general pattern of biofilm formation includes initial adherence of cells to a solid surface, followed by multiplication of the adherent cells, formation of microcolonies, production of an extracellular polysaccharide matrix, and finally, differentiation into a three-dimensional biofilm (28, 33). Some “mature” biofilms are characterized by the formation of mushroom- or tower-shaped structures with intervening water channels that allow the flow of nutrients into and waste products out of the biofilms (8, 23). Recent works have discovered that cell-cell communication through quorum sensing influences the formation of mature biofilms by P. aeruginosa (11, 12, 35). A mutation that blocked the generation of the signal molecules hindered the differentiation of P. aeruginosa biofilms and resulted in a biofilm lacking differentiated structure. Such an undifferentiated biofilm may be more susceptible to treatment by antimicrobial agents (11). Thus, at least some bacteria must not only be able to sense surfaces and nutritional conditions for the transition to biofilm life but also apparently require cell-to-cell signaling and other coordinated activities to form differentiated, mature communities. Despite the significant advances that have been made in our understanding of quorum sensing in biofilm differentiation and maturation of gram-negative bacteria, little is known of cell-to-cell signaling systems or networks in biofilm formation by gram-positive organisms.
The data from our study show that the architectural change in the comC mutant biofilms may be associated with a defect in cell separation. Because of this defect, the comC mutant tends to form extremely long chains that result in the formation of large aggregates or web-like biofilms. The molecular mechanisms that regulate cell separation modulated by the CSP gene remain unclear, since the comD and comE mutants defective in sensing and responding to the synthetic CSP do not exhibit a phenotype similar to that of the comC mutant. These data suggest that there may be a second receptor that also responds to the same signal peptide (CSP) as the comD sensor protein but apparently affects a process responsible for other phenotypes, such as cell separation. The mechanisms governing cell separation in gram-positive bacteria are unresolved, yet it has been suggested that separation involves both lytic and mechanical processes (15, 38). The partial restoration of the wild-type phenotype by CSP in the comC mutant may have resulted from depletion of an effective CSP concentration in the mutant cultures; only one dose of the CSP was used throughout the 16-h incubation, whereas wild-type cells presumably continuously secrete CSP. This idea was strengthened when we compared the growth of the genetically complemented comC mutant with that of the parent strain; little difference was observed (data not shown).
Predictably, the comX mutant displayed a phenotype similar to that of the comD and comE mutants, providing evidence for a link in S. mutans between the signal peptide and signal transduction via comX. comX may encode an alternate sigma factor that directs transcription of a number of late competence-specific genes, similar to its homolog in S. pneumoniae (25). S. mutans comX appears to be related to S. pneumoniae comX in that inactivation leads to a competence-defective phenotype (data not shown). However, there are apparent differences between the species since S. pneumoniae has two copies of comX, both of which must be inactivated to abolish competence. In S. mutans strain NG8, we also found two copies of comX (unpublished data); yet inactivation of one copy (comX1) rendered the resultant mutant transformation defective. DNA microarray analyses of expression patterns of CSP-induced genes in S. pneumoniae revealed that a number of genes apparently unrelated to competence are under the regulatory influence of comX (13, 37). Most of these genes have a “com-box” TACGAATA sequence located in their promoter region (25). We have discovered a similar com-box 5′ of and proximal to many S. mutans open reading frames, including several that encode late-competence gene homologs (unpublished data). S. mutans also appears to have multiple CSP-mediated phenotypes, including genetic competence (30), acid tolerance (29), and, as we have just demonstrated, biofilm formation. Identification of the genes encompassing the Com regulon is under way.
It was previously shown that inactivation of the genes (comD or comE) encoding a TCS resulted in concomitant defects in genetic competence and growth yield (30). Both comD and comE mutants grown in fluid cultures had decreased cell densities compared to that of the wild-type strain, suggesting that the TCS plays an important role in the detection of population density. This phenomenon is also apparent with the comD and comE mutants that formed biofilms with decreased biomasses compared to that of the wild-type strain. Previous studies showed that biofilm formation by S. mutans grown in the controlled environment of our model system was characterized by a rapid accumulation phase following initial adherence to surfaces (28, 30). During this active accumulation phase, the number of biofilm cells increased exponentially, which accounted for most of the biomass in the first 20 h of cell accumulation. A similar active accumulation phase of S. gordonii in an in vivo model was also noted by Bloomquist et al. (2) and Liljemark et al. (31), who found that density-dependent cell multiplication contributes to 90% of the biomass in the first 24 h of dental plaque formation. These authors also showed that a cell-free supernatant taken from growing S. gordonii cultures could induce an exponential increase in the incorporation of [3H]methyl-thymidine into biofilm cells, suggesting that a cell-cell signaling mechanism was activated. More recently, Loo et al. (32) observed that a Tn916 insertion in the comD homolog of S. gordonii resulted in a phenotype defective in biofilm formation similar to that which we observed with S. mutans. Those authors suggested that cell-cell signaling involving the TCS involved in genetic competence might be important in the formation of S. gordonii biofilms. Thus, evidence from both in vivo and in vitro studies supports the concept that signal peptide-mediated cell-cell signaling functions to regulate the formation of biofilms by gram-positive streptococci.
Inactivation of any gene encoding a component of the quorum-sensing system was expected to produce the same phenotypic effect on biofilm initiation and formation. However, the mutants unable to produce or secrete the CSP formed biofilms that differed from those formed by the mutants defective in the comD or comE gene. After analysis of the S. mutans comC, comD, and comE loci, we found that the orientation of the genes in this region was clearly different from that of previously described orthologs in other streptococci. In S. pneumoniae, Streptococcus mitis, Streptococcus oralis, S. gordonii, and Streptococcus sanguinis, the comC, comD, and comE genes are organized as an operon in which the genes are arranged beginning with comC and followed immediately 3′ by comD and comE (6). In S. pneumoniae, all three genes are transcribed together and phosphorylated ComE acts as a transcriptional activator that binds to its own promoter region 5′ of and proximal to the comC gene (42). In S. mutans, however, the comC gene is encoded divergently on the strand complementary to the comD and comE genes (30). There are also promoter-like sequences observed 5′ of and proximal to both the comC and comE genes (30). Therefore, it is possible that transcription of comC in S. mutans may not only be coregulated with comDE for the induction of genetic competence but may also be divergently regulated to signal an unknown pathway related to other cell density-dependent phenotypes. Investigation of the regulation of these genes is under way in our laboratory.
Based on the results of previous work (29) and our results from the present study, we propose a two-receptor cell-cell signaling model to illustrate how the quorum-sensing system in S. mutans functions to regulate both genetic competence, acid tolerance, and biofilm formation (Fig. 1). The principle of this model is that the signal peptide (CSP) encoded by comC can simultaneously interact with two cognate receptors, one encoded by comD and the other encoded by an unknown gene. These receptors likely transfer the input signal through two different pathways. Environmental cues, such as nutrient concentration, pH, and adherence to a surface, may modulate the level of the CSP or the effectiveness of CSP interaction with the receptors. As cell density in a growing culture or biofilm increases, basal expression of comC may cause the absolute concentration of CSP outside the cells to increase and reach a threshold concentration at which CSP can effectively activate the cognate sensor proteins. In S. pneumoniae, the activated HK sensor protein (ComD) activates its cognate regulator (ComE) via phosphorylation (20). Phosphorylated comE, in turn, is known to activate at least two competence-specific operons, comAB and comCDE, as well as the comX gene, resulting in the induction of genetic competence (20, 25). This forms a positive feedback loop for quorum sensing. In S. mutans, the CSP may interact with the HK product of comD and a second receptor that can activate a signal cascade that is apparently associated with cell separation. The genes encoding the components involved in the second pathway are still unknown. Their identification could provide insight into the mechanism of S. mutans biofilm formation and potentially facilitate the development of therapeutic agents to control biofilm-mediated infections by S. mutans.
Acknowledgments
We thank Robert Chernecky for SEM technical services and Donald Morrison for S. pneumoniae CSP-1.
Our work was supported by PHS grant DE 013230-02 from the National Institute of Dental and Craniofacial Research, grant MT-15431 from the Canadian Institutes of Health Research, and infrastructure grants from the Canadian Foundation for Innovation and The Ontario Innovation Trust. D.G.C. is supported by a Canada Research Chair. R. P. Ellen is a member of the CIHR group in Matrix Dynamics.
REFERENCES
- 1.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 2.Bloomquist, C. G., B. E. Reilly, and W. F. Liljemark. 1996. Adherence, accumulation, and cell division of a natural adherent bacterial population. J. Bacteriol. 178:1172-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckley, N. D., L. N. Lee, and D. J. LeBlanc. 1995. Use of a novel mobilizable vector to inactivate the scrA gene of Streptococcus sobrinus by allelic replacement. J. Bacteriol. 177:5028-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burne, R. A., Y. Y. Chen, and J. E. Penders. 1997. Analysis of gene expression in Streptococcus mutans in biofilms in vitro. Adv. Dent. Res. 11:100-109. [DOI] [PubMed] [Google Scholar]
- 5.Carlsson, J., G. Soderholm, and I. Almfeldt. 1969. Prevalence of Streptococcus sanguis and Streptococcus mutans in the mouth of persons wearing full-dentures. Arch. Oral Biol. 14:243-249. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, Q., E. A. Campbell, A. M. Naughton, S. Johnson, and H. R. Masure. 1997. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 23:683-692. [DOI] [PubMed] [Google Scholar]
- 7.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 9.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cvitkovitch, D. G., D. A. Boyd, T. Thevenot, and I. R. Hamilton. 1995. Glucose transport by a mutant of Streptococcus mutans unable to accumulate sugars via the phosphoenolpyruvate phosphotransferase system. J. Bacteriol. 177:2251-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 12.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 15.Giesbrecht, P., T. Kersten, H. Maidhof, and J. Wecke. 1997. Two alternative mechanisms of cell separation in staphylococci: one lytic and one mechanical. Arch. Microbiol. 167:239-250. [DOI] [PubMed] [Google Scholar]
- 16.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 17.Gutowski-Eckel, Z., C. Klein, K. Siegers, K. Bohm, M. Hammelmann, and K.-D. Entian. 1994. Growth phase-dependent regulation and membrane localization of SpaB, a protein involved in biosynthesis of the lantibiotic subtilin. Appl. Environ. Microbiol. 60:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Håvarstein, L. S., and D. A. Morrison. 1999. Quorum sensing and peptide pheromones in streptococcal competence for genetic transformation, p. 9-26. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 21.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleerebezem, M., L. E. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal- transduction systems in gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, J. R., D. R. Korber, B. D. Hoyle, J. W. Costerton, and D. E. Caldwell. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeBlanc, D. J., L. N. Lee, and A. Abu-Al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, S. F., Y. H. Li, and G. H. Bowden. 1996. Detachment of Streptococcus mutans biofilm cells by an endogenous enzymatic activity. Infect. Immun. 64:1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. F., A. Progulske-Fox, G. W. Erdos, D. A. Piacentini, G. Y. Ayakawa, P. J. Crowley, and A. S. Bleiweis. 1989. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II). Infect. Immun. 57:3306-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y. H., and G. H. Bowden. 1994. Characteristics of accumulation of oral gram-positive bacteria on mucin-conditioned glass surfaces in a model system. Oral Microbiol. Immunol. 9:1-11. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y. H., M. N. Hanna, G. Svensäter, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Y.-H., P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liljemark, W. F., C. G. Bloomquist, B. E. Reilly, C. J. Bernards, D. W. Townsend, A. T. Pennock, and J. L. LeMoine. 1997. Growth dynamics in a natural biofilm and its impact on oral disease management. Adv. Dent. Res. 11:14-23. [DOI] [PubMed] [Google Scholar]
- 32.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 34.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 35.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen, F. C., and A. A. Scheie. 2000. Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral Microbiol. Immunol. 15:329-334. [DOI] [PubMed] [Google Scholar]
- 37.Peterson, S., R. T. Cline, H. Tettelin, V. Sharov, and D. A. Morrison. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 182:6192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronda, C., J. L. Garcia, E. Garcia, J. M. Sanchez-Puelles, and R. Lopez. 1987. Biological role of the pneumococcal amidase. Cloning of the lytA gene in Streptococcus pneumoniae. Eur. J. Biochem. 164:621-624. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro, J. A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81-104. [DOI] [PubMed] [Google Scholar]
- 40.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 41.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ween, O., P. Gaustad, and L. S. Håvarstein. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33:817-827. [DOI] [PubMed] [Google Scholar]