Abstract
Open reading frame orf192, which is located immediately upstream of the aerobic repressor gene crtJ, was genetically and biochemically demonstrated to code for a second aerobic repressor (AerR) of photosynthesis gene expression in Rhodobacter capsulatus. Promoter-mapping studies indicate that crtJ has its own promoter but that a significant proportion of crtJ expression is promoted by read-through transcription of orf192 (aerR) transcripts through crtJ. Disruption of aerR resulted in increased photopigment biosynthesis during aerobic growth to a level similar to that of disruption of crtJ. Like that reported for CrtJ, β-galactosidase assays of reporter gene expression indicated that disruption of aerR resulted in a two- to threefold increase in aerobic expression of the crtI and pucB operons. However, unlike CrtJ, AerR aerobically represses puf operon expression and does not aerobically repress bchC expression. Gel mobility shift analysis with purified AerR indicates that AerR does not bind to a bchC promoter probe but does bind to the crtI, puc, and puf promoter probes. These results indicate that AerR is a DNA-binding protein that targets genes partially overlapping a subset of genes that are also controlled by CrtJ. We also provide evidence for cooperative binding of AerR and CrtJ to the puc promoter region.
Oxygen tension is an important factor regulating synthesis and assembly of the photosynthetic apparatus in photosynthetic bacteria (7). Oxygen suppression of photosystem synthesis is mainly attributed to the activation or inactivation of many transcription factors that regulate photosynthesis gene expression. One well-characterized, oxygen-regulated transcription factor is the aerobic repressor CrtJ, which in the presence of oxygen represses bacteriochlorophyll (bch), carotenoid (crt), and respiratory (29) and light harvesting-II (puc) (22) gene expression. It has recently been demonstrated that exposure of CrtJ to oxygen results in the formation of an intramolecular disulfide bond and that formation of this bond is needed for binding of CrtJ to its target promoters (S. Masuda, C. Dong, D. Swem, and C. E. Bauer, unpublished data). Another well-characterized redox regulator is the sensor kinase RegB, which, together with its cognate response regulator RegA, is responsible for the global control of many aerobically and anaerobically regulated cellular processes (3). This includes photosynthesis (17, 26), nitrogen fixation (8), hydrogen utilization (8), carbon fixation (32), respiration (29, 30), and cytochrome biosynthesis (29, 30).
In this study, we have identified a new transcription factor coded by an open reading frame (orf192) located just upstream of crtJ in the previously sequenced Rhodobacter capsulatus photosynthesis gene cluster (1). Disruption of orf192 indicates that it codes for an aerobic repressor (AerR) of carotenoids and bacteriochlorophyll and for the reaction center and light-harvesting apoproteins. The results of gel mobility shift assays indicate that AerR binds to the pufQ, crtA-crtI, and pucB promoters. Additionally, we show that AerR and CrtJ cooperatively interact with a subset of promoters that are aerobically repressed by both of these repressors.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The wild-type R. capsulatus strain SB1003 (33), the regA-disrupted strain MS01 (26), and the crtJ-disrupted strain CD2-4 (Dong et al., unpublished) have been described previously (33). Escherichia coli strain DH5α (Novagen) was used for routine DNA cloning, and BL21(DE3)/pLysE was used for protein overproduction. E. coli strains S17-1 λ-pir (27) and HB101/pDPT51 (31) were used to mobilize plasmids into R. capsulatus.
R. capsulatus strains were cultured photosynthetically under anaerobic conditions or aerobically at 34°C in PYS medium (34). E. coli strains were grown in Luria broth (24) or in Terrific broth (24) at 37°C. Appropriate antibiotics were used for plasmid and strain selections, with concentrations of 50 μg/ml for ampicillin and for kanamycin in E. coli. For R. capsulatus strains, antibiotic concentrations were 10 μg/ml for kanamycin, 10 μg/ml for spectinomycin, 10 μg/ml for gentamicin, and 50 μg/ml for rifampin.
Construction of aerR and crtJ reporter plasmids.
Several lacZ-based reporter plasmids were constructed to assay the transcription of aerR and crtJ. The initial construction involved PCR amplification of a 3.2-kb aerR-crtJ DNA segment using primers Ose5 (5′-GAGCTCATCGCGTCCCCCGTT-3′) and Ose6 (5′-GGTGGAAACGGTCCTGGAGCA-3′). The amplified product was cloned into the SrfI site of pPCR-Script SK(+) vector (Stratagene), creating the plasmid pES3. Fragments (0.64-kb NruI-MscI, 1.13-kb EcoRV-MscI, 0.74-kb MscI-HincII and 1.9-kb EcoRV-HincII fragments) were subcloned from pES3 and into plasmid pUC119 at the SmaI site by blunt-end ligation, yielding plasmids pES11, pES12, pES13, and pES14, respectively. Reporter plasmids pES15 and pES16 were constructed by insertion of the 0.64-kb KpnI-XbaI DNA fragment from pES11 and the 1.13-kb KpnI-XbaI fragment from pES12, respectively, into the KpnI and XbaI sites of the lacZ reporter plasmid pPHU236 (11). Reporter plasmids pES17 and pES18 were constructed by inserting the 0.74-kb KpnI-XbaI fragment from pES13 and the 1.9-kb KpnI-XbaI fragment from pES14 into KpnI-XbaI sites of pPHU234 (11). Reporter plasmids pES16 and pES18 also underwent deletion of a 0.9-kb KpnI-XhoI fragment, treated with T4 DNA polymerase and then blunt-end ligated, to construct reporter plasmids pESM16 and pESM18, respectively. The plasmids were introduced into the wild-type strain SB1003 by triparental mating as described by Nickens and Bauer (19).
Gene disruptions.
A deletion of aerR, as well as a deletion of both aerR and crtJ, was constructed by the homologous recombination of a kanamycin resistance (Kanr) cassette into the chromosome. For constructing aerR deletion strains, a 1.1-kb BamHI fragment from pES3 was cloned into a BamHI site in pGEM-7Zf(+) (Promega), yielding plasmid pES7/8. A 346-bp BglII-StyI fragment from pES7/8 was blunt-end replaced with a 1.4-kb SmaI Kanr cassette that was isolated from pUC4KIXX (2) (Pharmacia Fine Chemicals). One resulting plasmid, pES7, has Kanr in the same orientation as aerR-crtJ, and the second plasmid, pES8, has the Kanr cassette in the opposite orientation to that of aerR-crtJ.
An aerR-crtJ double-deletion strain was made by replacing both genes with the Kanr cassette. In brief, a 567-bp fragment upstream of aerR was PCR amplified by using primers 5′-GGAGTTCGGCATAGTGGC A-3′ and 5′-GGAGATCTCCAAGCTCCGGTCCCG-3′, which introduced a BglII site (restriction sites are underlined) to the primer that was closest to aerR. A 729-bp fragment downstream of crtJ was also PCR amplified by using primers 5′-CCAGATCTCAACGCCTGCTCCAGGAC-3′ and 5′-CCTCTAGATAGATCGACGGCGTGATCGA-3′, with a BglII site present in the primer closest to the crtJ coding region and an XbaI added to the primer furthest from crtJ (underlined). The two DNA segments were subcloned into pBluescript II SK(±) separately to generate pBluescript::uporf19 2 and pBluescript::downcrtJ. The BglII-HindIII fragment from pBluescript::downcrtJ and a Kanr DNA fragment flanked by BamHI-cut sites that was isolated from pBSL15 were together inserted into pBluescript::uporf192 between BglII and XbaI sites to generate the plasmid pCD3, which has the Kanr cassette in the same orientation as aerR-crtJ. This construct has the Kanr DNA fragment in the same orientation as aerR-crtJ.
For disruption of the chromosomal copy of aerR, plasmids pES7 and pES8 were introduced into HB101/pDPT51 and then transferred into SB1003 by biparental conjugation as described earlier by Young et al. (34). Allelic replacement of the aerR::Kanr DNA segments from plasmids pES7 and pES8 into the genome of SB1003 generated strains ES7 and ES8, respectively. The aerR-crtJ deletion strain was constructed by homologous recombination using gene transfer agent-mediated allelic replacement (25) of the aerR::Kanr::crtJ DNA segment from plasmid pCD3 into the chromosome of SB1003, resulting in strain CD3. Each of the gene disruptions was confirmed by PCR amplification and DNA sequence analysis.
Immunodetection analysis.
In vivo levels of CrtJ in the wild-type strain SB1003 and in the aerR-disrupted strains ES7 and ES8 were measured by the addition of an in-frame FLAG epitope to the carboxyl terminus of CrtJ (Scientific Imaging Systems; Eastman Kodak Co.). For epitope plasmid construction, the CrtJ coding sequence was PCR amplified using primers, 5′-CAAGCTTATGCGACGGGAGGCCTTG-3′ and 5′-GCAGATCTCCGCGTCCTTCGACAACAG-3′, which contain HindII and BglII sites, respectively (underlined). The amplified segment was cloned into pBluescript II SK(±), from which a HindIII-BglII fragment was then subcloned into HindIII and BamHI sites in pJM23 (12). The resulting construct had crtJ fused in frame with a 7-amino-acid-long FLAG epitope followed by a stop codon. A CrtJ-FLAG DNA fragment was then isolated as a HindIII-SpeI fragment, blunt ended, and subcloned into an SmaI site of the suicide vector pZJD3 (12). The resulting plasmid was then recombined into the chromosomes of strains SB1003, ES7, and ES8 by homologous recombination by selecting for gentamicin resistance. Proper integration was confirmed by PCR amplification and DNA sequence analysis.
The expression levels of FLAG-tagged CrtJ in the recombinant strains SB1003-FLAG, ES7-FLAG, and ES8-FLAG were examined by Western blot analysis by using the FLAG epitope-specific monoclonal antibody M2 as a primary antibody as described by the supplier (Sigma) and a chemiluminescence-based horseradish peroxidase-linked secondary antibody (ECL Western blotting detection system; Amersham Pharmacia Biotech). Cells were harvested at a culture density of 100 Klett units by centrifugation at 7,649 × g for 5 min. The cell pellets were resuspended in a buffer containing 50 mM Tris-HCl (pH 8.0), with equal volumes of cell suspensions subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis.
β-Galactosidase assay and spectral analysis.
Cells were grown photosynthetically or aerobically in PYS medium to a cell density of approximately 65 Klett units. For analysis of crtI, bchC, puc, and pufQ expression, cell extracts were prepared and assayed for β-galactosidase activity as described by Young et al. (34). For measuring crtJ and aerR expression, β-galactosidase activity was measured as described by Miller (16) using modifications described by Elsen et al. (10). Protein concentration was determined by the method of Bradford (6) (Bio-Rad).
For spectral analysis, cells were grown to a density of approximately 50 Klett units, chilled to 0°C, harvested by centrifugation at 8,000 × g for 10 min, and resuspended in 1/10 volume of 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA. The cells were then disrupted by sonication and clarified by centrifugation at 8,000 × g for 10 min, and the supernatant was scanned for absorbency.
AerR and CrtJ purification.
Forward and reverse primers 5′-CCATGGACCTGCTGTTCGACG-3′ and 5′-GGATCCCAGTCATACCAGAGA-3′ were used to amplify the aerR coding sequence that contains NcoI and BamHI sites (underlined), respectively. The amplified fragment was cloned into pBluescript II SK(±) and was subsequently introduced into pET32(a) (+) (Novagen Corp.) between NcoI and BamHI to generate the overexpression plasmid pET32(a) (+)::aerR. The plasmid was transformed into E. coli strain BL21(DE3) pLysE (28), and a His6-Trx-AerR fusion protein was overexpressed in 4 liters of Terrific broth by induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 25°C for 4 h. Cells were harvested and resuspended in 50 ml of ice-cold 1× loading buffer (5 mM imidazole, 0.5 M NaCl, and 20 mM Tris-HCl, pH 7.9) and were lysed by three passages through a chilled French pressure cell at 18,000 lb/in2. The lysate was clarified by centrifugation at 26,890 × g for 30 min, after which the supernatant was filtered through a 0.45-μm-pore-size Acrodisc (HT Tuffryn membrane; Gelman Sciences). The clarified supernatant was loaded on an Ni2+ column, washed with at least 40 volumes of 1× loading buffer, and eluted with 1× washing buffer (60 mM imidazole, 0.5 M NaCl, and 20 mM Tris-HCl, pH 7.9). Elution fractions were pooled and dialyzed overnight at 4°C against TPAE buffer composed of 50 mM Tris-HCl (pH 8.0), 200 mM potassium acetate, and 1 mM EDTA. The AerR protein was further purified by incubation with S-protein agarose beads at 4°C overnight and was eluted with 3 M MgCl2. The purified protein was dialyzed against TPAE buffer containing 20% glycerol overnight at 4°C and was stored at −80°C.
A His6 derivative of CrtJ that was used in gel mobility shift assays in this study was isolated from E. coli as described by Ponnampalam and Bauer (21). The percent active protein fraction was assayed as described by Ponnampalam and Bauer (21).
Gel mobility shift assay.
32P-labeled DNA probes containing either the bchC, pucB, pufQ, or crtIA promoter region were prepared by PCR amplification as previously described (21). Primers 5′-CAATTCGACCTGAAAATTCC and 5′-AAGCTCTCTCGGTTAAGTTC-3′ were used to amplify a 464-bp pucB promoter region containing two TGT-N12-ACA CrtJ-binding palindromes. Primers 5′-CGGACATTATGACGACTTGCG-3′ and 5′-TTCACCAAGGTGTCGAAACCG-3′ were used to amplify a 262-bp bchC promoter region, and primers 5′-CGCGGTTTGATCCGCCAA A-3′ and 5′-GCAGCACTGTCCATTTCGGAA-3′ were used to amplify a 263-bp pufQ promoter region. A 300-bp crtI-crtA promoter region containing two CrtJ palindromic sequences was amplified by using primers 5′-CGGCGGGACAGGATCATCT-3′ and 5′-GCCAAGGCCGGCACCGAT-3′. For testing the cooperation between CrtJ and AerR, two additional DNA fragments of each of the pucB promoter were amplified, with each of them containing only one TGT-N12-ACA palindrome. Primers 5′-CCATCGCCCGATCTGCGA-3′ and 5′-CCAGACGCATCTTTGGGCGA-3′ were used to amplify a 221-bp pucB promoter segment containing the upstream TGT-N12-ACA palindrome, and primers 5′-GCGCCACCGGCC CCGGAAT-3′ and 5′-GCTGCCGGACCGTCTGTGAT-3′ were used to amplify a 181-bp pucB promoter segment containing the downstream palindrome. All of the DNA probes described above were purified by electrophoresis in a 5% nondenaturing polyacrylamide gel and were recovered by electroelution.
For gel mobility shift assays, various amounts of purified AerR protein were added to 20 μl of reaction buffer composed of 4 fmol of 32P-labeled DNA probe, 0.6 μg of heparin as a competitor (800- to 1,000-fold excess by weight relative to the DNA probe), 5 mM HEPES (pH 7.8), 4 μg of bovine serum albumin/ml, 2% glycerol, 0.2 mM dithiothreitol, 2 mM MgCl2, and 20 μM EDTA. The reaction mixture was incubated at room temperature for 30 min and was subjected to a nondenaturing 4% Tris-glycine-EDTA-buffered polyacrylamide gel and was electrophoresed at room temperature for 2 h at 20 mA.
For cooperative binding of CrtJ and AerR, different amounts of purified His-tagged CrtJ were incubated with a 32P-labeled DNA segment of pucB or crtI promoter in the presence or absence of Trx-AerR. The relative amount of radioactivity in each shifted band was quantitated using a PhosphorImager (Molecular Dynamics) with the percentage of DNA probes bound to protein plotted versus the CrtJ concentration. Curves were plotted with a SigmaPlot program using the equation F = Y0 + a/{1 + exp[− (x − x0)/b]} (c) from which the 50% effective concentration (EC50) was estimated.
RESULTS
Expression patterns of aerR and crtJ.
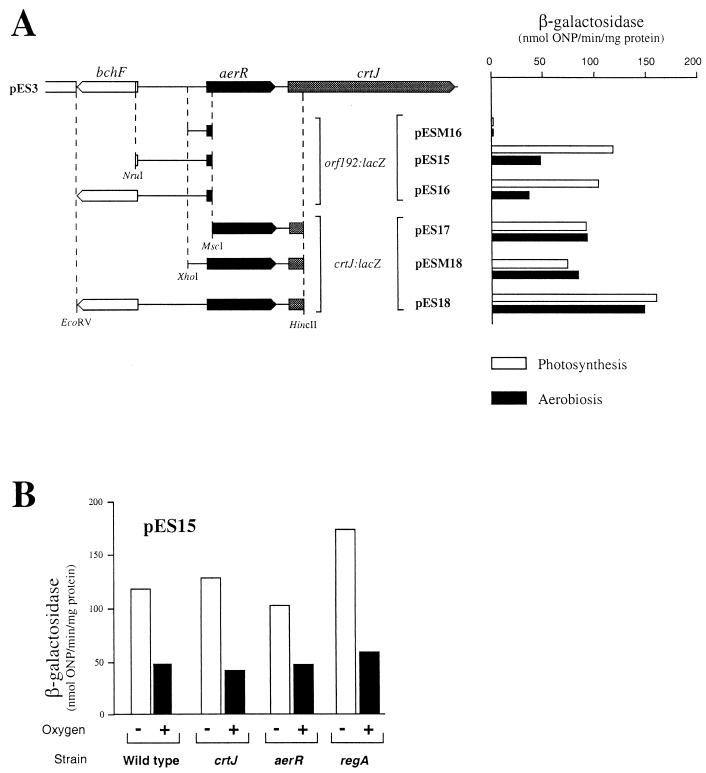
Series of plasmid-encoded lacZ translational fusions to aerR and to crtJ were constructed to assay the aerobic versus anaerobic expression pattern as well as to address the issue of promoter location (Fig. 1). The location of the aerR promoter was determined by constructing three plasmids, pESM16, pES15, and pES16. Plasmid pESM16 contains 162 bp of DNA upstream of the aerR coding region. SB1003 cells harboring this plasmid do not exhibit β-galactosidase activity above that which was observed with the vector alone, indicating that this segment does not contain the aerR promoter region. In contrast, SB1003 cells containing plasmids pES15 and pES16, which contain 590 and 1,080 bp of DNA upstream of aerR, respectively, exhibit significant amounts of β-galactosidase activity. The expression pattern of both strains is the same, with 2.5-fold-higher levels of aerR::lacZ expression in photosynthetically (anaerobically) grown cells over that observed with aerobically grown cells (Fig. 1A). We also addressed whether AerR, CrtJ, or RegA may be responsible for oxygen-regulated expression of aerR by assaying expression of the aerR::lacZ reporter plasmid pES15 in various regulatory mutants. As seen in the bar graph in Fig. 1B, oxygen-regulated expression of aerR is retained in strains that are disrupted for AerR, CrtJ, and RegA. This indicates that there is an additional unidentified oxygen-responding regulator that controls expression of aerR.
FIG. 1.
Expression patterns of aerR and crtJ. (A) β-Galactosidase activities of various aerR and crtJ reporter plasmids in the wild-type strain SB1003 cells grown under aerobic (black bars) and anaerobic (open bars) conditions. ONP, o-nitrophenyl-β-d-galactopyranoside. (B) Anaerobic and aerobic β-galactosidase activities present in various regulatory mutant strains containing plasmid pES15. Wild type is strain SB1003, crtJ is strain CD2-4, aerR is strain ES8, and regA is strain MS01. β-Galactosidase values represent the average of at least three independent assays.
Plasmids pES17, pESM18, and pES18 were constructed to measure the expression pattern of crtJ. Plasmid pES17, which contains a crtJ::lacZ fusion preceded by the aerR-crtJ intergenic region as well as a large portion of the aerR coding region, exhibits significant crtJ expression. This indicates that crtJ has its own promoter, which is located either in the intergenic region between aerR and crtJ or within the aerR coding sequence (Fig. 1A). Unlike the aerR promoter, the presence or absence of oxygen (Fig. 1A) does not affect the crtJ promoter. aerR transcripts clearly affect crtJ expression, since the β-galactosidase activity of the crtJ::lacZ fusion plasmid pES18, which contains both the aerR and crtJ promoter regions, is reproducibly higher than the level of expression observed with plasmids pES17 and pESM18, which contain only the crtJ promoter region. This suggest that read-through of the aerR transcript may impart an important contribution to crtJ expression.
Phenotypic effects of aerR disruption.
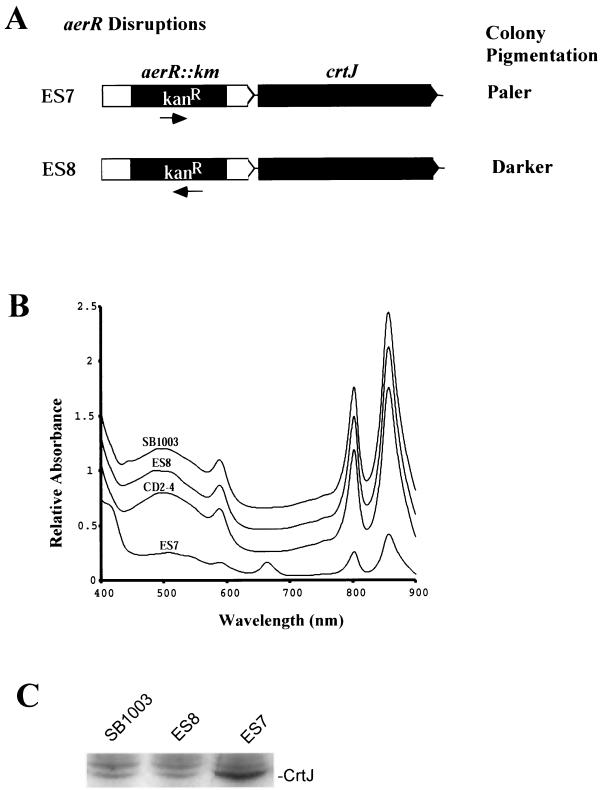
The phenotypes of two aerR-disrupted strains were characterized. Strain ES7 was constructed; it replaced codons 59 to 175 of aerR with a DNA fragment coding for a Kanr gene that is in the same orientation as aerR and crtJ. Strain ES8 has the same aerR DNA segment (codons 59 to 175) replaced with the same Kanr gene, which is in the orientation opposite to that of aerR and crtJ (Fig. 2A).
FIG. 2.
Assays for polarity of the aerR::Kanr insertion mutations. (A) Strains ES7 and ES8 contain the same Kanr gene insertion in two different orientations. (B) Spectral analysis of membrane fractions from the wild-type strain SB1003, the two aerR-disrupted mutants ES7 and ES8, and the crtJ-disrupted strain CD2-4, grown under anaerobic conditions. (C) Immunoblot analysis of the amounts of FLAG CrtJ synthesized in strains SB1003-FLAG, ES8-FLAG, and ES7-FLAG.
Phenotypic differences between aerobically grown colonies of strains ES7 and ES8 are striking. Strain ES7 exhibits pale, poorly pigmented colonies, whereas strain ES8 produces colonies that are slightly more pigmented in the central region of the colony than are observed with wild-type cells (data not shown). Spectral analysis of photopigments present in membrane fractions of dark anaerobically grown cells indicated that strain ES7 synthesized significantly lower amounts of bacteriochlorophyll and carotenoid photopigments than did wild-type SB1003 cells (Fig. 2B). Indeed, pigment production in ES7 is reduced to the extent that this strain is incapable of photosynthetic growth under low-illumination conditions of 6.8 μM/m2. Strain ES7 also accumulates significant amounts of an uncharacterized pigment at 666 nm. In contrast to the spectral alterations observed with strain ES7, anaerobically grown ES8 cells exhibited wild-type levels of photopigment production (Fig. 2B) and normal photosynthetic growth capabilities under anaerobic conditions.
To address the different phenotypes exhibited by the aerR-disrupted strains, we explored the possibility of polar effects on expression of crtJ. The in vivo expression levels of CrtJ were directly assayed by constructing epitope-tagged chromosomal versions of crtJ in the wild-type strain SB1003 as well as in the aerR-disrupted strains ES7 and ES8, leading to strains SB1003-FLAG, ES7-FLAG, and ES8-FLAG, respectively. Western blot analysis was then performed on whole-cell extracts to measure CrtJ-FLAG levels using a monoclonal antibody to the FLAG epitope tag. Strain ES8-FLAG, which has the Kanr cassette in the orientation opposite to that of crtJ, had a level of CrtJ-FLAG that was indistinguishable from that of SB1003-FLAG, which had no disruption of aerR (Fig. 2C). This indicates that elevated aerobic pigment biosynthesis observed in strain ES8 is not a consequence of polarity on crtJ expression. In contrast, the amount of CrtJ in strain ES7-FLAG, which has the Kanr gene inserted in aerR in the same orientation as is crtJ, is approximately 20-fold higher, as measured by densitometric scanning of the autoradiograph, than the level observed in strain SB1003-FLAG (Fig. 2C). Presumably the Kmr gene promoter inserted into aerR in strain ES7 drives increased transcription of crtJ, which leads to its overexpression and subsequent reduction of pigment levels.
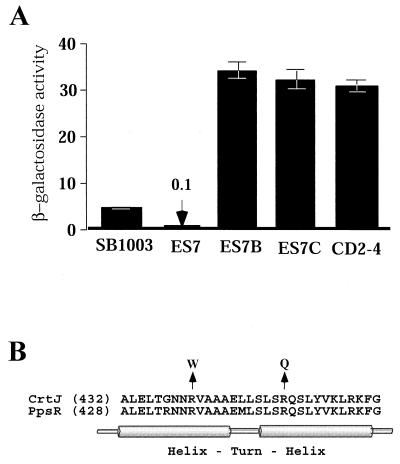
As discussed below, genetic evidence indicates that bchC expression is regulated by CrtJ but not by AerR. Thus, the effect of increasing CrtJ concentration on bch gene expression can readily be assayed by measuring β-galactosidase activity in SB1003 and ES7 cells that harbor the bchC::lacZ expression plasmid pDAY23Ω (34). The β-galactosidase levels in the bar graph in Fig. 3A show that overexpression of CrtJ in strain ES7 resulted in a 50-fold reduction in bchC::lacZ expression. This also indicates that reduced pigment biosynthesis observed in ES7 is likely caused by increased repression of bch gene expression by overexpressed CrtJ.
FIG. 3.
(A) β-Galactosidase analysis of bchC::lacZ expression from aerobically grown SB1003, ES7, ES7B, ES7C, and CD2-4 cells containing plasmid pDAY23Ω. The β-galactosidase activities are the means of three independent analyses, with the values representing micromoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein. Standard deviations are indicated by the error bars. (B) Alignment of the helix-turn-helix DNA-binding region of CrtJ and PpsR. The lengths (in amino acid numbers) of the two proteins are indicated in brackets. The mutated codons that are found in the photosynthetically competent suppressors of strain ES7 are indicated with an arrow.
To definitively test whether overexpression of CrtJ superrepresses bch gene expression, we selected for photosynthetically competent (PS+) suppressors of ES7 and then sequenced the crtJ coding sequence from two randomly selected PS+ suppressors. As shown in Fig 3B, both PS+ suppressors of ES7 contained point mutations in the helix-turn-helix DNA-binding motif of CrtJ at bases that are conserved with the Rhodobacter sphaeroides CrtJ homolog PpsR (20). One suppressor (ES7B) converted the Arg codon (Cgg) at position 440 to a Trp codon (Tgg); the other suppressor (strain ES7C) converted an Arg codon (cGg) at position 451 to a Gln codon (cAg). When grown aerobically, both point mutation strains exhibited a dark red colony phenotype indistinguishable from that of the crtJ deletion strain CD2-4 (5, 22).
We also assayed bchC::lacZ expression in the two suppressor strains ES7B and ES7C, which harbored the bchC expression plasmid pDAY23Ω. The bar graph in Fig. 3A shows that bchC::lacZ expression was significantly elevated in the two suppressor strains relative to that observed in ES7. Indeed, the level of bchC::lacZ expression observed in the suppressor strains is comparable with that observed for strain CD2-4, which contains a deletion of crtJ (Fig 3A). We can conclude from these suppressor studies that reduced pigment production in strain ES7 is indeed a consequence of constitutive suppression of bch and crt gene expression, which is caused by overproduction of CrtJ.
Disruption of aerR leads to elevated photosynthesis gene expression.
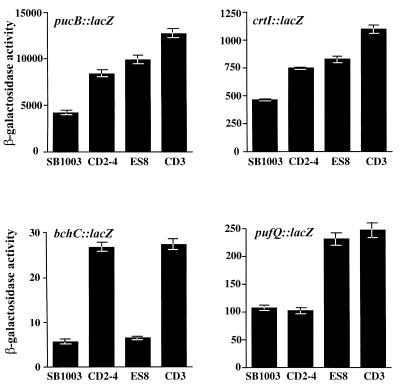
β-Galactosidase activity was analyzed from aerobically grown derivatives of strain ES8, which contained plasmids with lacZ translational fusions to different photosynthesis genes. For comparison, we assayed in parallel the gene expression patterns of wild-type cells, cells of the crtJ-disrupted strain CD2-4 (5), and cells of strain CD3, which contains a deletion of both aerR and crtJ. As shown in Fig. 4, disruption of aerR in strain ES8 resulted in elevated expression of the puc::lacZ and crtI::lacZ reporter genes to a level that is slightly above that of the crtJ-disrupted strain CD2-4. Deletion of both aerR and crtJ in strain CD3 resulted in additional elevation of puc and crtI gene expression beyond that observed with disruption of aerR or crtJ alone. Interestingly, disruption of aerR shows no effect on expression of the bchC gene as assayed with a bchC::lacZ reporter plasmid (Fig. 4). However, bchC expression was elevated in the crtJ mutant to a level similar to that previously reported, with no additional elevation in gene expression upon disruption of both aerR and crtJ. We also observed that aerR is involved in aerobic repression of the puf operon (Fig. 4), which codes for the light harvesting-I and reaction center structural genes, as based on elevated expression of a pufQ::lacZ reporter plasmid in strain ES8. This is an operon that is not under the control of CrtJ (Fig. 4) (22). Expression of the puf operon is also not enhanced in the double-deletion strain over that observed with the single disruption of aerR. Based on the different gene expression patterns exhibited by the aerR and the crtJ deletion strains, we can conclude that the phenotype exhibited by disrupting aerR is not a consequence of affecting crtJ expression.
FIG. 4.
Measurement of photosynthesis gene expression in the wild-type strain SB1003, in the crtJ-disrupted strain CD2-4, in the aerR-disrupted strain ES8, and in the strain CD3 with a deletion of aerR-crtJ. The four graphs represent aerobic expression levels of the pucB::lacZ fusion plasmid pLHIIZ (26), the crtI::lacZ fusion plasmid pCrtI:ZΩ, the bchC::lacZ fusion plasmid pDAY23Ω, and the pufQ::lacZ fusion plasmid pCB532Ω (4). β-Galactosidase activities are as described for Fig. 3.
Purified AerR exhibits DNA-binding activity.
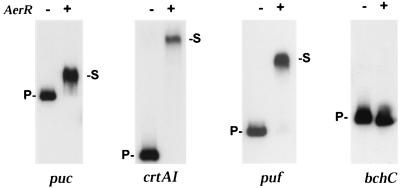
The DNA-binding activity of AerR was examined through gel mobility shift assays using affinity-purified AerR. For this analysis, we used 2.0 μM affinity-purified AerR, 4 fmol of 32P-labeled DNA probes, and a 1,000-fold excess of heparin as a competitor. The results shown in Fig. 5 indicate that AerR exhibits good in vitro binding to DNA fragments containing the pufQ, pucB, and crtA-crtI promoter regions (the crtA-crtI intergenic region contains divergent, CrtJ-regulated promoters that are located 116 bp apart). There was also no observable stable interaction of AerR with the bchC promoter region. These in vitro DNA-binding results are consistent with the in vivo β-galactosidase assays (Fig. 4), which indicate that AerR represses crtI, pufQ, and pucB expression but not bchC expression. Taken together, the in vivo and in vitro data indicate that AerR is a transcriptional repressor that directly interacts with a specific subset of photosynthesis gene promoters.
FIG. 5.
Gel mobility shift assays with purified AerR. The first lane of each gel had only 32P-labeled probe and heparin (1,000-fold excess), while the second lane had the same probe and heparin preincubated with 2 μM AerR. puc, crtAI, puf, and bchC represent the various promoter probes that were used with each probe containing two CrtJ recognition sequences. P- represents the mobility of the unshifted probe, while -S represents the mobility of the AerR-shifted probe.
Cooperative interactions between AerR and CrtJ.
To explore whether CrtJ and AerR cooperatively interact with each other, we performed in vitro gel mobility assays with puc promoter DNA probes containing various amounts of both repressor proteins. For this analysis, we used three different puc DNA probes, the lengths of which were designed based on previous studies that indicated that CrtJ binds cooperatively to two distant binding sites. The upstream CrtJ DNA-binding site is located at −279 to −296 bp, and the downstream one is located at −39 to −56 bp relative to the start site of puc transcription, as described by Elsen et al. (9). For the cooperative studies, we PCR amplified one probe containing both CrtJ palindromes that extends from −392 to +72 bp relative to the puc transcription initiation site. A second probe encompassing only the upstream palindrome was PCR amplified; it extends from −332 to −112 bp. The third probe extends from −149 to +29 bp and contains only the downstream CrtJ DNA-binding site.
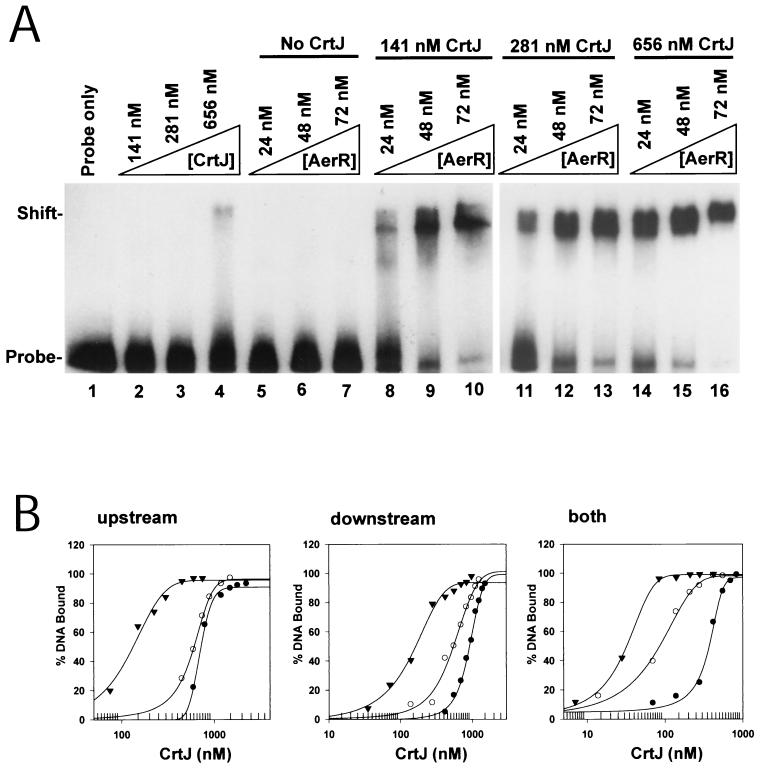
Figure 6A shows the results of a typical gel mobility shift cooperativity experiment for the puc promoter probe that contains only the downstream CrtJ DNA-binding site. Lanes 2 through 4 demonstrate that CrtJ does not bind to this probe at relatively low concentrations of 141 and 281 nM and binds only a slight percentage of shifted probe at a CrtJ concentration of 656 nM. Lanes 5 to 7 demonstrate that there is also no shift of the probe in the presence of AerR at a concentration of 24, 48, or 72 nM. However, lanes 8 to 10 show that incubation of 141 nM CrtJ, with as little as 24 nM AerR, results in a noticeable gel shift. There is nearly a complete shift of the DNA probe when 141 nM CrtJ is incubated with 72 nM AerR (lane 10). In the presence of 281 and 656 nM CrtJ (lanes 11 to 16), even less AerR is required to observe a shift of the probes. Therefore, the presence of both proteins clearly enhanced their individual DNA-binding affinities.
FIG. 6.
Cooperation between AerR and CrtJ in binding to the puc promoter region. (A) Gel mobility shift of CrtJ binding to the downstream puc palindrome in the absence or presence of AerR. Lane 1 is of probe only, while lanes 2 to 4 contain probe plus CrtJ at 141, 281, and 656 nM, respectively. Lanes 5 to 7 contain AerR at 24, 48, and 72 nM, respectively. Lanes 8 to 10 contain CrtJ at 141 nM in all lanes, as well as AerR at 24, 48, and 72 nM, respectively. Lanes 11 to 13 contain 281 nM CrtJ in each lane as well as AerR at 24, 48, and 72 nM, respectively. Lanes 14 to 16 contain 656 nM CrtJ in each lane as well as AerR at 24, 48, and 72 nM, respectively. (B) The graphs show a plot of the percentage of shifted 32P-labeled pucB probe versus the amount of CrtJ in the assay. Filled circles represent the DNA-binding isotherm of CrtJ with no AerR present, empty circles represent the CrtJ DNA-binding isotherm obtained in the presence of 24 nM AerR, and filled inverted triangles represent the CrtJ DNA-binding isotherm obtained in the presence of 48 nM AerR. The left, middle, and right graphs represent the binding curves obtained with a probe containing only the upstream CrtJ recognition palindrome, with a probe containing only the downstream recognition palindrome, and with a probe containing both recognition palindromes, respectively.
To quantitate cooperation between AerR and CrtJ, different amounts of CrtJ were incubated with the three pucB promoter probes in the presence or absence of AerR. For each of the probes, the percentage of the probe that was shifted was calculated using a PhosphorImager with the results plotted with a SigmaPlot graph. The DNA-binding curves obtained with the probe containing just the upstream CrtJ DNA-binding site (−332- to −112-bp probe) (Fig. 6B, upstream) and with the probe containing just the downstream CrtJ DNA-binding site (probe, −149 to +29 bp) (Fig. 6B, downstream) exhibited very similar EC50s for CrtJ binding to these probes in the presence or absence of AerR (EC50 is defined as the effective concentration of CrtJ needed to obtain a shift of 50% of the probe). When no AerR was added (graphs of filled circles), the upstream probe had an EC50 of 710 nM and the downstream probe had an EC50 of 875 nM CrtJ. With the addition of 24 nM AerR (graphs of open circles), the EC50s decreased to 580 and 530 nM for the upstream and downstream probes, respectively. With the addition of 48 nM AerR (graphs of inverted filled triangles), there was a further decrease in the EC50s to 185 and 155 nM for the upstream and downstream probes, respectively. The enhancing effect of AerR on CrtJ binding was increased to 15-fold for the pucB probe containing both palindromes (Fig. 6B, both). For this probe, the EC50 was reduced from 355 nM with no AerR (filled circles) to 85 and 31 nM in the presence of 24 and 48 nM AerR (empty circles and solid inverted triangles, respectively). Consistent with the β-galactosidase assay, which showed that AerR did not regulate expression from the bchC promoter (Fig. 4), the presence of AerR did not enhance CrtJ binding to the bchC probe (data not shown). This implies that AerR may cooperate with CrtJ for binding to a specific subclass of CrtJ-controlled promoters.
DISCUSSION
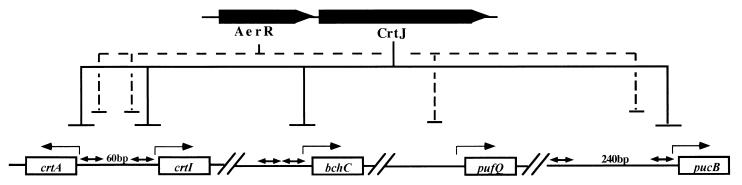
This study demonstrates that the open reading frame that is located immediately upstream of the aerobic repressor CrtJ (orf192) also codes for a repressor of photosynthesis gene expression. Expression studies indicate that some of the genes repressed by AerR are also aerobically repressed by CrtJ. For example, as shown in Fig 7, the puc and crtI promoters are both corepressed by AerR and CrtJ. In contrast, expression of the puf operon is repressed only by AerR, while the bchC operon is repressed only by CrtJ. Indeed, the different photosynthesis gene expression patterns exhibited by the aerR-disrupted strain ES8 (Fig. 4), relative to those of the crtJ-disrupted strain CD2-4, indicate that the aerR::Kanr disruption in ES8 does not exert a polar effect on crtJ expression. This conclusion is also supported by our immunoblot data (Fig. 2C), which indicate that ES8 has normal amounts of CrtJ expression.
FIG. 7.
A diagram depicting CrtJ and AerR circuits that repress individual transcripts. Both proteins affect transcription initiation (lines with a single arrowhead) at promoters that contain distantly removed CrtJ recognition palindromes (lines with double arrows). Only CrtJ represses the bchC promoter that contains two closely spaced palindromes (8 bp apart), and only AerR represses the puf promoter, which does not contain CrtJ recognition palindromes.
In contrast to the lack of polarity observed in strain ES8, strain ES7 (which contains the Kanr gene in the same orientation as crtJ) does have a significant polar effect on crtJ expression. In this strain, the amount of CrtJ is increased approximately 20-fold, which results in a significant reduction in photosystem biosynthesis. Since the difference in DNA-binding affinities between oxidized and reduced CrtJ to its target promoters is approximately fivefold (21), it seems likely that the 20-fold increase of CrtJ observed in strain ES7 is causing constitutive repression of photosynthesis gene expression. This conclusion is supported by the observation that photosynthetically competent suppressors of ES7 map to the helix-turn-helix DNA-binding motif of CrtJ, which presumably abolishes or significantly reduces the DNA-binding activity of CrtJ.
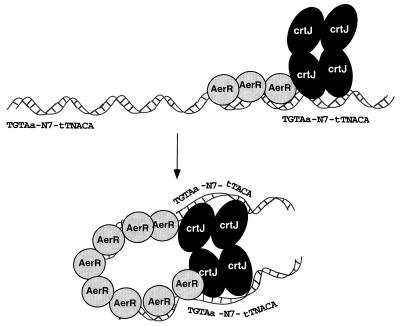
Previous studies have indicated that CrtJ-repressed promoters fall into two classes. One class, represented by the bchC promoter, has two closely spaced CrtJ recognition palindromes located just 8 bp apart, with one overlapping the −35 promoter region and the other the −10 promoter region (23). Mutational studies have demonstrated that binding of CrtJ to the bchC promoter involves cooperative interactions between CrtJ bound at the −35 palindrome and CrtJ bound at the −10 palindrome (23). Mutations that increase or decrease the spacing between these palindromes disrupt cooperative interactions between CrtJ repressors that are bound to these two sites. AerR does not appear to affect CrtJ-mediated repression of this class of promoters. The second class of CrtJ-repressed promoters also has two CrtJ recognition palindromes, with the difference being that these palindromes are distantly removed (9). In the puc operon, one CrtJ palindrome is located at the −35 promoter region and the second palindrome is located 240 bp upstream (9). In the crtA-crtI promoter region, the palindromes are 76 bp apart, with one palindrome overlapping the −35 region of the crtI promoter and the other overlapping the −10 region of the crtA promoter (9). One model is that AerR may have a role in stabilizing the binding of CrtJ, which is known to exist in solution as a tetramer, to one of the distant binding sites. Previous mutational and in vitro DNA-binding studies have indicated that CrtJ cooperatively binds to the distantly removed palindromes in the puc and crtA-crtI promoters (9). This suggests that the DNA must loop around, allowing coordinate interaction of CrtJ to these distant sites. Figure 8 depicts a model that we favor, in which AerR may have a role in bending the DNA that would facilitate the interaction of CrtJ tetramers to distantly removed recognition sites. The DNA-binding curves for CrtJ binding to the puc probe in the presence and absence of AerR initially indicate that AerR is capable of stimulating CrJ binding to the DNA probes at substoichiometric levels. However, studies have indicated that E. coli-overexpressed, purified CrtJ contains a significant fraction that is inactive for DNA binding. When correcting for the percent active fraction of CrtJ that is capable of DNA binding (Table 1 [21]), we find that an excess of AerR is actually needed to promote maximal binding of CrtJ to the full-length puc probe that contains both palindromes. Thus, it is possible that AerR may be binding to the intervening sequence between the CrtJ recognition palindromes to facilitate DNA bending. Additional DNA-binding and DNase I footprint studies are ongoing with AerR and AerR plus CrtJ to specifically address aspects of this model.
FIG. 8.
A model depicting possible interactions between CrtJ and AerR at promoters that contain distantly spaced palindromes. The number of AerR symbols drawn relative to that of CrtJ symbols is arbitrary.
TABLE 1.
Corrected EC50s for CrtJ binding to the puc promoter region in presence and absence of AerRa
| Probe | Corrected CrtJ EC50 (nM) with:
|
||
|---|---|---|---|
| 48 nM AerR | 24 nM AerR | No AerR | |
| Upstream palindrome | 14.8 | 46.4 | 56.8 |
| Downstream palindrome | 12.4 | 42.4 | 70.0 |
| Both palindromes | 2.48 | 6.8 | 28.4 |
Values are corrected for percent active fraction of CrtJ as described by Ponnampalam and Bauer (21).
In addition to functioning as a corepressor of puc expression with CrtJ, AerR also functions on its own to repress light harvesting-I and reaction center (puf) gene expression. Mutational analysis of the puf promoter region has indicated that this promoter may be under control of a repressor, as evidenced by increased aerobic expression of some promoter point mutations (18). There has also been a report of an uncharacterized protein that binds to the puf operon promoter in an oxygen-dependent manner (13, 14, 15). The identification of AerR as an aerobic repressor of the puf operon confirms that puf expression undergoes complex regulation involving aerobic repression by AerR as well as anaerobic activation by RegA. Indeed, the puf and puc operons are both aerobically repressed and anaerobically activated by unique sets of regulatory proteins. Presumably this allows for differential expression of light harvesting-I and puc under various oxygen and light intensities. Forthcoming details of how AerR interacts with its target promoters should provide an understanding of the mechanism of repression that is affected by AerR.
Acknowledgments
We thank James Smart, Danielle Swem, and Shinji Masuda for comments regarding the manuscript.
This work was supported by National Institutes of Health grant GM53940 to C.E.B.
REFERENCES
- 1.Alberti, M., D. H. Burke, and J. E. Hearst. 1995. Structure and sequence of the photosynthesis gene cluster, p. 1083-1106. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 2.Alexeyev, M. F. 1995. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques 18:52-56. [PubMed] [Google Scholar]
- 3.Bauer, C. E., and T. H. Bird. 1996. Regulatory circuits controlling photosynthesis gene expression. Cell 85:5-8. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, C. E., D. A. Young, and B. L. Marrs. 1988. Analysis of the Rhodobacter capsulatus puf operon. Location of the oxygen-regulated promoter region and the identification of an additional puf-encoded gene. J. Biol. Chem. 263:4820-4827. [PubMed] [Google Scholar]
- 5.Bollivar, D. W., J. Y. Suzuki, J. T. Beatty, J. M. Dobrowolski, and C. E. Bauer. 1994. Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J. Mol. Biol. 237:622-640. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Cohen-Bazire, G., W. R. Sistrom, and R. Y. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Comp. Physiol. 49:25-68. [DOI] [PubMed] [Google Scholar]
- 8.Elsen, S., W. Dischert, A. Colbeau, and C. E. Bauer. 2000. Expression of uptake hydrogenase and molybdenum nitrogenase in Rhodobacter capsulatus is coregulated by the RegB-RegA two-component regulatory system. J. Bacteriol. 182:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsen, S., S. N. Ponnampalam, and C. E. Bauer. 1998. CrtJ bound to distant binding sites interacts cooperatively to aerobically repress photopigment biosynthesis and light harvesting II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 273:30762-30769. [DOI] [PubMed] [Google Scholar]
- 10.Elsen, S., P. Richaud, A. Colbeau, and P. M. Vignais. 1993. Sequence analysis and interposon mutagenesis of the hupT gene, which encodes a sensor protein involved in repression of hydrogenase synthesis in Rhodobacter capsulatus. J. Bacteriol. 175:7404-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubner, P., J. C. Willison, P. M. Vignais, and T. A. Bickle. 1991. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 173:2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang, Z. Y., H. Gest, and C. E. Bauer. 1997. Chemosensory and photosensory perception in purple photosynthetic bacteria utilize common signal transduction components. J. Bacteriol. 179:5720-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klug, G. 1991. A DNA sequence upstream of the puf operon of Rhodobacter capsulatus is involved in its oxygen-dependent regulation and functions as a protein binding site. Mol. Gen. Genet. 226:167-176. [DOI] [PubMed] [Google Scholar]
- 14.Klug, G., N. Gad'on, S. Jock, and M. L. Narro. 1991. Light and oxygen effects share a common regulatory DNA sequence in Rhodobacter capsulatus. Mol. Microbiol. 5:1235-1239. [DOI] [PubMed] [Google Scholar]
- 15.Klug, G., and S. Jock. 1991. A base pair transition in a DNA sequence with dyad symmetry upstream of the puf promoter affects transcription of the puc operon in Rhodobacter capsulatus. J. Bacteriol. 173:6038-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Mosley, C. S., J. Y. Suzuki, and C. E. Bauer. 1994. Identification and molecular genetic characterization of a sensor kinase responsible for coordinately regulating light harvesting and reaction center gene expression in response to anaerobiosis. J. Bacteriol. 176:7566-7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narro, M. L., C. W. Adams, and S. N. Cohen. 1990. Isolation and characterization of Rhodobacter capsulatus mutants defective in oxygen regulation of the puf operon. J. Bacteriol. 172:4549-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nickens, D. G., and C. E. Bauer. 1998. Analysis of the puc operon promoter from Rhodobacter capsulatus. J. Bacteriol. 180:4270-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 176:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponnampalam, S. N., and C. E. Bauer. 1997. DNA binding characteristics of CrtJ. A redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 272:18391-18396. [DOI] [PubMed] [Google Scholar]
- 22.Ponnampalam, S. N., J. J. Buggy, and C. E. Bauer. 1995. Characterization of an aerobic repressor that coordinately regulates bacteriochlorophyll, carotenoid, and light harvesting-II expression in Rhodobacter capsulatus. J. Bacteriol. 177:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponnampalam, S. N., S. Elsen, and C. E. Bauer. 1998. Aerobic repression of the Rhodobacter capsulatus bchC promoter involves cooperative interactions between CrtJ bound to neighboring palindromes. J. Biol. Chem. 273:30757-30761. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 25.Scolnik, P. A., and R. Haselkorn. 1984. Activation of extra copies of genes coding for nitrogenase in Rhodopseudomonas capsulata. Nature 307:289-292. [DOI] [PubMed] [Google Scholar]
- 26.Sganga, M. W., and C. E. Bauer. 1992. Regulatory factors controlling photosynthetic reaction center and light-harvesting gene expression in Rhodobacter capsulatus. Cell 68:945-954. [DOI] [PubMed] [Google Scholar]
- 27.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposition mutagenesis in gram negative bacteria. BioTechnology 1:784-791. [Google Scholar]
- 28.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:61-89. [DOI] [PubMed] [Google Scholar]
- 29.Swem, D. L., and C. E. Bauer. 2002. Coordination of ubiquinol oxidase and cytochrome cbb3 oxidase expression by multiple regulators in Rhodobacter capsulatus. J. Bacteriol. 184:2815-2820. [DOI] [PMC free article] [PubMed]
- 30.Swem, L. R., S. Elsen, T. H. Bird, D. L. Swem, H.-G. Koch, H. Myllykallio, F. Daldal, and C. E. Bauer. 2001. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J. Mol. Biol. 309:121-138. [DOI] [PubMed] [Google Scholar]
- 31.Taylor, D. P., S. N. Cohen, W. G. Clark, and B. L. Marrs. 1983. Alignment of genetic and restriction maps of the photosynthesis region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J. Bacteriol. 154:580-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vichivanives, P., T. H. Bird, C. E. Bauer, and F. Robert Tabita. 2000. Multiple regulators and their interactions in vivo and in vitro with the cbb regulons of Rhodobacter capsulatus. J. Mol. Biol. 300:1079-1099. [DOI] [PubMed] [Google Scholar]
- 33.Yen, H. C., and B. Marrs. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 126:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young, D. A., C. E. Bauer, J. C. Williams, and B. L. Marrs. 1989. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment-binding proteins in Rhodobacter capsulatus. Mol. Gen. Genet. 218:1-12. [DOI] [PubMed] [Google Scholar]