Abstract
The lytic salmonella phage SP6 encodes a tail protein with a high degree of sequence similarity to the tail protein of the biologically unrelated lysogenic salmonella phage P22. The SP6 tail gene is flanked by an upstream region that contains a promoter and a downstream region that contains a putative Rho-independent transcription terminator, giving it a cassette or modular structure almost identical to the structure of the tail genes of coliphages K1E, K5, and K1-5. It now appears that SP6, K1-5, K5, and K1E are very closely related but have different tail fiber proteins, giving them different host specificities.
SP6 is a lytic double-stranded salmonella phage specific for Salmonella enterica serovar Typhimurium LT2 and is morphologically and biologically similar to coliphage T7. K1-5 is a recently isolated dual-specificity coliphage that can infect and replicate on the K1 and K5 strains of Escherichia coli and encodes two different tail fiber proteins that confer this extended host range (11). One tail protein is a K5 lyase that is almost identical to that of phage K5 which allows the phage to recognize and degrade the K5 polysaccharide capsule (5). The other K1-5 tail protein is an endosialidase almost identical to that of phage K1E that similarly allows the phage to attach to and degrade the K1 polysaccharide capsule (8). SP6, K1-5, K1E, and K5 are all lytic linear double-stranded DNA phages with genomes of around 40 kb and have very similar virion morphologies. Two striking features of the tail gene regions of K1-5, K1E, and K5 are a common upstream sequence that contains an SP6-like promoter and an 85-base intergenic region that contains a putative Rho-independent transcription terminator. This modular structure of the tail genes suggests that host specificity can be changed by horizontal exchange of complete tail genes.
SP6 encodes a P22-like tail protein
On the basis of the common upstream region of the tail genes of phages K1E, K5, and K1-5 and our belief that SP6 is very similar to these three phages, we speculated that SP6 may also share this similarity in the tail gene structure, so we designed a primer that would hybridize to SP6 DNA to sequence the SP6 tail fiber gene. As predicted, the primer did hybridize, and we were able to sequence the tail genes by primer walking. A 550-amino-acid open reading frame was found starting at the exact analogous position (with respect to the promoter) as the tail genes of K1-5, K5, and K1E. A BLAST (1) search found that this open reading frame shares a high degree of similarity (58% amino acid identity and 73% similarity) to the C-terminal 550-amino-acid portion of the 667-amino-acid salmonella phage P22 tail spike protein (10). The structure of the P22 tail spike has been studied extensively and serves as a model for protein folding. Like the K1-5 tail proteins, it has an enzymatic (endorhamnosidase) activity involved in degradation of the Salmonella O antigen (for reviews, see references 3 and 15).
The N-terminal 124-amino-acid sequence portion of the P22 tail spike protein was identified as the head-binding domain (12). This domain is conspicuously missing from the putative SP6 protein (Fig. 1A). Recently, a truncated version of the P22 tail spike missing the first 115 amino acids (resulting in a protein whose first amino acid corresponds with the second amino acid of the putative SP6 protein) has been used in studies of catalysis and receptor binding because it is more accessible to crystallographic analysis (2). Note that the SP6 protein is almost identical in size and contains the same regions as this stable, catalytically active, truncated P22 tail spike.
FIG. 1.
(A) Comparison of the putative SP6 tail protein and the P22 tail spike. The SP6 protein shares 73% amino acid similarity with the P22 protein but is missing the head-binding domain. (B) Comparison of the K1E/K1-5 endosialidase compared to the K1F endosialidase. Like the SP6 tail protein, the K1E/K1-5 protein is missing a potential head-binding domain compared to the K1F endosialidase. The N- and C-terminal portions of the proteins are indicated.
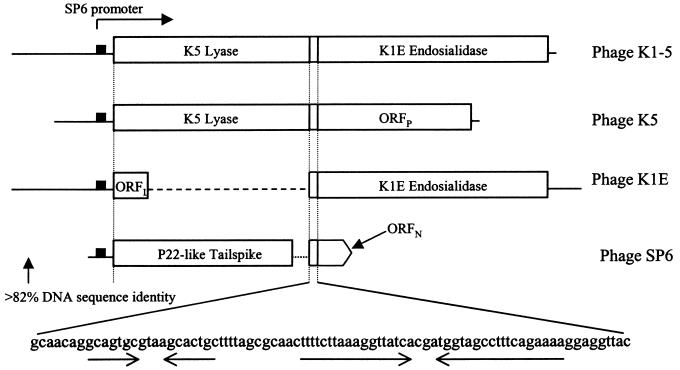
Immediately following the termination codon of the SP6 tail gene is an 84-base region very similar to that of the 85-base intergenic region of K1-5, K5, and K1E that contains two symmetrical elements (Fig. 2). Immediately downstream is the beginning of another open reading frame (ORFN). The first 11 amino acids of the ORFN protein are identical to the first 11 amino acids of the K1-5 endosialidase, but the 60-amino-acid sequence that follows is not similar to any sequence in the databases. We also determined the DNA sequence 138 bases upstream of the putative tail gene. This region is 82% identical to the upstream region of K1-5 and contains a promoter very similar to other SP6 promoters (4). When regions encoding the tail genes of these four phages are compared, we find that the SP6 gene is in a position analogous to that of the lyase genes of K1-5 and K5 and the 111-amino-acid ORFL of K1E (Fig. 2).
FIG. 2.
Arrangement of the tail gene regions of the K1-5, K1E, K5, and SP6 phages. All phages have a common upstream region with an SP6 promoter. Immediately downstream of this region, phages K1-5 and K5 encode the K5 lyase, K1E encodes the ORFL protein, and SP6 encodes a P22-like tail spike. All phages then have an intergenic region of 85 (84 for SP6) bases just after the stop codon. Immediately following this region, K1-5 and K1E encode an endosialidase, K5 encodes the ORFP protein, and SP6 encodes the small ORFN protein.
Evidence that the putative SP6 tail protein is present in SP6 virions
The putative SP6 tail protein was expressed as an N-terminal His-tagged fusion product in E. coli (BL21) using the pET-15b expression vector (Novagen). The expressed protein had a molecular mass of approximately 58 kDa (predicted molecular mass, 59.5 kDa) and was partially purified by binding a crude extract to a Ni-nitrilotriacetic acid column and eluting with 250 mM imidazole.
Sodium dodecyl sulfate-polyacrylamide gel analysis of cesium chloride-purified SP6 virions showed a protein with a molecular mass of about 58 kDa that comigrated with the overexpressed His-tagged protein (Fig. 3A). Western blotting was performed, using rabbit antiserum to phage P22 virions with the assumption that P22 antiserum would contain some antibodies to the tail spike that would cross-react with the SP6 tail protein due to the sequence similarity. In this experiment a band of the predicted molecular weight of the SP6 tail protein was seen in SP6 virions (Fig. 3B). The expressed SP6 protein also reacted with the rabbit anti-P22 serum. When P22 virions were run, a band of the predicted molecular mass of P22 tail spike (72 kDa) was detected. Since the antiserum was raised against whole phage P22 virions, a strong signal is seen for the major protein that we presume to be capsid protein. A strong signal is also seen in SP6 virions that we believe is a major capsid protein (based on its abundance). This SP6 protein may also be similar to the P22 counterpart, if not at the sequence level then perhaps at a structural level.
FIG. 3.
(A) Tris-glycine-sodium dodecyl sulfate-10% polyacrylamide gel stained with colloidal Coomassie blue (ICN). Lane 1, semipurified SP6 tail protein; lane 2, 5 × 1010 cesium chloride-purified SP6 phage particles. The arrow points to purified SP6 tail protein. (B) Western blot on nitrocellulose (Schleicher and Schuell) blocked with a solution of Tris-buffered saline, 0.02% Tween 20, and 5% nonfat dried milk. The primary antibody was rabbit anti-P22 serum, and the secondary antibody was horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Pierce). The signal was detected with Pierce SuperSignal extended signal substrate. Lane 1, 1010 SP6 particles; lane 2, 5 × 109 P22 particles; lane 3, partially pure SP6 tail protein. The top arrow points to P22 tail spike, and the bottom arrow points to SP6 tail protein.
Phage K1-5 encodes both an RNA polymerase and a helicase that are very similar to those of SP6
Sequences of two other genes of SP6 have been published. One is the RNA polymerase (7), and the other is a primase/helicase (14). We are currently in the process of sequencing the entire K1-5 genome, and part of our sequence analysis included both of these genes. A putative 874-amino-acid K1-5 RNA polymerase shares 85% amino acid identity and 93% similarity to the 875-amino-acid SP6 RNA polymerase, which is not surprising since both phages have very similar promoters. The primase/helicase genes are even more similar, with 92% identity and 96% similarity at the amino acid level (the K1-5 protein is composed of 662 amino acids, and the SP6 protein is made up of 661 amino acids). Both of these proteins are major factors in replication, so it seemed clear that the two phages probably have nearly identical life cycles.
Conclusions
SP6, K1-5, K5, and K1E are all lytic phages with very similar virion morphology, have similar-sized linear double-stranded DNA genomes with the tail genes located at one end, and encode similar and highly specific RNA polymerases. We believe that these phages are all recently derived from a common ancestor and differ mainly in the tail proteins, giving them different host specificities. Furthermore, the cassette-like structure of the tail genes suggests that the phages can rapidly change or extend host specificity by acquiring new tail genes horizontally. Juhala et al. (6) describe morons, which may be mobile genetic elements in lambdoid phages. Morons are characteristically single genes flanked by a promoter and a Rho-independent terminator. The fact that the tail genes of these phages also have potential Rho-independent terminators suggests that this may be a common characteristic of mobile modules that are important for rapid phage evolution.
Since SP6 and P22 both infect the same host, it may not be surprising that they encode similar tail proteins. However, these two phages are biologically very different; P22 is a lambdoid phage that is capable of forming lysogens, whereas SP6 is a lytic phage. The entire P22 sequence is known, and no other sequence similarities exist between it and any known SP6 sequence. It is quite likely that the tail genes were acquired horizontally by one or both phages.
One question that needs to be addressed in the future is how the tail proteins attach to the head in the SP6 and K1-5 family. The SP6 tail is completely missing the head-binding domain of the P22 protein. Similarly, the K1-5 and K1E endosialidases are missing the N-terminal 205-amino-acid region compared to the endosialidase of phage K1F, a region that shares amino acid similarity with the T7 tail fiber protein which is also known to be involved in head binding (9, 13) (Fig. 1B). Since K1-5 was shown to have two different tail proteins attached to the head (11), it is not unreasonable to assume that head attachment is based on a different mechanism. It is possible that two polypeptides are required, one of which plays more of a structural role. It appears that there is another coding region downstream of the tail gene in SP6 (ORFN) that is identical to the first 11 amino acids of the K1-5 endosialidase (Fig. 4). This could be yet another tail protein giving SP6 the ability to infect some strain still to be identified.
FIG. 4.
Sequence comparison of the ORFN protein of SP6 and the endosialidase (ENDO) of K1-5. The first 11 amino acids are identical. Identical amino acids (|), amino acids that are highly similar (:), and amino acids with lower similarity (.) are indicated between the two sequences. Gaps introduced to maximize sequence alignment (.) are indicated. It is possible that the ORFN protein is a tail fiber protein and/or could still play a structural role in attachment of the tail to the capsid.
Nucleotide sequence accession numbers
The nucleotide sequences reported here have been deposited in GenBank and assigned the following accession numbers: SP6 tail, AF478465; K1-5 RNA polymerase, AF478466; and K1-5 primase/helicase, AF478467.
Acknowledgments
We thank Anthony Poteete for kindly supplying anti-P22 serum.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baxa, U., S. Steinbacher, A. Weintraub, R. Huber, and R. Seckler. 1999. Mutations improving the folding of phage p22 tail spike protein affect its receptor binding activity. J. Mol. Biol. 293:693-701. [DOI] [PubMed] [Google Scholar]
- 3.Betts, S., and J. King. 1999. There's a right way and a wrong way: folding/misfolding and subunit assembly of the p22 tail spike. Structure Fold. Des. 7:R31-R39. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. E., J. F. Klement, and W. T. McAllister. 1986. Sequences of three promoters for the bacteriophage SP6 RNA polymerase. Nucleic Acids Res. 14:3521-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, B. R., F. Esumah, and I. S. Roberts. 2000. Cloning, expression, and purification of the K5 capsular polysaccharide lyase (KflA) from coliphage K5A: evidence for two distinct K5 lyase enzymes. J. Bacteriol. 182:3761-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 7.Kotani, H., Y. Ishizaki, N. Hiraoka, and A. Obayashi. 1987. Nucleotide sequence and expression of the cloned gene of bacteriophage SP6 RNA polymerase. Nucleic Acids Res. 15:2653-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long, G. S., J. M. Bryant, P. W. Taylor, and J. P. Luzio. 1995. Complete nucleotide sequence of the gene encoding bacteriophage E endosialidase: implications for K1E endosialidase structure and function. Biochem. J. 309:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petter, J. G., and E. R. Vimr. 1993. Complete nucleotide sequence of the bacteriophage K1F tail gene encoding endo-N-acylneuraminidase (Endo-N) and comparison to an Endo-N homolog in bacteriophage PK1E. J. Bacteriol. 175:4354-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauer, R. T., W. Krovatin, A. R. Poteete, and P. B. Berget. 1982. Phage P22 tail protein: gene and amino acid sequence. Biochemistry 21:5811-5815. [DOI] [PubMed] [Google Scholar]
- 11.Scholl, D., S. Rogers, S. Adhya, and C. R. Merril. 2001. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steinbacher, S., S. Miller, U. Baxa, N. Budisa, A. Weintraub, R. Seckler, and R. Huber. 1997. Phage P22 tail spike protein: crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267:865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steven, A. C., B. L. Trus, J. V. Maizel, M. Unser, D. A. D. Parry, J. S. Wall, J. F. Hainfield, and W. F. Studier. 1988. Molecular substructure of a viral receptor-recognition protein. J. Mol. Biol. 200:351-365. [DOI] [PubMed] [Google Scholar]
- 14.Tseng, T. Y., D. N. Frick, and C. C. Richardson. 2000. Characterization of a novel DNA primase from Salmonella typhimurium bacteriophage SP6. Biochemistry 39:1643-1654. [DOI] [PubMed] [Google Scholar]
- 15.Villafane, R. J., and K. Baksi. 1999. A tail of protein folding. P. R. Health Sci. J. 18:105-115. [PubMed] [Google Scholar]