Abstract
The proteins under the control of the two-component system VirR/VirS in Clostridium perfringens were analyzed by using two-dimensional gel electrophoresis of the culture supernatant from the wild type and the virR mutant. Based on matrix-assisted laser desorption ionization-time of flight/mass spectrometry, seven positively regulated proteins and eight negatively regulated proteins were identified. Transcriptome analysis confirmed that 7 of the 15 proteins were regulated by the VirR/VirS system at the transcriptional level, but the remaining proteins were modified with a VirR/VirS-directed protease at the posttranslation and secretion levels. We purified and characterized the VirR/VirS-directed protease from the culture supernatant and identified it as a kind of clostripain. Because this proteolytic activity was strongly inhibited by leupeptin and antipain, it was concluded that this protease was a member of the family of cysteine proteases of C. perfringens.
Clostridium perfringens is the primary causative agent of clostridial myonecrosis, also known as gas gangrene (25). This anaerobic organism invades traumatized or ischemic tissue, and although the infection is relatively localized, the bacteria produce numerous extracellular toxins that are responsible for the extensive tissue destruction and necrosis seen in classical cases of gas gangrene (1, 9, 30).
Two-component regulatory systems, consisting of a membrane sensor (histidine kinase) and a cytoplasmic response regulator, enable bacteria to sense and respond to environmental conditions. In response to an appropriate signal, autophosphorylation occurs at a conserved histidine residue in the cytoplasmic domain of the sensor. The phosphate group is then transferred to an aspartate residue on the response regulator, which in turn stimulates or represses target genes at the transcriptional level. The importance of these two-component systems, in control of both metabolism and virulence factor regulation, has been demonstrated in a wide range of bacterial species (8).
Previous works have indicated that one of the two-component systems, VirR/VirS, of C. perfringens globally controls the production of the virulence factors alpha-toxin (phospholipase C), theta-toxin (perfringolysin O), kappa-toxin (collagenase), sialidase, protease, and hemagglutinin (18, 27). Studies on virR mutants have also disclosed that the VirR/VirS system regulates the expression of the genes plc (alpha-toxin gene), pfoA (theta-toxin gene), and colA (kappa-toxin gene) at the transcriptional level (5). Banu et al. (3) identified the other genes that were regulated either positively or negatively by the VirR/VirS system by means of a differential display method using the wild type and the virR mutant of C. perfringens.
Proteome analysis is an excellent tool for analyzing the final products of these regulated genes. The profiles of proteome and transcriptome should be different, based on differences in the posttranscriptional regulation that control the translation rate (12) and half-lives of proteins or mRNAs (33), their intracellular location, and their molecular association with other proteins (32).
In this study, we analyzed the secreted proteins that are regulated with the two-component system VirR/VirS by using two-dimensional gel electrophoresis. By comparing the proteome profile of the C. perfringens wild type with that of the virR mutant, 15 proteins were identified as members of the family of VirR/VirS-dependent proteins. One of the VirR/VirS-dependent proteins which showed proteolytic activity was purified and characterized.
MATERIALS AND METHODS
Bacterial strains and media.
C. perfringens strain 13 (19) and the virR mutant TS133 (27) were grown in RPMI 1640 (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) in all experiments. These strains have been shown to produce more extracellular proteins in this medium than in TSF medium (T. Shimizu, unpublished data).
Preparation of extracellular protein fraction.
C. perfringens cells were grown in RPMI 1640 medium with and without protease inhibitors at 37°C under anaerobic conditions and harvested at the late exponential growth phase. The cells were harvested by centrifugation at 10,000 × g for 5 min at 4°C. The extracellular proteins in supernatant were precipitated with 10% (wt/vol) trichloroacetic acid overnight at 4°C and centrifuged at 10,000 × g for 5 min at 4°C. The resulting protein precipitate was washed with cold acetone and air-dried.
Two-dimensional electrophoresis.
The proteins were treated with a mixture containing 9 M urea, 4% 3-[(3-cholamidylpropyl)-dimethylammoni]-1-propanesulfonate (CHAPS), 100 mM dithiothreitol, and 0.2% (wt/vol) Bio-Lytes 3/10 (Bio-Rad Laboratories, Hercules, Calif.) to obtain completely denatured and reduced proteins. The protein samples were separated using an immobilized pH gradients Ready Strip system (Bio-Rad Laboratories, Hercules, Calif.) in the pH range of 5 to 8 or 3 to 10. For database construction and the identification of proteins by mass spectrometry (MS), 70 to 80 μg of protein was applied. The proteins were silver stained using a Silver Stain II kit (Wako Pure Chemical Industries, Ltd., Osaka, Japan). After the gels were scanned with an imaging system, analysis of the two-dimensional images was performed with the PDQuest software package (Bio-Rad Laboratories, Hercules, Calif.). More than three separate gels of each condition were analyzed, and only spots displaying the same pattern in all parallels were selected for further characterization.
Peptide mass fingerprinting.
Peptide mass fingerprinting was performed by the method of Jensen et al. (14) with slight modification. The protein spots were excised with a scalpel and cut into pieces (1 mm by 1 mm). The gel pieces were placed in a microtube and washed in distilled water for 10 min. The gel pieces were washed twice in 25 mM ammonium bicarbonate-50% acetonitrile for 10 min and then once in acetonitrile for 5 min. A minimum volume of 100 mM ammonium bicarbonate was added to totally immerse the gel pieces, followed by incubation for 5 min. An equal volume of acetonitrile was added, incubated for 15 min, and discarded. The gel pieces were dried in a vacuum centrifuge for 30 min.
For enzyme digestion, 1 μg of lysylendopeptidase (Wako Pure Chemical Industries, Ltd., Osaka, Japan) was dissolved in 100 μl of 100 mM Tris-HCl (pH 9.0) and gradually added to the dried gel pieces, followed by incubation for 45 min on ice. After swelling of the gel pieces, the supernatant was discarded, and a minimum volume of 100 mM Tris-HCl (pH 9.0) was added to immerse the gel pieces. Samples were incubated at 37°C for 20 h. After digestion, 40 to 60 μl of 25 mM ammonium bicarbonate was added and mixed for 15 min. Then an equal volume of acetonitrile was added, followed by mixing for 15 min and recovery of the supernatant. From 40 to 60 μl of 5% trifluoroacetic acid-50% acetonitrile was added, the solution was mixed for 15 min, and the supernatant was recovered twice. The extracts were combined, concentrated to about 20 μl by vacuum centrifugation, and supplemented with 80 μl of 0.1% trifluoroacetic acid. The extracts were then concentrated to about 20 μl by an additional vacuum centrifugation.
Before measuring the mass of the peptide mixture, the peptides were purified using a ZipTip C18 (Millipore, Bedford, Mass.) according to the manufacturer's instructions. Purified peptide solution (0.6 μl) was prepared with equal volumes of saturated α-cyano-4-hydroxycinnamic acid in 50% acetonitrile-0.1% trifluoroacetic acid to create a sample template for matrix-assisted laser desorption ionization-time of flight/mass spectrometry (MALDI-TOF/MS) (Voyager Linear DE and Voyager DE RP; PE Biosystems, Foster City, Calif.). Peptide mass fingerprints were analyzed by using Mascot software (Matrix Science, Ltd., London, United Kingdom).
Northern hybridization.
Northern hybridization was performed as described previously (5, 16). Chromosomal DNA from C. perfringens strain 13 was used as a template to amplify each DNA fragment by PCR with each primer set (Table 1), except the CPE0163 and CPE0178 genes, according to the DNA sequence data of the C. perfringens genome sequence (28). Plasmids pTS302 (29) and pKY3135 (20) were used as the templates to amplify each DNA fragment by PCR with a universal primer set in the CPE0163 and CPE0178 genes, respectively. Each DNA fragment was labeled with alkaline phosphatase using an AlkPhos Direct kit (Amersham Pharmacia Biotech United Kingdom, Ltd., Buckinghamshire, England) according to the manufacturer's instructions. The qualitative examination of the Northern hybridization analysis data was validated by scanning the blots with a densitometer and determining the relative amounts of each specific transcript with a Quantity One software package (Bio-Rad Laboratories, Hercules, Calif.).
TABLE 1.
Oligonucleotide primers used for PCRa
| Primer | Sequencea | Location |
|---|---|---|
| 1951-1 | ATACTACCAATACCAGTTAAA | Internal CPE1951; forward primer |
| 1951-2 | TGGATCTCCTTGTACCCCTT | Internal CPE1951; reverse primer |
| 1529-1 | TCTACTGGTGCTGGAACAGA | Internal CPE1529; forward primer |
| 1529-2 | AAGAACCACTTCCAGTTGGT | Internal CPE1529; reverse primer |
| 0846-1 | AATTTCATCTTGGTACTTACCTTC | Internal CPE0846; forward primer |
| 0846-2 | CTATTATATCCACCAAGTGAAGTA | Internal CPE0846; reverse primer |
| 1231-1 | TGCTGCTAAAGCCTTTAACA | Internal CPE1231; forward primer |
| 1231-2 | TGATGCATACCATGGTGTTGT | Internal CPE1231; reverse primer |
| 0220-1 | AAGATGTGGTAGCAACGCCA | Internal CPE0220; forward primer |
| 0220-2 | TGGTGAACCAGTCTGTGGTA | Internal CPE0220; reverse primer |
| 1428-1 | AGCGGCACAATCATCAGGGGT | Internal CPE1428; forward primer |
| 1428-2 | TTCTTATCATAGCACCAGCT | Internal CPE1428; reverse primer |
| 0202-1 | AAGCAAGAGATATTAGGCGA | Internal CPE0202; forward primer |
| 0202-2 | TGAAGCCACCACCATGATCT | Internal CPE0202; reverse primer |
| 1785-1 | TGGACTACTCAGCAACTACA | Internal CPE1785; forward primer |
| 1785-2 | TCTTGACCACCACCGTGGA | Internal CPE1785; reverse primer |
| 1632-1 | TAGAGCCAACAGGAATGGCT | Internal CPE1632; forward primer |
| 1632-2 | AGCTATCTCCAGCAGCTGTA | Internal CPE1632; reverse primer |
| 2297-1 | TGGTGCAGGTACAATGGGT | Internal CPE2297; forward primer |
| 2297-2 | ATCCTGGTGCTTCAGCAACT | Internal CPE2297; reverse primer |
| 1350-1 | AGTATCAGAAGGTGCTGCT | Internal CPE1350; forward primer |
| 1350-2 | AGGGATTCCAGTACCACCGT | Internal CPE1350; reverse primer |
| 2408-1 | AGGCAGACAAGTACGGCGT | Internal CPE2408; forward primer |
| 2408-2 | TGAAGCTCACCCATACCAGCT | Internal CPE2408; reverse primer |
| 0278-1 | TCGCAACTCCACTTACAGAT | Internal CPE0278; forward primer |
| 0278-2 | TCTGCATCTACCTCTGGAGT | Internal CPE0278; reverse primer |
All primers are given in the 5′ to 3′ orientation and are based on nucleotide sequence data for the C. perfringens genome (AP003185 to AP003194 and AP003515).
Proteolytic activity assay.
Proteolytic activities were determined by using the insoluble proteolytic substrate azocoll (Wako Pure Chemical Industries, Ltd., Osaka, Japan) as previously described (10) with slight modification. The substrate (4 mg/ml) was suspended in 100 mM phosphate buffer (pH 7.0) containing 5 mM dithiothreitol. Then 500 μl of a sample was added to 500 μl of substrate suspension. The mixture was incubated for 2 h at 37°C with shaking. After incubation, the assay mixtures were centrifuged, and the absorbances at 520 nm were determined.
Proteases were distinguished through the use of different protease inhibitors. The samples were placed on ice, supplemented with the inhibitors, and left to react for 45 min before adding the proteolytic substrate azocoll. For determination of the proteolytic activity of purified protein, 0.1 mg of bovine serum albumin was added to the assay mixture. One unit of protease activity was arbitrarily defined as proteolysis that resulted in the release of 0.001 absorbance unit of dye for 2 h. Protease specific activities were expressed as units per milligram of protein. Protein concentrations were determined by the Bradford method (Bio-Rad Laboratories, Hercules, Calif.).
Purification of protease.
C. perfringens wild-type strain 13 cells were grown in RPMI 1640 medium under anaerobic conditions and harvested at the late exponential growth phase. The cells were harvested by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was ultracentrifuged at 100,000 × g for 1 h at 4°C. Proteins in the supernatant were concentrated with an Amicon stirred-cell 8050 system and supplemented with a 1/10 volume of 1 M Tris-HCl (pH 7.5)-1.5 M NaCl. Concentrated supernatant solution was applied to a benzamidine-Sepharose 6B (Amersham Pharmacia Biotech United Kingdom, Ltd., Buckinghamshire, England) affinity column (bed volume, 2 ml) equilibrated with 50 mM Tris-HCl (pH 8.0) containing 0.5 M NaCl. The column was washed with 20 ml of the equilibration buffer and eluted with 0.1 M glycine-HCl (pH 3.0). Fractions of 1 ml were collected in a tube that contained 0.3 ml of 0.1 M Tris-HCl (pH 9.0), and the protease activity was assayed.
RESULTS
Analysis of culture supernatant proteins of wild type and virR mutant by two-dimensional gel electrophoresis.
Because proteins normally have a wide range of isoelectric points, we used two gel systems: (i) pH 5 to pH 8 with 10% polyacrylamide and a pH 5 to 8 gel and (ii) pH 3 to pH 10 with 10% polyacrylamide and a pH 3 to 10 gel.
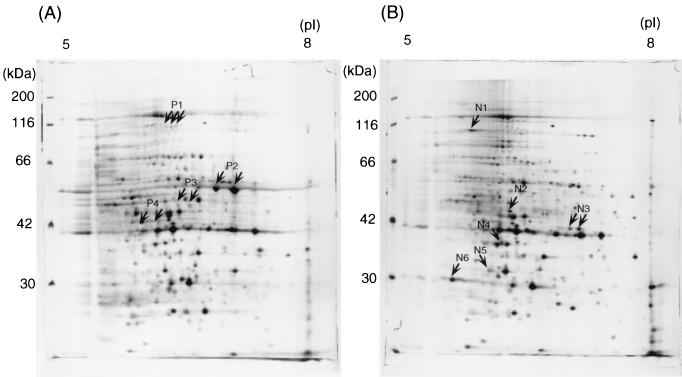
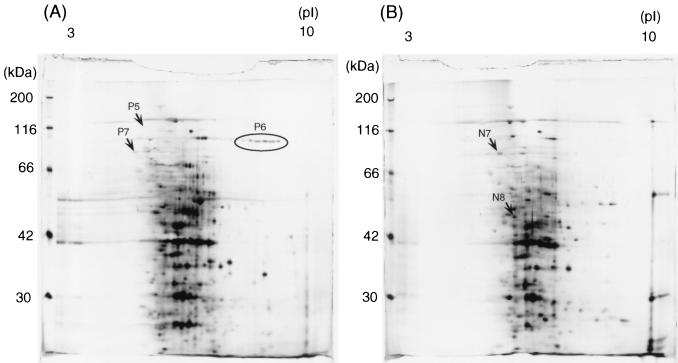
The culture supernatant of the wild-type strain 13 and the virR mutant TS133 were collected at the late exponential growth phase, and the proteins were separated by two-dimensional gel electrophoresis. The virR mutant expressed a pattern different from that of the wild type in the pH 5 to 8 gel, as shown in Fig. 1. The virR mutant showed six intensified and four reduced spots compared with the wild strain. This profile was reproducible. The density of the P2 spot was remarkably enhanced in the wild type but absent in the virR mutant. The P4 spot appeared transparent after silver staining in the wild type but was absent in the virR mutant. In the pH 3 to 10 gel, the virR mutant pattern showed three spots with reduced density and two enhanced spots (Fig. 2).
FIG. 1.
Two-dimensional silver-stained gel of proteins in the culture supernatant of C. perfringens strain 13 (A) and the virR mutant TS133 (B). Proteins were separated in the first dimension by a pH 5 to 8 immobilized pH gradient gel and then in the second dimension by a 10% polyacrylamide gel. Reproducible differences in the density of protein spots were examined by at least three independent experiments. Spots were excised, and the corresponding proteins were identified by MALDI-TOF/MS and database searches. The spots are labeled on the gel according to the numbers presented in Table 2.
FIG. 2.
Two-dimensional silver-stained gel of proteins in the culture supernatant of C. perfringens wild-type strain 13 (A) and the virR mutant TS133 (B). Proteins were separated in the first dimension by pH 3 to 10 immobilized pH gradients gel and then in the second dimension by a 10% polyacrylamide gel. Reproducible differences in the density of protein spots were examined by at least three independent experiments. Spots were excised, and the corresponding proteins were identified by MALDI-TOF/MS and database searches. The spots are labeled on the gel according to the numbers presented in Table 2.
Those protein spots which were different between the two strains were subjected to in-gel digestion and MALDI-TOF/MS analysis. Table 2 summarizes the results. Seven protein spots were expressed exclusively in the wild type, while eight spots were restricted to the virR mutant. The proteins were further analyzed by peptide mass fingerprinting using MALDI-TOF/MS. The identified proteins were assigned the appropriate CPE gene numbers by reference to the C. perfringens strain 13 genome (28).
TABLE 2.
Summary of proteome analysis, Northern hybridization analysis, and effect of protease inhibitors on the two-dimensional pattern
| Spot no. | CPE no. | Product (homologous protein) (reference) | Patterna | Densityb
|
Northernc | Effect of protease inhibitord
|
||
|---|---|---|---|---|---|---|---|---|
| Wild type | Mutant | L, A | B | |||||
| P1 | CPE1951 | 2′,3′-cyclic nucleotide 2′-phosphodiesterase | W | 726 | 474 | P (0.26) | +/− | +/− |
| P2 | CPE0163 | Perfringolysin O | W | 22,167 | 732 | P (0.22) | +/− | − |
| P3 | CPE1529 | Hypothetical protein (ydaL gene product from Bacillus subtilis [29.3%]) (17) | W | 2,017 | 0 | P (0.83) | +/− | +/− |
| P4 | CPE0846 | Cysteine protease (alpha-clostripain from Clostridium histolyticum [58.3%]) (7) | W | N.D. | N.D. | P (0.32) | +/− | − |
| P5 | CPE0173 | Collagenase | W | 530 | 0 | P (0.30) | +/− | +/− |
| P6 | CPE1231 | Probable surface protein (Aas surface protein from Staphylococcus saprophyticus [25.4%]) (13) | W | 11,243 | 0 | I (1.09) | − | − |
| P7 | CPE0220 | Hypothetical protein (chitinase A from Clostridium paraputrificum [38%]) (24) | W | 1,158 | 0 | I (1.04) | +/− | +/− |
| N1 | CPE1428 | ClpB protein (endopeptidase Clp ATP-binding chain B1 from Synechocystis sp. [55.8%]) (15) | M | 0 | 4,780 | I (1.00) | + | + |
| N2 | CPE0202 | Probable cell wall-binding protein (cell wall-binding protein from Bacillus halodurans [32.4%]) (31) | M | 0 | 3,839 | N (1.51) | + | + |
| N3 | CPE1785 | Iron-sulfur cofactor synthesis protein NifS | M | 3,015 | 13,371 | I (1.05) | + | + |
| N4 | CPE1632 | Ribokinase | M | 0 | 5,352 | I (1.08) | + | + |
| N5 | CPE2297 | β-Hydroxybutyryl-coenzyme A dehydrogenase, NAD dependent | M | 0 | 6,403 | I (1.00) | +/− | +/− |
| N6 | CPE1350 | Fructose-bisphosphate aldolase | M | 0 | 10,109 | N (1.46) | + | + |
| N7 | CPE2408 | Elongation factor G | M | 1,792 | 3,525 | I (0.95) | +/− | +/− |
| N8 | CPE0278 | Conserved hypothetical protein (p45 from Listeria monocytogenes [26%]) (26) | M | 0 | 8,698 | I (0.98) | +/− | +/− |
W, exclusive to the wild type, M, exclusive to the virR mutant.
Values represent the ratio of spot density level (in ppm) to the total of all valid spots using PDQuest software (Bio-Rad). N.D., not determined.
P, positively regulated; I, independently regulated; N, negatively regulated. Values represent the ratio of specific mRNA level in the mutant to that in the wild type.
+/−, not altered; +, induced; −, reduced. L, leupeptin; A, antipain; B, benzamidine.
Transcription of VirR/VirS-regulated proteins.
To investigate whether the expression of these proteins was directly regulated by the VirR/VirS system at the transcriptional level, Northern hybridization analysis was performed at the mid-exponential growth phase (Fig. 3). The results clearly demonstrated that the VirR/VirS system positively regulated CPE0163 (pfoA) and CPE0846 (Table 2 and Fig. 3). CPE1951, CPE1529, and CPE0173 were positively regulated by the VirR/VirS system at the transcriptional level (Table 2 and Fig. 3). However, CPE1231 and CPE0220 were not positively regulated by the VirR/VirS system at the transcriptional level (Table 2 and Fig. 3).
FIG. 3.
Northern hybridization using various gene probes. The calculated sizes (in kilobases) of the mRNAs are shown at right. Values represent the ratios of specific mRNA levels to those in the wild type. These values are derived from three independent experiments. Lanes: 1, strain 13/pJIR418; 2, strain TS133/pJIR418; 3, strain TS133/pBT404 (virR+) (27). These results are summarized in Table 2.
On the other hand, the products of CPE1428, CPE0202, CPE1785, CPE1632, CPE2297, CPE1350, CPE2408, and CPE0278 appeared to be negatively regulated by proteome analysis. However, six transcripts (CPE1428, CPE1785, CPE1632, CPE2297, CPE2408, and CPE0278) of these genes were not negatively regulated by the VirR/VirS system (Table 2 and Fig. 3). The transcription of CPE0202 and CPE1350 appeared to be negatively regulated at the transcriptional level (Table 2 and Fig. 3).
Analysis of VirR/VirS-regulated proteolytic activity of culture supernatant.
Proteolytic activity of the culture supernatant of C. perfringens was reported to be positively regulated by the VirR/VirS system (18). In the present study, the proteolytic activity of the culture supernatant of the C. perfringens wild type and the virR mutant were determined at the late exponential growth phase. The activity of the culture supernatant of the wild type (2.8 × 105 U/mg) was much higher than that of the virR mutant (1.2 × 103 U/mg). The proteolytic activity of complemented strain TS133(pBT404) was 2.9 × 105 U/mg. In the former, the activity was inhibited by benzamidine, leupeptin, antipain, and EDTA, but it was not affected by phenylmethylsulfonyl fluoride (PMSF), pepstain, phosphoramidon, E-64, or soybean trypsin inhibitor (Table 3).
TABLE 3.
Effect of inhibitors on proteolytic activitya
| Inhibitor | Concn | % Inhibition
|
|
|---|---|---|---|
| Supernatant | Purified | ||
| PMSF | 0.5 mM | 1 | 21 |
| Leupeptin | 10 μM | 84 | 100 |
| Antipain | 10 μM | 82 | 100 |
| E-64 | 0.7 μM | 0 | 16 |
| Pepstatin | 1 μM | 0 | 14 |
| EDTA | 10 mM | 82 | 73 |
| Benzamidine | 5 mM | 74 | 87 |
| SBTI | 0.1 mg/ml | 0 | 17 |
| Phosphoramidon | 20 μM | 0 | 16 |
Proteolytic activities of the culture supernatant of wild-type and purified protease were determined with azocoll in the presence of inhibitors. Activities are expressed as percentages of the respective controls. E-64, l-trans-epoxysuccinyl-leucylamido(4-guanidino)butane; SBTI, soybean trypsin inhibitor.
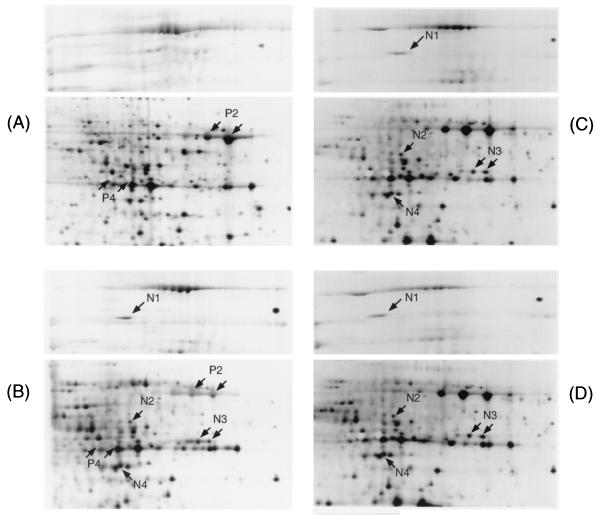
We suspected that the protease in the supernatant might affect the proteome. To examine the effect of the proteolytic activity on the two-dimensional pattern, the culture supernatants were examined after the addition of various kinds of protease inhibitors. The protein spots N1, N2, N3, N4, and N6 increased in intensity in the wild-type strain after the addition of benzamidine (5 mM), leupeptin (10 μM), and antipain (10 μM) (Fig. 4). However, the addition of PMSF (0.5 mM), pepstatin (1 μM), phosphoramidon (20 μM), or E-64 (0.7 μM) did not cause any change (data not shown). Protein spot P6 of the wild type disappeared after addition of benzamidine, leupeptin, and antipain (data not shown), while protein spots P2 and P4 were reduced by benzamidine (Fig. 4), PMSF, and phosphoramidon (data not shown). This result suggested that proteins N1, N3, N4, and P6 were not directly regulated by the VirR/VirS system at the transcriptional level, but rather were degraded by the VirR/VirS-directed protease(s) during culture, which activity was inhibited by benzamidine, leupeptin, and antipain. Proteins N2 and N6 were under the control of the VirR/VirS system at the transcriptional level and then also were degraded by the VirR/VirS-directed protease(s) during culture.
FIG. 4.
Portion of the two-dimensional pattern of the culture supernatant proteins of the wild type (A), the wild type with benzamidine (5 mM) (B), the wild type with leupeptin (10 μM) (C), and the wild type with antipain (10 μM) (D). The spots are labeled on the gel according to the numbers presented in Table 2.
Purification and characterization of VirR/VirS-regulated protease.
In order to characterize the VirR/VirS-directed protease(s) in the culture supernatant of C. perfringens strain 13, we purified the protein(s) from the culture supernatant at the late exponential growth phase using a benzamidine-Sepharose column. The 40-kDa single band was visualized by Coomassie brilliant blue R-250 staining on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions (Fig. 5). The 40-kDa band was cut out and subjected to in-gel digestion, followed by peptide mass fingerprinting using MALDI-TOF/MS. The protein was identified as a product of CPE0846 and shown to have 58.3% homology with clostripain of C. histolyticum, to be identical to the P4 protein, and to be positively regulated by the VirR/VirS system.
FIG. 5.
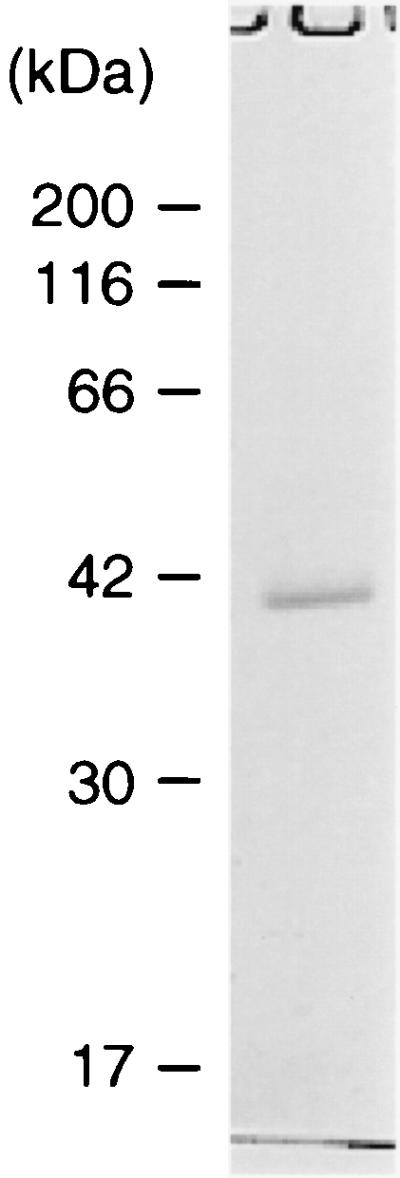
SDS-PAGE analysis of purified protease. Concentrated culture supernatant was applied to a benzamidine-Sepharose 6B column (2 ml) in 50 mM Tris-HCl (pH 8.0)-0.5 M NaCl. The retained enzyme was eluted by applying 0.1 M glycine-HCl (pH 3.0). Fractions of 1 ml were collected and monitored for enzyme activity. Enzyme activity was determined by using azocoll as a substrate. Fractions showing proteolytic activity were pooled. Protein concentration was determined by the Bradford method, and this fraction was subjected to SDS-PAGE analysis under reducing conditions.
The specific activity of purified protease was 4.5 × 106 U/mg. Table 3 shows the effects of protease inhibitors on the activity of the purified protease. Leupeptin and antipain, which are cysteine protease inhibitors, strongly inhibited the proteolytic activity. However, E-64, which is also a cysteine protease inhibitor, did not inhibit the activity. PMSF, a serine protease inhibitor, and pepstatin, an aspartate protease inhibitor, did not inhibit the proteolytic activity. Benzamidine, a trypsin-like inhibitor, inhibited the proteolytic activity, but soybean trypsin inhibitor did not. EDTA, a metalloprotease inhibitor, partially inhibited the proteolytic activity. These results are consistent with the general properties of a cysteine protease except for the effects of E-64 and EDTA.
The activity of the purified protease was diminished during 1 week of storage in phosphate-buffered saline at −20°C. The autodigestive degradation of the peptide during storage was also confirmed by SDS-PAGE.
DISCUSSION
Approximately 200 protein spots were detectable in the culture supernatant of C. perfringens at the late exponential growth phase by two-dimensional gel electrophoresis and silver staining. Although these proteins were derived from the culture supernatant, the supernatant also contained intracellular proteins that had no putative signal sequences, such as ribokinase, β-hydroxybutyryl-coenzyme A dehydrogenase, fructose-bisphosphate aldolase, and elongation factor G (Table 2). Little difference was detected in the pattern according to the growth phases. This indicated that the growing cells in liquid medium undergo autolysis at a relatively high rate during growth.
Among the spots that were apparently different in two-dimensional gel electrophoresis of the wild type and the mutant, 15 spots were found to be the proteins regulated under the VirR/VirS system, and some spots were identified as fragments of the P2 and P5 proteins when analyzed by peptide mass fingerprinting using MALDI-TOF/MS. These fragments are probably derived from cleavage of P2 and P5 by a VirR/VirS-directed protease. The rest of the spots were not so consistently reproducible and therefore were not included in our analysis.
Among these 15 genes, CPE1951, CPE0163, CPE1529, CPE0846, and CPE0173 were positively regulated by the VirR/VirS system at the transcriptional level. The CPE0163 gene was found to be identical to the pfoA gene, and the CPE0846 gene had a consensus sequence that bound to VirR (6, 28). However, the CPE0173 gene, which was identified as the colA gene, and the CPE1951 and CPE1529 genes (28) did not have the consensus sequences to bind VirR. It is probable that the CPE0173, CPE1951, and CPE1529 genes are regulated by a secondary regulator, as in the case of the hyp7 gene, whose expression has been shown to be regulated by the VirR/VirS system (4).
The CPE1231 gene was independent of the VirR/VirS system. The product of CPE1231 had homology with Aas, a surface protein of Staphylococcus saprophyticus (13). This product may be located on the cell surface of C. perfringens and could be released by the VirR/VirS-dependent protease (a product of CPE0846) into the culture supernatant. This may be supported by the following evidence: the P6 protein spot on the two-dimensional pattern disappeared when the culture supernatant was treated with benzamidine, leupeptin, or antipain (data not shown), and the molecular mass of the P6 protein (97 kDa), as estimated by two-dimensional gel electrophoresis, was smaller than that of the product of CPE1231 (122 kDa) (28).
Lyristis et al. (18) measured the proteolytic activity of the culture supernatants of C. perfringens using azocoll as the substrate and found that the activity was regulated by the VirR/VirS system. Award et al. (2) reported that a collagenase gene-defective mutant had very little azocoll activity compared with the wild type. These results suggested that the proteolytic activity of the culture supernatant of C. perfringens using azocoll as the substrate was mostly derived from the collagenase. However, in this study, leupeptin and antipain, which are cysteine protease inhibitors, inhibited 80% of the proteolytic activity in the culture supernatant and did not inhibit the collagenase activity. In the presence of benzamidine, leupeptin, and antipain, the two-dimensional pattern of the wild-type culture supernatant was similar to that of the virR mutant. These results indicated that the culture supernatant contained a VirR/VirS-regulated protease other than the collagenase. In this study, C. perfringens was grown in RPMI 1640, but the other studies used Gifu anaerobic medium (GAM) broth, so the difference is probably due to the use of different culture media. This inference was supported by the increased cysteine protease activity observed in the RPMI 1640 culture supernatant (data not shown).
We used a benzamidine-Sepharose column to purify this protease. The purified protein was homologous with clostripain of C. histolyticum, which is a cysteine protease. The clostripain is a trypsin-like cysteine protease that was specific for the cleavage of arginyl bonds and susceptible to various trypsin inhibitors (23). The purified protease from C. perfringens was also susceptible to trypsin inhibitors such as benzamidine but not to soybean trypsin inhibitor. The substrate specificity of clostripain was similar to that of trypsin; however, clostripain has been shown to preferentially cleave to the carboxyl-terminal side of arginine residues (21). E-64, which inactivates most cysteine proteases (4), did not affect the clostripain. The protease of C. perfringens was also not affected by E-64.
The seven amino acids (FDACLMG) of the active site of the clostripain of C. histolyticum (11) were identical to those of the protease of C. perfringens. The clostripain of C. histolyticum requires calcium ions to enhance its proteolytic activity (22). Similar to this, the protease activity of C. perfringens was reduced with 10 mM EDTA. The presence of the protease in C. perfringens was predicted by a homology search study on the whole genome sequence of the wild strain, using the VirR binding motif as the target sequence (28). It was confirmed in the present study by purification of the clostripain-like protease in C. perfringens.
Acknowledgments
We are grateful to William Ba-Thein for critical reading of the manuscript.
This work was supported by a Grant-in-Aid for Encouragement of Young Scientists from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Awad, M. M., A. E. Bryant, D. L. Stevens, and J. I. Rood. 1995. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15:191-202. [DOI] [PubMed] [Google Scholar]
- 2.Awad, M. M., D. M. Ellemor, A. E. Bryant, O. Matsushita, R. L. Boyd, D. L. Stevens, J. J. Emmins, and J. I. Rood. 2000. Construction and virulence testing of a collagenase mutant of Clostridium perfringens. Microb. Pathog. 28:107-117. [DOI] [PubMed] [Google Scholar]
- 3.Banu, S., K. Ohtani, H. Yaguchi, T. Swe, S. T. Cole, H. Hayashi, and T. Shimizu. 2000. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol. Microbiol. 35:854-864. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, A. J., A. A. Kembhavi, M. A. Brown, H. Kirschke, C. G. Knight, M. Tamai, and K. Hanada. 1982. L-trans-epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H, and L. Biochem. J. 201:189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ba-Thein, W., M. Lyristis, K. Ohtani, I. T. Nisbet, H. Hayashi, J. I. Rood, and T. Shimizu. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J. Bacteriol. 178:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, J. K., and J. I. Rood. 2000. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 182:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dargatz, H., T. Diefenthal, V. Witte, G. Reipen, and D. von Wettstein. 1993. The heterodimeric protease clostripain from Clostridium histolyticum is encoded by a single gene. Mol. Gen. Genet. 240:140-145. [DOI] [PubMed] [Google Scholar]
- 8.Dziejman, M., and J. J. Mekalanos. 1995. Two-component signal transduction and its role in expression of bacterial virulence factors, p. 305-317. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 9.Ellemor, D. M., R. N. Baird, M. M. Awad, R. L. Boyd, J. I. Rood, and J. J. Emmins. 1999. Use of genetically manipulated strains of Clostridium perfringens reveals that both alpha-toxin and theta-toxin are required for vascular leukostasis to occur in experimental gas gangrene. Infect. Immun. 67:4902-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilles, A. M., A. De Wolf, and B. Keil. 1983. Amino-acid sequences of the active-site sulfhydryl peptide and other thiol peptides from the cysteine proteinase alpha-clostripain. Eur. J. Biochem. 130:473-479. [DOI] [PubMed] [Google Scholar]
- 12.Harford, J. B., and D. R. Morris. 1997. Post-transcriptional gene regulation. Wiley-Liss, Inc., New York, N.Y.
- 13.Hell, W., H. G. Meyer, and S. G. Gatermann. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871-881. [DOI] [PubMed] [Google Scholar]
- 14.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, T. Ashford, M. Mann, G. Griffiths, and J. Krijnse Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, T., T. Shimizu, and H. Hayashi. 1995. Transcriptional analysis of the beta-galactosidase gene (pbg) in Clostridium perfringens. FEMS Microbiol. Lett. 133:65-69. [DOI] [PubMed] [Google Scholar]
- 17.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 18.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 19.Mahony, D. E., and T. I. Moore. 1976. Stable L-forms of Clostridium perfringens and their growth on glass surfaces. Can. J. Microbiol. 22:953-959. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita, O., K. Yoshihara, S. Katayama, J. Minami, and A. Okabe. 1994. Purification and characterization of Clostridium perfringens 120-kilodalton collagenase and nucleotide sequence of the corresponding gene. J. Bacteriol. 176:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, W. M. 1977. Cleavage at arginine residues by clostripain. Methods Enzymol. 47:165-170. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell, W. M., and W. F. Harrington. 1970. Clostripain. Methods Enzymol. 19:635-642. [Google Scholar]
- 23.Mitchell, W. M., and W. F. Harrington. 1968. Purification and properties of clostridiopeptidase B (clostripain). J. Biol. Chem. 243:4683-4692. [PubMed] [Google Scholar]
- 24.Morimoto, K., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 1999. Sequencing, expression, and transcription analysis of the Clostridium paraputrificum chiA gene encoding chitinase ChiA. Appl. Microbiol. Biotechnol. 51:340-347. [DOI] [PubMed] [Google Scholar]
- 25.Rood, J. I., and S. T. Cole. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55:621-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schubert, K., A. M. Bichlmaier, E. Mager, K. Wolff, G. Ruhland, and F. Fiedler. 2000. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch. Microbiol. 173:21-28. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu, T., W. Ba-Thein, M. Tamaki, and H. Hayashi. 1994. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J. Bacteriol. 176:1616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu, T., A. Okabe, J. Minami, and H. Hayashi. 1991. An upstream regulatory sequence stimulates expression of the perfringolysin O gene of Clostridium perfringens. Infect. Immun. 59:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, D. L., R. K. Tweten, M. M. Awad, J. I. Rood, and A. E. Bryant. 1997. Clostridial gas gangrene: evidence that alpha and theta toxins differentially modulate the immune response and induce acute tissue necrosis. J. Infect. Dis. 176:189-195. [DOI] [PubMed] [Google Scholar]
- 31.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urlinger, S., K. Kuchler, T. H. Meyer, S. Uebel, and R. Tampe. 1997. Intracellular location, complex formation, and function of the transporter associated with antigen processing in yeast. Eur. J. Biochem. 245:266-272. [DOI] [PubMed] [Google Scholar]
- 33.Varshavsky, A. 1996. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93:12142-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]