Abstract
Many Helicobacter pylori isolates carry cryptic plasmids of extremely variable size. In this study we analyzed two H. pylori plasmids, pHel4 and pHel5, from H. pylori strains P8 and P29, respectively. Plasmid pHel4 consists of 10,970 bp, constituting 15 putative open reading frames (ORFs), whereas pHel5 consists of 18,291 bp, constituting 17 ORFs. The findings that both plasmids encode a conserved RepA protein and that both have an origin of replication containing an iteron place them in the group of theta plasmids. In pHel4, the products of the overlapping orf4C, orf4D, orf4E, and orf4F sequences are homologous to MobA, MobB, MobC, and MobD, encoded by colicinogenic plasmids, suggesting that pHel4 might be mobilizable. A further putative operon consists of orf4B and orf4A, the products of which are homologous to microcin C7 (MccC7) biosynthesis and secretion proteins MccB and MccC, respectively. Plasmid pHel5 carries putative genes encoding proteins with homology to an endonuclease and gene products of an H. pylori chromosomal plasticity zone. Both plasmids contain repeat sequences, such as the previously identified R2 repeat, which are considered preferred recombination sites. In pHel4, a new repeat sequence (R4 repeat), which seems to act as a hot spot for site-specific recombination, was identified. All H. pylori plasmids characterized so far have a modular structure. We suggest a model that explains the existing plasmids by insertions and deletions of genetic elements at the repeat sequences. A genetic exchange between plasmids and the bacterial chromosome, combined with plasmid mobilization, might add a novel mechanism to explain the high genetic macrodiversity within the H. pylori population.
Helicobacter pylori is a highly motile, microaerophilic, gram-negative bacterium colonizing the human stomach mucosa (44). H. pylori is well recognized as a major cause of several gastroduodenal pathologies, including chronic active gastritis, peptic ulcer disease, mucosa-associated lymphoid tissue lymphoma, and gastric adenocarcinoma (6). Several bacterial factors are known to be essential for colonization in animal models, including a bundle of sheathed flagella and a potent urease (13, 14). Other bacterial virulence factors only present in a subset of H. pylori strains, the type I strains, include the vacuolating cytotoxin (VacA) and the cytotoxin-associated antigen A (CagA). Genes such as vacA and cagA and 32 genes encoding putative outer membrane proteins (OMPs) show characteristics of mosaic genes (4), resulting in the existence of several alleles in different H. pylori strains (microdiversity). Genes involved in lipopolysaccharide biosynthesis or restriction-modification systems are prone to frameshift mutations by slipped-strand mispairing during replication (38), a means by which overproduction or nonproduction of the corresponding proteins is determined (16).
Comparison of the two available genome sequences of H. pylori 26695 (42) and J99 (1) reveals that differences between the strains are the result of intragenomic rearrangements, resulting in deletion, inversion, or translocation of larger genome fragments (macrodiversity) (1). Many H. pylori strains possess several copies of the insertion sequences IS605 and IS606. Furthermore, H. pylori 26695 contains five so-called plasticity zones with G+C contents of 33% (zone 1), 35% (zone 2), 33% (zone 3), 43% (zone 4), and 33% (zone 5), which differ from the chromosomal G+C content of 39% (42). Strain H. pylori J99 (1) carries several different plasticity zones which are not present in H. pylori 26695. It is speculated that H. pylori received the plasticity zones by horizontal gene transfer.
Transduction, conjugation, and natural transformation are the three common mechanisms of horizontal gene transfer in bacteria. Natural transformation competence was first described for H. pylori by Nedenskov-Sorensen et al. (32). Several genes that are involved in the transformation process of H. pylori have been identified (2, 24, 39, 40). We demonstrated recently that natural transformation of H. pylori is mediated by basic components of a type IV secretion system (23). Plasmid conjugation has not been proven for H. pylori, but there is some evidence that horizontal transfer of genes in H. pylori could take place via a DNase-resistant, conjugation-like mechanism (3, 28).
Although about 50% of H. pylori strains carry cryptic plasmids ranging in size from 2 to about 100 kb (35), the role of these plasmids is not well understood. Several H. pylori plasmids of up to 6 kb have been analyzed in detail in the last few years (20, 27, 30). They can be grouped into at least two separate classes. Plasmid pHPK255 (27) reveals homology to plasmids of gram-positive bacteria replicating via the “rolling-circle” mechanism, whereas other plasmids (pHPM180, pHel1, and pHPS1) fall into the group of iteron-containing plasmids and replicate via the theta mechanism (10).
In this study, we investigated which genetic information is encoded by larger H. pylori plasmids (6 to 20 kb) and whether plasmids might be a source for horizontal gene transfer and for generation of macrodiversity in H. pylori. The sequencing and comparison of two H. pylori plasmids, pHel4 and pHel5, identified both common and unique open reading frames (ORFs), suggesting that individual plasmids express distinct features. From database comparisons we identified putative genes involved in conjugative transfer (mob) as well as gene sequences homologous to that of an operon present in certain Escherichia coli strains responsible for microcin production. Furthermore, the H. pylori plasmids described in this study show a modular structure, which apparently enables them to integrate or delete complete functional modules, thus making them ideal candidates for an efficient gene-shuffling mechanism.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. pylori strains were grown on GC agar plates (Difco) supplemented with horse serum (8%), vancomycin (10 mg liter−1), trimethoprim (5 mg liter−1), and nystatin (1 mg liter−1) (serum plates) and incubated for 2 to 3 days in a microaerobic atmosphere (85% N2, 10% CO2, 5% O2) at 37°C. For H. pylori liquid cultures Brucella medium (Becton Dickinson) supplemented with 10% fetal calf serum was used; the medium was inoculated with a bacterial suspension with an optical density at 550 nm of 0.1.
For selection of H. pylori mutant strains, serum plates supplemented with chloramphenicol (6 mg liter−1) were used. E. coli strains HB101 (8) and DH5α (Bethesda Research Laboratories) were grown on Luria-Bertani (LB) agar plates or in LB liquid medium (37) supplemented with ampicillin (100 mg liter−1), chloramphenicol (30 mg liter−1), or tetracycline (15 mg liter−1), as appropriate. The H. pylori strains were isolated from patients undergoing endoscopy in different university gastroenterology units in Germany (Munich, Hamburg, and Erlangen). Thus, most of the analyzed strains were from Germany or from other European countries.
DNA manipulations and plasmid constructions.
Cloning and DNA analysis procedures were performed as described by Sambrook et al. (37). H. pylori chromosomal DNA was isolated with the QIAamp tissue kit (Qiagen). Plasmid DNA was purified from E. coli by the boiling procedure, and electroporation-competent E. coli cells were prepared according to the protocol recommended for the Gene Pulser (Bio-Rad).
Plasmid preparation and plasmid library construction for DNA sequencing.
Plasmid DNA of different Helicobacter strains was extracted from cells growing on plates or from cell pellets after cultivation in liquid media. The latter method resulted in general in plasmid DNA of better quality. Plasmid DNA was isolated by using either the Wizard Plus SV Minipreps purification system (Promega) according to the manufacturers protocol or the XSP buffer extraction method described by De Ungria et al. (11). In brief, cell pellets were resuspended in 100 μl of TES (10 mM Tris-HCl [pH 7.4], 1 mM EDTA [pH 8.0], 15 mM NaCl) by adding 1.5 ml of prewarmed (65°C) XSP buffer. XSP buffer contains XS buffer (1% potassium ethyl xanthogenate, 100 mM Tris-HCl [pH 7.4], 20 mM EDTA [pH 8.0], 1% sodium dodecyl sulfate [SDS], and 800 mM ammonium acetate) with an equal volume of phenol. The resuspended pellets were incubated for 30 min at 65°C. After a short vortexing, incubation on ice for 5 min, and centrifugation at 14,000 × g for 15 min the aqueous phase was transferred to a fresh Eppendorf tube. The samples were purified with two phenol-chloroform-isoamyl alcohol (25:24:1) extractions, and RNA was digested with RNase A for 30 min at 37°C. One volume of isopropanol and 0.1 volume of 3 M sodium acetate, pH 4.8, were used to precipitate the DNA, which was washed with 70% ethanol once. After being dried the plasmid DNA was resuspended in 50 μl of distilled water.
The isolation of plasmid DNA from H. pylori by the XSP method resulted in minor contamination with chromosomal DNA, which was removed by digestion with an exonuclease (Plasmid-Safe ATP-dependent DNase; Epicentre). Purified plasmids pHel4 and pHel5 were mechanically dissected into 1.2- to 1.4-kb fragments, and the plasmid fragments were cloned into the Topo-blunt-II vector (Invitrogen) for DNA sequencing of 96 subclones. Every section of the plasmids was covered with at least two contigs. The analysis of the sequences with MAPSORT resulted in HindIII fragments corresponding in size to the restriction patterns, verifying the sequence assembly.
Tetracycline susceptibility determination of H. pylori.
Tetracycline resistance of H. pylori strains was determined by a plate diffusion test. Approximately 108 H. pylori cells were plated on serum plates, and an antibiotic disk with a tetracycline concentration of 30 μg/ml was placed in the middle of the plates. After incubation under microaerobic conditions for 5 days at 37°C the size of the zone of resistance around the antibiotic disk was measured. For quantitative tetracycline resistance determination an E-test strip with a tetracycline gradient of 0.016 to 256 mg/ml was used.
Southern hybridization for detection of mob genes in H. pylori plasmids.
Southern blotting and hybridizations with DNA fragments were performed using the ECL labeling and detection system according to the manufacturer's protocol (Amersham). For hybridization 0.5 M NaCl was used; the washing buffer contained 0.5× SSPE (180 mM NaCl, 10 mM sodium phosphate [pH 7.5], 1 mM EDTA), 6 M urea, and 0.4% SDS at 42°C. Filters were washed with 2× SSPE-0.05% SDS at 42°C.
Computer analyses.
Predictions of ORFs were performed by the programs MAP of Genetics Computer Group (GCG) software (12), ARTEMIS (http://www.sanger.ac.uk/Software/Artemis), with a cutoff of 100 bp, and BLASTX (http://www.sanger.ac.uk/Software/Artemishttp://www.ncbi.nlm.nih.gov/BLAST). The GC contents were determined by COMPOSITION (GCG), the GC profiles were created by ARTEMIS, and the terminator structures were identified with TERMINATOR (GCG). Physical properties of proteins (Mr, pI, and net charge) were calculated with PEPTIDESORT (GCG), and structural properties were determined with PREDICTPROTEIN (http://www.embl-heidelberg.de/predictprotein/). Membrane associations were determined by the programs PEPTIDESTRUCTURE (GCG), PSORT (http://psort.nibb.ac.jp), TMPRED (http://www.ch.embnet.org/software/TMPRED), and TMHMM (http://www.cbs.dtu.dk/services/TMHMM). The program COILS (http://www.ch.embnet.org/software/COILS) was used for finding α-helical coiled-coil domains, SIGNALP (http://www.cbs.dtu.dk/services/SignalP) was used for identification of putative s-dependent signal sequences, and SMART (http://smart.embl-heidelberg.de) was used for determining additional structural properties and domains. Protein motifs were searched with the programs SCANPROSITE (http://www.expasy.org/tools/scnpsit1.html) and MOTIF (http://motif.genome.ad.jp). Homology searches were performed with BLASTP/N (http://www.ncbi.nlm.nih.gov/BLAST) and FASTA3 (http://www2.ebi.ak.uk/fasta3), and identities were calculated with BESTFIT (GCG). Sequence alignments were performed with PILEUP (GCG) and BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html).
Nucleotide sequence accession numbers.
The complete plasmid sequences of pHel4 and pHel5 have been submitted to GenBank, and the following accession numbers have been provided: pHel4, AF469112; pHel5, AF469113.
RESULTS AND DISCUSSION
Nucleotide sequence and gene organization of plasmids pHel4 and pHel5.
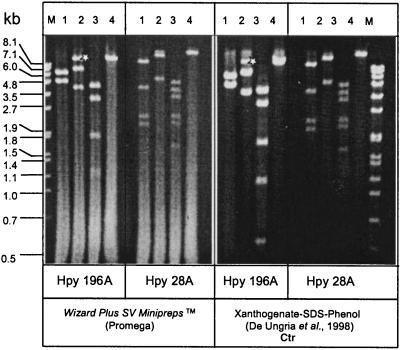
Agarose gel electrophoresis of total H. pylori DNA isolated from a set of 10 H. pylori strains revealed, in addition to the chromosomal DNA, the presence of plasmids of different sizes in five H. pylori strains (P3, P8, P12, P26, and P29) (data not shown). Of these strains only plasmid pHel1 of strain P3 has been studied in detail before (20). Plasmid pHel4 from strain P8 was estimated to be approximately 11 kb in size, and plasmid pHel5 from P29 was estimated to be about 19 kb (Fig. 1). The xanthogenate-SDS-phenol method of plasmid extraction (11) produced better-quality plasmids and less contamination with chromosomal DNA than the Promega Wizard Plus SV Minipreps method (Fig. 1), which was employed in earlier studies (20, 21). Plasmid pHel6, isolated from strain P12, was difficult to isolate by either method, probably due to a high DNase activity in this strain (data not shown).
FIG. 1.
Purification and analysis of H. pylori plasmids by restriction enzyme digestion. Plasmids pHel4 and pHel5 were isolated from H. pylori strains P8 and P29, respectively, by the Wizard Plus SV Minipreps (Promega) and the xanthogenate-SDS-phenol methods. Plasmids were digested with restriction endonucleases BglII (lane 1), EcoRI (lane 2), HindIII (lane 3), and SacI (lane 4) and separated on a 0.8% agarose gel. ∗, partially digested EcoRI fragment. Lane M, marker. kb, kilobases.
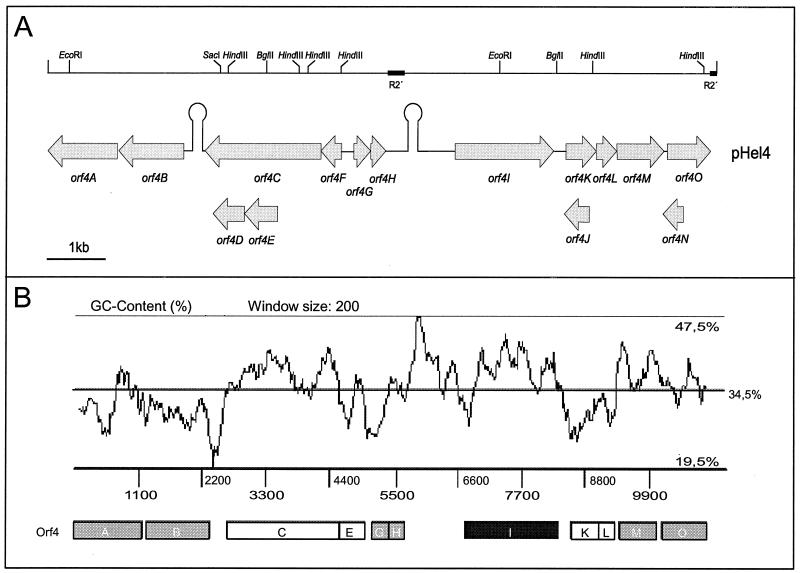
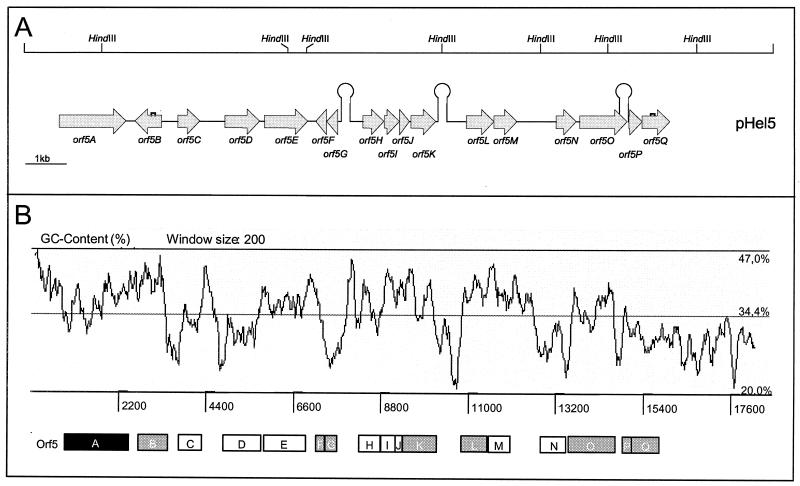
A shotgun approach was used to clone random gene segments for sequencing pHel4 and pHel5. Plasmid pHel4 consists of a total of 10,970 bp, whereas pHel5 consists of 18,291 bp and represents the largest H. pylori plasmid sequence determined to date. The annotation revealed a total of 15 putative ORFs in plasmid pHel4 and 17 in plasmid pHel5 (Fig. 2A and 3A). In addition to ATG, GTG and TTG were identified as putative translation start codons (Tables 1 and 2), a feature that is also described for chromosomal H. pylori genes (1, 42). The overall G+C contents of 34.5 (pHel4) and 34.4% (pHel5) (Fig. 2B and 3B) are significantly lower than that of H. pylori chromosomes of strains 26695 and J99 (39%) (1, 42), though they are comparable to those of other H. pylori plasmids.
FIG. 2.
Physical and genetic map and graphic display of the G+C content of plasmid pHel4. (A) The restriction map (top), as deduced from the plasmid sequence, for BglII, EcoRI, HindIII, and SacI corresponds to the restriction digestion in Fig. 1. The location and orientation of putative ORFs in pHel4 as well as putative terminator sequences (Ω-like symbols), as determined by computer program TERMINATOR, are given below. (B) G+C content plotted over the whole plasmid sequence by using the computer program ARTEMIS. Black box, ORF encoding RepA; grey boxes, putative ORFs with homology to the H. pylori chromosome; white boxes, other putative genes.
FIG. 3.
Physical and genetic map and graphic display of the G+C content of plasmid pHel5. (A) Restriction map (top), as deduced from the plasmid sequence, for HindIII, which corresponds to the restriction digestion in Fig. 1. The location and orientation of putative ORFs in pHel5 as well as putative terminator sequences (Ω-like symbols), as determined by the computer program TERMINATOR, are given below. Half-squares mark shifts in the reading frame for putative genes orf5B and orf5Q, which result in premature termination of the putative proteins. (B) G+C content plotted over the whole plasmid sequence with ARTEMIS. Black box, ORF encoding RepA; grey boxes, putative ORFs with homology to the H. pylori chromosome; white boxes, other putative genes.
TABLE 1.
List of identified ORFs of plasmid pHel4
| ORF | Codon position (start-stop) | Sequencea | Molecular mass (kDa) | pI | Net charge | Propertiesb | Homology (BLASTP)c |
|---|---|---|---|---|---|---|---|
| orf4A | 1166-3 | AATTTATAGAGTTTCTTAATATG | 43.9 | 9.3 | 9 | TM; SP; α-β | HP1165, 1e-92; JHP1092, 2e-92; BB126, 7e-34; TetA(P), 2e-06; MccC, 5e-5 |
| orf4B | 2254-1178 | AAAGTTTTATCGGAGTAGAGTTG | 41.0 | 6.6 | −2 | α-β | MccB, 1e-18 |
| orf4C | 4488-2506 | TTAGAACAACTAAGAGCTAAATG | 78.1 | 10.4 | 26 | Mix; cc | MbeAy, 7e-30; MbeA, 23-27; MobA, 5e-24; BdrC3, 0.026 |
| orf4D | 3245-2739 | CCAAGAAATAAGGGGAAAACATG | 19.6 | 5.3 | −4 | Leucine zipper; α | |
| orf4E | 3785-3255 | ATTAAAAGGATTTTACTACCATG | 20.4 | 10.1 | 8 | TM; cc; α | MobB2, 0.65 |
| orf4F | 4822-4478 | TAAGGGATAGCCAAAAAAATATG | 13.2 | 10.6 | 8 | Mix | MbeCy, 0.021 |
| orf4G | 5020-5298 | GATTGAGTTAAGGAGATAAGATG | 10.8 | 10.1 | 5 | cc; α | JHP0828, 2e-16; HP0316, 0.002 |
| orf4H | 5285-5551 | ATGAGTTAAAAAGAGAGTATATG | 10.7 | 8.7 | 2 | Mix | HP0894, 4e-26; HP0892, 1e-21; JHP0831, 4e-21; JHP0825, 2e-18; ORF10, 1e-10; YafQ, 6e-09 |
| orf4I | 6671-8296 | AAGCATTAAAAGGTGCTTAAATG | 64.1 | 10.2 | 19 | cc; mix | RepA, 0.0 |
| orf4J | 8854-8483 | GAAAAACAAGGGGATCACTAATG | 13.6 | 8.5 | 4 | TM; α-β | |
| orf4K | 8490-8990 | TTGTAATAGGAGTTTAAAAAATG | 20.0 | 9.8 | 6 | α (cc) | |
| orf4L | 8990-9322 | AGTTATGTAAAGAGCATGTAGTG | 13.4 | 9.6 | 3 | TM; cc; mix | |
| orf4M | 9326-10099 | TCAATCGTTTGGAGTAGCATGTG | 29.6 | 6.5 | −5 | Mix | Orf6-pHPM8, 4e-57; JHP0651, 5e-33; HP0712, 7e-22; pHPM180-Orf2, 1e-08; HP0713, 5e-07 |
| orf4N | 10406-10086 | TATTCTAAACTCAAAGGTTCTTG | 12.2 | 9.2 | 1 | TM; α-β | |
| orf4O | 10149-10856 | ACAGGTGAAAAGACAGATGCATG | 27.6 | 7.9 | 1 | Mix | pHPM180-ORF2, e-115; JHP0651, 6e-11; HP0713, 7e-09; ZK593.8, 5e-08; Tou1, 5e-06; ORF56-TP901-1, 0.20; ORF178-wss virus, 0.23 |
The putative Shine-Dalgarno sequence is underlined; the start codon is in boldface.
TM, transmembrane domain; SP, signal peptide; α, α-β, and mix, secondary structure elements as predicted by PREDICTPROTEIN; cc, coiled-coil structure.
BB126, AAC66191; TetA(P), BAA19230; MccC, CAA40810; MccB, CAA40809; MbeAy, AAB05464; MbeA, CAA33883; MobA, AAA69498; BdrC3, AAF19116; BobB2, AAD02406; MbeCy, AAB05463; ORF10, AAF05106; YafQ, Q47149; ZK593.8, T27927; Tou1, AAF08816; ORF56-TP901-1, AAK38073; ORF178-white spot syndrome (wss) virus, AAK77847.
TABLE 2.
List of ORFs of plasmid pHel5
| ORF | Codon positions (start-stop) | Sequencea | Molecular mass (kDa) | pI | Net charge | Propertiesb | Homology (BLASTP) |
|---|---|---|---|---|---|---|---|
| orf5A | 865-2481 | CCAAAAGATAAGGAGTATAGAGTG | 47.0 | 10.25 | 19 | cc; mix | RepA, 0.0 |
| orf5B | 3352-2706 | GCAACTTACACAGGAAAAACAATG | 25.2 | 10.36 | 13 | SP; mix | JHP1295, 2e-37; HP1382, 2e-05 |
| orf5C | 3761-4279 | AAAGGAAAGGAAAATGCAACCATG | 19.9 | 7.81 | 1 | α | |
| orf5D | 4898-5749 | CAACACAAGGAGATTTTAAAAATG | 34.1 | 9.73 | 7 | cc; α | |
| orf5E | 5862-6929 | CACAAGCAAGAAGGACCAAAAATG | 41.6 | 9.28 | 7 | Mix | |
| orf5F | 7387-7121 | ATGAACTAAAAAGAGAGATACATG | 10.8 | 9.24 | 3 | Mix | HP0894, 4e-22; JHP831, 2e-17; HP0892, 3e-17; JHP0825, 5e-13 |
| orf5G | 7652-7374 | AGCTTAATACAAGGAAATGAGATG | 10.8 | 10.19 | 5 | cc; | JHP0828, 4e-11; HP0316, 0.030 |
| orf5H | 8271-8783 | ACTATTTTTATCAATGTCGCAATG | 19.8 | 8.49 | 3 | cc; α | |
| orf5I | 8795-9196 | AGAAGAATAGGAACAAAAAGAATG | 15.0 | 8.50 | 3 | SP; cc; α-β | |
| orf5J | 9193-9405 | GAAAGAACAATTGGATGACTTATG | 8.1 | 9.99 | 4 | Mix | |
| orf5K | 9440-10048 | TTAGACACTAGGAACAAAGTGATG | 23.4 | 5.60 | −10 | Mix | JHP0651, 3e-31; HP0712, 2e-22 |
| orf5L | 10800-11474 | CATTAAATATAAAGGAACAGAATG | 25.5 | 5.96 | −2 | Mix | HP1000, 4e-53; JHP0935, 1e-36 |
| orf5M | 11471-12037 | ACCCAACGAAGAAAGGAAAGTATG | 21.7 | 9.76 | 3 | cc; mix | |
| orf5N | 12898-13491 | CACATAAAAAGGATAAAAATTATG | 23.2 | 9.18 | 5 | Mix | HP0879, 7e-08; JHP812, 8e-08; HP1334, 2e-06 |
| orf5O | 13571-14731 | ATTTTTCCACAAAGGATCGCAATG | 46.7 | 8.98 | 5 | cc; mix | |
| orf5P | 14834-15085 | AAATTTACTAGGAATAGTAAAATG | 9.8 | 4.67 | −4 | Mix | HP0993, 7e-30 |
| orf5Q | 15057-15769 | TCCCTATGGAAAATATTCAGTATG | 27.8 | 6.52 | −4 | Mix | HP0994, 2e-94 |
The putative Shine-Dalgarno sequence is underlined; the start codon is in boldface.
TM, transmembrane domain; SP, signal peptide; α, α-β, and mix, secondary structure elements as predicted by PREDICTPROTEIN; cc, coiled-coil structure.
The best homologies of the putative ORFs to database entries and some important features of the deduced proteins are summarized in Tables 1 and 2. Based on these searches it appears that plasmids pHel4 and pHel5 consist of genes for plasmid maintenance, bacteriocin production, and conjugal transfer and chromosomal homologues of H. pylori, organized in distinct regions. Whether these regions are functional or not has to be demonstrated in the future. Moreover, pHel4 and pHel5 harbor border sequences of IS elements and conserved repeat sequences also found in other plasmids of H. pylori, estimated to act as hot spots for DNA recombination.
Plasmid maintenance regions of pHel4 and pHel5.
Orf4I, encoded by pHel4, and Orf5A, encoded by pHel5, were identified as replication initiation proteins (Rep) showing significant sequence identity to the corresponding RepA proteins encoded by different H. pylori plasmids (Table 3). This group of H. pylori Rep proteins supports replication according to the theta replication mechanism (10). The upstream region of orf4I is highly conserved compared to the pHel1 repA gene, indicating that the same transcriptional start site as that experimentally determined for pHel1 (20) might be used in pHel4. The upstream region of orf5A (pHel5) differs significantly from the corresponding regions of pHel1 and pHel4 but is identical to the upstream region of repA of pHPS1, suggesting that the repA genes of pHel5 and pHPS1 use the same transcriptional start site (data not shown).
TABLE 3.
Comparison of products of ORFs in plasmids pHel4 and pHel5 with products of the H. pylori chromosome
| Chromosome-encoded protein, % identity to pHel4- encoded protein | pHel4-encoded protein | % Identity between pHel4- and pHel5-encoded proteinsa | pHel5-encoded protein | Chromosome-encoded protein, % identity to pHel5- encoded protein |
|---|---|---|---|---|
| Orf41 (RepA) | 78.3 | Orf5A (RepA) | ||
| HP1165, 53.8 | Orf4A | |||
| JHP1092, 52.3 | ||||
| Orf5B | JHP1295, 50.3 | |||
| JHP0828, 67.8 | Orf4G | 91.3 | Orf5G | JHP0828, 73.5 |
| HP0316, 35.6 | HP0316, 35.6 | |||
| HP0892, 61.4 | Orf4H | 96.6 | Orf5F | HP0892, 59.1 |
| HP0894, 60.3 | HP0894, 58.0 | |||
| JHP0831, 60.2 | JHP0831, 59.1 | |||
| JHP0825, 55.7 | JHP0825, 53.4 | |||
| HP0712, 46.0 | Orf4M | 95.0 | Orf5K | HP712, 46.0 |
| JHP0651, 40.6 | JHP0651, 44.0 | |||
| HP0713, 38.1 | HP0713, 44.3 | |||
| JHP0651, 30.1 | Orf4O | 30.7 | Orf5K | JHP0651, 44.0 |
| Orf5L | HP1000, 49.5 | |||
| JHP0935, 45.5 | ||||
| Orf5O | HP0879, 26.7 | |||
| JHP0812, 33.5 | ||||
| HP1334-36.2 | ||||
| Orf5P | HP0993, 85.5 | |||
| Orf5Q | HP0944, 79.7 |
pHel4-encoded RepA identities to RepA of plasmids pHel1, pHPM180, pHPS1: 98.0, 89.3, and 74.8, respectively; corresponding values for pHel5-encoded RepA: 78.7, 81.8, and 76.3%, respectively.
As described for pHel1 and pHPM180, we find a region of 97 bp, consisting of 22-bp iterons repeated 4 times and located about 350 bp upstream of the start codon of the repA gene (data not shown). The sequences of the iterons of pHel4 and pHel1 are identical, whereas the iteron region of pHel5 shows best homology to the iteron sequence of pHPS1 (84%). The iteron regions of pHel4 and pHel5 are 77% identical.
For pHel1 we demonstrated a low copy number of about 4 to 10 plasmid copies per cell in H. pylori (21). In addition, pHel4 and pHel5 replicate stably in H. pylori growing in vitro on agar plates without any selection pressure. The putative gene product of Orf5L has significant homology to a group of ParA proteins involved in the correct partitioning of plasmids to the bacterial daughter cells during cell division, which is necessary for the maintenance of low-copy-number plasmids (17). A protein identical to Orf5L, encoded by plasmid pHPM186 (the unpublished plasmid sequence is available in the database under accession no. AF077006), and one encoded by plasticity zone 3 in H. pylori 26695 (HP1000; 49.5% identity) were described. We also identified JHP0935, homologous to ParA and encoded by a plasticity zone of H. pylori J99; this protein is 45.5% identical to Orf5L. HP1000 was recently grouped in the new ParF subgroup of the ParA superfamily (19). Further studies need to be conducted to show whether or not plasmids without parA or -F homologues, for example, pHel1, pHel4, pHPM180, and pHS1, use the chromosomally encoded plasmid partitioning systems for plasmid maintenance.
Microcin MccC7 homology region.
Orf4A (pHel4) revealed the best sequence homology to a putative TetA(P) tetracycline efflux membrane transporter protein of H. pylori strains 26695 (HP1165) (42) and J99 (JHP1092) (1) (Table 1). Since H. pylori P8 did not tolerate higher concentrations of tetracycline than H. pylori strains 26695, J99, P1, P12, and P29 (0.05 to 0.6 μg/ml; data not shown), it was concluded that Orf4A and chromosomally encoded proteins HP1165 and JHP1092 are probably not involved in tetracycline resistance.
Orf4A displayed a lower homology to several transporter proteins involved in protein secretion, especially E. coli microcin secretion protein MccC (23% identity). Interestingly, the orf4B sequence of pHel4, located upstream of orf4A, seems to be organized with orf4A in an operon, and its product reveals in addition a sequence identity to the MccB protein (28% identity), encoded by the E. coli microcin operon (18). In the E. coli system, MccB is involved in modification of MccA (MccC7) (J. E. Gonzalez-Pastor, J. L. San Millan, and F. Moreno, Letter, Nature 369:281, 1994) and microcin is exported by MccC. The modified secreted MccA peptide is taken up by related bacteria and acts there as an inhibitor of translation if no corresponding immunity protein is expressed. In plasmid pHel4 a candidate microcin structural gene upstream of mccB was also identified (D. Hofreuter and R. Haas, unpublished data).
Conjugation-like ORFs.
Between two regions (bp 2380 to 2454 and bp 5043 to 5245) of pHel4 with very low G+C contents (21 and 25%, respectively) a gene cluster with homology to a conjugal mobilization (mob) region of colicinogenic plasmids was identified. The observed overlap of ORFs orf4C to orf4F (Fig. 4A) is very reminiscent of the structural organization of mobA, mobB, mobC, and mobD of colicin-encoding plasmids pColA (31), pColE1 (7), pColD157 (22), and pWQ799 (26). Orf4C shows significant homology in its N terminus to different MobA proteins and the RLX protein encoded by plasmid pC223 of Staphylococcus aureus, which is also a mobilizing protein (Fig. 4B). Orf4F, encoded by pHel4, reveals best homologies to MobC proteins encoded by colicinogenic plasmids.
FIG. 4.
Comparison of putative pHel4 mobilization (mob) genes with mob genes of other mobilized plasmids. (A) Schematic representation of the mob regions (mobA, mobB, mobC, and mobD) of colicinogenic plasmids such as pColE1, pColA, and pColK. The mob regions of colicinogenic plasmids are organized similarly to the putative mob genes of plasmid pHel4 (orf4C, orf4D, orf4E, and orf4F). (B) Comparison of Orf4C and MobA and MbeA (relaxase) proteins encoded by colicinogenic plasmids. Identical amino acids and conservative replacements compared to Orf4C are in boldface, and the amino acid identities of the corresponding proteins, compared to Orf4C, are shown after the sequences. P. multocida, Pseudomonas multocida; E. cloacae, Enterococcus cloacae; S. enterica, Salmonella enterica; E. stewartii, Erwinia stewartii. (C) Sequences of three conserved motifs with conserved tyrosine (Y, motif I), serine (S, motif II), and histidine (H, motif III) residues compared to the Orf4C amino acid sequence.
MobA proteins belong to the group of relaxases. Together with MobB and MobC they bind to a single cis-active site of a mobilizing plasmid, the origin of transfer (oriT) region. For RP4 TraI and other related relaxases three conserved motifs, an active tyrosine (motif I), a serine (motif II), and two histidines (motif III) are essential for the relaxase function (Fig. 4C) (33). Comparison of Orf4C with the TraI protein and other relaxases identified the conserved tyrosine (motif I) and the serine (motif II) at the expected positions but did not identify the histidine motif, which is also absent in several other relaxases (Fig. 4C).
Although primary sequence homologies between putative proteins encoded by orf4E and orf4D and MobB and MobD proteins are not obvious, there are several conserved motifs in both groups of proteins, indicating a conserved function. In the orf4D-encoded protein there are two leucine zipper motifs (L-X[6]-L-X [6]-L-X[6]-L), which are also found in MobD encoded by colicinogenic plasmids. For Orf4E best homologies with several viral and eucaryotic proteins with coiled-coil structures, as well as an identity of 21% with a mobilizing protein (MobB2) of Staphylococcus epidermidis, were found. Thus, similar structural and physical properties of these proteins imply similar functional properties of the Orf4C, Orf4D, and Orf4F proteins with mobilization proteins encoded by conjugative plasmids. We investigated the distribution of the mob region in H. pylori by analyzing plasmids from clinical H. pylori isolates for the presence of the mobA gene by (i) PCR amplification with specific primers DHO104 and DHO105 and (ii) Southern hybridizations with a mob-specific (orf4C) DNA probe (data not shown). According to these data, about 35% of H. pylori plasmids investigated possess the putative mob region.
A putative origin of transfer (oriT) but no tra functions.
Essential for the mobilization of a plasmid is the presence of an origin of transfer (oriT) in the plasmid itself. The Mob and Tra proteins are usually active in trans and might be encoded by other conjugative plasmids or the bacterial chromosome. The oriT sequences in colicinogenic plasmids are usually located upstream of the mob regions, and the nic sites for plasmids pColE1 and pColA are identical. A corresponding sequence for pHel4 was not found at this position. A putative nic sequence identical to the well-characterized nic sequence of IncP plasmid RP4 was found in orf4M of pHel4 (TATCCTG/C [consensus sequence in bold type]); this sequence might act as a functional nic site of the plasmid. P-type nic sites in conjugative colicinogenic plasmids have not been described yet (29).
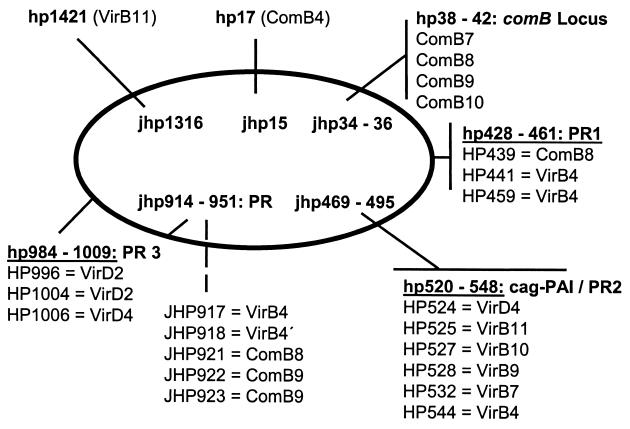
A further question relates to the tra functions for a conjugative transfer of pHel4, which are missing in the plasmid. Several putative proteins, which might be involved in DNA conjugation, are encoded by the genome sequence, especially in plasticity zones of H. pylori 26695 (42) and J99 (1). They show significant homology to VirB and Trb proteins (Fig. 5). The identification of putative IncP type relaxases encoded by hp0996 and hp1004 (5, 41) also supports the conjugative transfer of plasmids in H. pylori.
FIG. 5.
Distribution of genes encoding putative Mob- and Tra-like proteins in the genome of H. pylori 26695 and J99. The tra-like genes are also homologues to the Agrobacterium tumefaciens virB genes, encoding the type IV secretion system for Ti plasmid mobilization and transferred-DNA transfer. In the chromosomes of H. pylori 26695 (HP) and J99 (JHP) virB-like genes are located either in plasticity zones (PR1 to PR3) as operons, such as the comB operon, or as single genes, such as hp1421 and hp17. cag-PAI, cag pathogenicity island.
ORFs in pHel4 and pHel5 whose products are homologous to H. pylori plasmid-encoded and chromosomally encoded gene products.
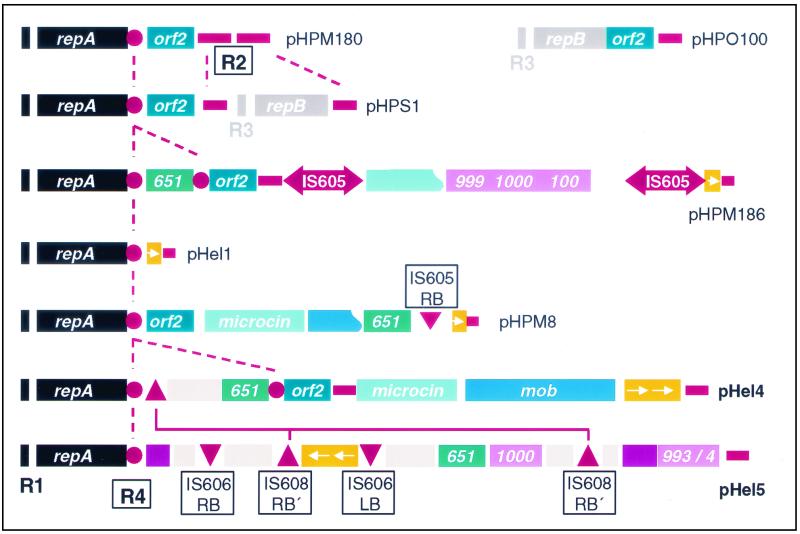
Five ORFs of pHel4 and eight of pHel5 show best homologies to chromosomal genes of strains 26695 and J99. Three ORFs of this group are common to both plasmids, and the deduced pairs of proteins (Orf4G and Orf5G, Orf4H and Orf5F, and Orf4M and Orf5K) show a remarkable sequence identity, ranging between 91.3 and 96.6% (Table 3). We also identified in pHel1, pHPM186, and pHPM8 homologues to orf4H and orf5F; however, in these plasmids the genes appear to be truncated and possess only the conserved 3′ ends, probably as a result of recombination or integration events. These fragments are flanked at the 5′ end by IS elements or border sequences of IS elements (Fig. 6).
FIG. 6.
Schematic comparison of the modules represented on individual H. pylori plasmids. Each plasmid carries a repA or a repB gene or both. The R1, R2, and R3 repeats and the novel R4 repeat are located at the borders of sequence modules. Complete IS605 elements are only present in pHPM186; pHel4 and pHel5 carry border sequences of IS elements, such as IS606 and IS608. Genes with homology to the H. pylori chromosome have the corresponding hp or jhp ORF number, and homologues have the same color. All putative elements involved in recombination or transposition events are red or orange. R1 to R4, repeats R1 to R4; IS, IS element; LB, left border sequence; RB, right border sequence.
The deduced Orf4G and Orf5G proteins reveal best homologies to proteins JHP0828 and HP0316 of H. pylori J99 (1) and 26695, respectively (42), both of which have unknown functions (Tables 1 to 3). The homology between orf4G and orf5G and the chromosomal region of jhp0828 is not restricted to the coding region but extends into a 43-bp upstream region (93% identity). The N-terminal regions of Orf4G and Orf5G show a nearly 100% identity to that of JHP0828, whereas their C termini are most homologous to that of HP0316.
The second pair of homologous proteins, Orf4H and Orf5F, show best homologies to HP0892 and HP0894 of H. pylori 26695, as well as to JHP0825 and JHP0831 of H. pylori J99 (Tables 1 to 3). These hypothetical proteins have well-conserved C termini. The BLASTP search also identified significant homology among Orf4H and Orf5F and gene products of other bacterial species, encoded either on plasmids or in the chromosome (Tables 1 and 2).
Most unexpected, orf4G and orf4H of plasmid pHel4 and orf5F and orf5G of pHel5 have the same organization as the corresponding chromosomal genes of H. pylori 26695 and H. pylori J99, suggesting an exchange of the gene clusters between chromosome and plasmid. For the chromosomal genes it was shown that the members of these two families interact with each other in a two-hybrid screen (36). Homologies by BLASTP are as follows: HP0316 and HP0895, 1e-57; HP0894 and HP0895, 1e-71; HP0895 and HP0895, 1e-204. ORFs in the chromosomes of H. pylori 26695 and J99 homologous to orf4G and orf4H and orf5F and orf5G are clustered in regions with high genetic diversity (9).
A switch-inducing repeat 1 sequence located in plasmids pHel4 and pHel5.
Interestingly, genes homologous to orf4G and orf5G are part of the repeat 1 sequence (42), which is located at different positions in the chromosome and which is coupled with the 3′ regions of different OMPs. The repeat 1 sequence can be considered a switch-inducing sequence, which might be supported by the concerted activity of Orf4G and Orf5G or Orf4H and Orf5F, acting as recombination-inducing proteins.
Such a switch mechanism, which is related to an intrachromosomal recombination event at the repeat 1 sequence, is actually found when the 26695 and the J99 genomes are compared. This switch resulted in an exchange in the chromosomal locations of bacterial adhesin genes babA and babB (1, 42). This mechanism leads to variation in the C termini of the corresponding OMPs (25, 34). Such a genetic switch might result in an immune system escape mechanism by phase variation of immunodominant OMPs but might also result in functional variation of the adhesin binding properties of BabA or BabB (25).
The third pair of common genes found in pHel4 and pHel5 encode proteins with homology to products of chromosomal genes orf4M, orf4O, and orf5K (Table 3). Orf4M and Orf5K have identities of between 38 and 46% to hypothetical proteins JHP651 of H. pylori J99 and HP0712 and HP0713 of H. pylori 26695. JHP0651 might be a fusion between HP0712 (N terminus) and HP713 (C terminus). The BLASTP analysis also identified homology to eucaryotic proteins, for instance, ZK593.8 of Caenorhabditis elegans and human huntingtin interacting protein E, an approximately 350-kDa protein of unknown function involved in the neuropathology of Huntington's disease (15). Orf4M and Orf5K also possess homology to the Orf2 protein, described first as a product of H. pylori plasmid pHPM180 (30), and to Orf-2, encoded by the NBU1 element (nonreplicating Bacteroides units) (43). The homology is clustered mainly in the N-terminal parts of Orf4M and Orf5K, which contain the conserved amino acid sequence PFSDGNGRTGRALMF (data not shown). Thus, Orf4M, Orf4O, and Orf5K belong to a group of proteins whose coding sequences are widespread in bacteriophages, plasmids, and NBU-1 elements and also in eubacteria and some eucaryotic organisms.
Further plasmid-encoded proteins also encoded by the H. pylori chromosome.
As observed in pHPM186, plasmid pHel5 encodes proteins with homology to products of genes clustered in plasticity zone 3 of H. pylori 26695. As described above, Orf5L belongs to the group of ParA proteins, as do the homologous proteins HP1000 and JHP0935. Additionally, orf5P and orf5Q encode proteins with high identity to HP0993 and HP0994, hypothetical proteins encoded by genes in the H. pylori plasticity zones with unknown functions (Table 3). The sequence of orf5Q in pHel5 carries a frameshift, resulting in a truncated HP0994 gene product. Further proteins encoded by pHel4 and pHel5 with homology to products of genes widespread in the chromosomes of H. pylori 26695 and J99 are Orf5B (similar to endonuclease HP1295) and Orf5O (best homologue to hypothetical protein HP1334).
Repeat sequences and site-specific recombination events determine size variation and the modular structure of H. pylori plasmids.
The comparison of independent H. pylori plasmid sequences identified several sequence repeats termed R1, R2, and R3 (10). The R1 and R3 repeats correspond to iteron sequences, located upstream of genes encoding replication initiation proteins RepA and RepB. It has been suggested that the R2 repeat, which consists, in pHP180, of two copies of 232 noncoding nucleotides, might act as a target sequence for recombination events (10, 30). Our sequence analysis of pHel4 and pHel5 revealed for both plasmids incomplete sequences of the 232-bp R2 repeat, designated R2′ (Fig. 6). For pHel4 we found one nearly complete 232-bp sequence (bp 5665 to 5842) between orf4H and the putative origin of replication. This region also contains an intact 36-bp stretch (bp 5578 to 5613), which was first found between the two R2 repeats in pHPM180. A second incomplete copy of the R2 repeat in pHel4 is between orf4O and orf4A (bp 10845 to 10954). For pHel5, one identical stretch to the 232-bp sequence was found (bp 18120 to 18291). The comparison of the locations of repeat sequences in H. pylori plasmids suggests a modular structure of H. pylori plasmids with insertion and deletion of sequence modules at different repeat sequences, which might act as hot spots for recombination or site-specific integration events (Fig. 6).
A novel R4 repeat in H. pylori plasmids is associated with gene shuffling.
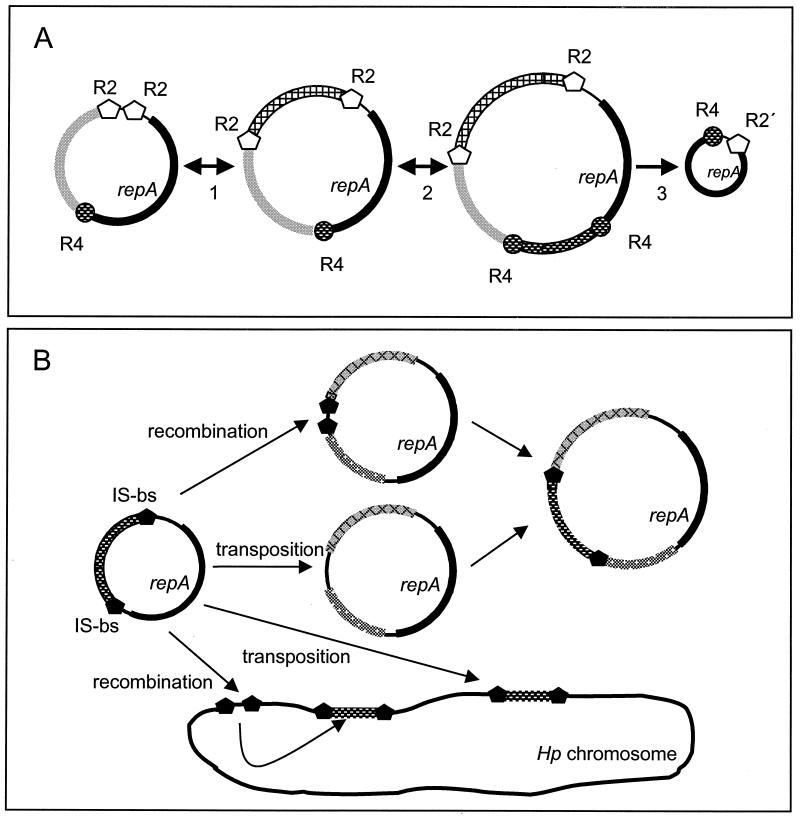
Further studies showed that in pHel4 a stretch of 36 bp located at the 3′ end of the repA gene occurred twice and that the two copies are separated by about 1.7 kb (Fig. 7A). We designated this 36-bp sequence (5′-CATTTGAAAAATTGGTTCAAGAAACACTACAGGTAA-3′), which codes for the C terminus of RepA (HLKNWFKKHYR), the novel R4 repeat. The R4 repeat is also well conserved in plasmids pHel5 (this study), pHel1 (20), pHPM180 (30), pHPS1 (10), and pHPM186 (accession no. AF077006) (Fig. 6). We were interested to test whether or not the duplication of the 36-bp sequence, as in pHel4 and pHPM186, is a common event in H. pylori plasmids. PCR amplification with oligonucleotides DHO106 and DHO107 (Fig. 7B) revealed fragments of 1.7 and 0.8 kb in pHel4 and four plasmids isolated randomly without prior screening from independent H. pylori isolates, respectively. Since the PCR fragments can only be generated if the R4 repeat is duplicated (Fig. 7A), we postulate the site-specific integration of a DNA fragment at the 3′ end of the repA gene for these plasmids, resulting in a duplication of the conserved R4 repeat. The 1.7-kb fragment of pHel4 codes for putative proteins Orf4J, Orf4K, Orf4L, and Orf4M. Orf4M has significant homology to HP712, HP713, and JHP651 (Table 3). Direct sequencing of two 800-bp PCR products revealed sequences with coding capacity for a protein with identity to Orf4M (100%), Orf5K (97%), Orf6-pHPM8 (86%), and JHP0651 (41%). These experimental data support our hypothesis that the R4 repeat might act as a site of integration of novel sequence modules into H. pylori plasmids (Fig. 7).
FIG. 7.
R4 repeats involved in gene shuffling. (A) Primer design for the detection of gene integration events by the R4 repeat in different H. pylori plasmids. The 3′ ends of repA genes of different H. pylori plasmids are shown. Primers DHO106 and DHO107 were designed to amplify a PCR product only after integration of novel DNA into the R4 repeat sequence, as is the case for pHPM186 and pHel4. (B) Demonstration of the integration of novel sequence modules by PCR amplification with primers DHO106 and DHO107 in plasmid pHel4 and four random H. pylori plasmids, pHP49, pHP57, pHP63, and pHP74, compared to the genetic organization of “empty-site” plasmid pHel1. 651, homologue to the chromosomal gene jhp0651. R4, R4 repeat.
From a comparison of all available plasmid sequences we strongly suggest that the R2 repeat sequence described first by Minnis et al. (30) and the R4 repeat identified in this study are hot spots for genetic recombination. This repeat recombination model could explain the observed size variation in the plasmids described so far. Some of the cryptic proteins encoded by the plasmid or the chromosome might be involved enzymatically in the specific recombination events.
Partial and complete IS elements on plasmids pHel4 and pHel5.
Plasmids pHel4 and pHel5 do not carry any complete IS elements, unlike pHPM186. Instead, we identified several regions in pHel5 identical to border sequences of IS elements, such as IS606 and IS608, and to the core sequence of mini-IS605 (42). A stretch of 35 bp between orf5G and orf5H is identical to the left border sequence of IS606. Between orf5C and orf5D there are 35 bp identical to the right border sequence of IS606. This region also has homology to mini-IS605. A second smaller stretch of this IS606 inverted repeat right (IRR) is located between orf5M and orf5N. This sequence is part of a sequence duplication (5′-TTTTGACATACTCCCCATAGCTAAAGCTAGAGACTTTGCGG-3′) in pHel5 (bp 6997 to 7037 and 12353 to 12380) since a second copy of that sequence was identified between orf5E and orf5F (Fig. 6). A single copy of this 41-bp sequence could also be found in pHel4 between the R4 repeat and orf4J.
Again, the repeat and IS border sequences are always located at defined positions, flanking certain genes or groups of genes, such as repA (R2 and R4 repeats), the mcc-mob region in pHel4 and pHP186 (R2 repeat), and the orf4J-orf4M region of plasmid pHel4 (R4 repeat). The advantage of such a modular organization is rather obvious (Fig. 8). H. pylori can distribute a high number of diverse genetic modules in the population. Different combinations of modules might be created by recombination (deletion and insertion) and selected for by the needs of the bacteria in their individual hosts. Due to the modular structure, plasmids might either pick up chromosomal genes of H. pylori or integrate sequence modules from foreign plasmids, which are taken up by the bacteria during its natural transformation competence (gene shuffling). For conjugative plasmids, as we postulate for pHel4, the novel sequences could be rapidly distributed within the H. pylori population and exchange novel plasmid sequences with the bacterial chromosome. Such events might help to explain the development of macrodiversity among H. pylori strains and the rapid generation of substrains.
FIG. 8.
Model for plasmid size variation and gene shuffling in H. pylori. (A) Integration and deletion events for modules at the R2 repeat (step 1) and the R4 repeat (step 2). The deletion of modules may occur at any stage and finally results in the basic replicon only (step 3). (B) Model for the exchange of gene sequences between different H. pylori plasmids and the H. pylori chromosome. The basic mechanisms might be recombination or transposition via the IS border sequences present in H. pylori plasmids, e.g., pHel4 and pHel5, and in the H. pylori (Hp) chromosome. Identical genes or elements are represented by the same patterns.
Acknowledgments
We thank E. Weiss for excellent technical assistance and B. P. Burns for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (HA2697/3-1).
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ando, T., Q. Xu, M. Torres, K. Kusugami, D. A. Israel, and M. J. Blaser. 2000. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 37:1052-1065. [DOI] [PubMed] [Google Scholar]
- 4.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. R. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 5.Backert, S., E. Von Nickisch-Rosenegk, and T. F. Meyer. 1998. Potential role of two Helicobacter pylori relaxases in DNA transfer? Mol. Microbiol. 30:673-674. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1996. The bacteria behind ulcers. Sci. Am. 274:104-107. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, A. C., J. A. Archer, and D. J. Sherratt. 1989. Characterization of the ColE1 mobilization region and its protein products. Mol. Gen. Genet. 217:488-498. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 9.Cao, P., and T. L. Cover. 1997. High-level genetic diversity in the vapD chromosomal region of Helicobacter pylori. J. Bacteriol. 179:2852-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Ungria, M. C., T. Kolesnikow, P. T. Cox, and A. Lee. 1999. Molecular characterization and interstrain variability of pHPS1, a plasmid isolated from the Sydney strain (SS1) of Helicobacter pylori. Plasmid 41:97-109. [DOI] [PubMed] [Google Scholar]
- 11.De Ungria, M. C., D. Tillett, B. A. Neilan, P. T. Cox, and A. Lee. 1998. A novel method of extracting plasmid DNA from Helicobacter species. Helicobacter 3:269-277. [DOI] [PubMed] [Google Scholar]
- 12.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton, K. A., C. L. Brooks, D. R. Morgan, and S. Krakowka. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 59:2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faber, P. W., G. T. Barnes, J. Srinidhi, J. Chen, J. F. Gusella, and M. E. MacDonald. 1998. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 7:1463-1474. [DOI] [PubMed] [Google Scholar]
- 16.Ge, Z., and D. E. Taylor. 1999. Contributions of genome sequencing to understanding the biology of Helicobacter pylori. Annu. Rev. Microbiol. 53:353-387. [DOI] [PubMed] [Google Scholar]
- 17.Gerdes, K., J. Moller-Jensen, and J. R. Bugge. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Pastor, J. E., J. L. San Millan, M. A. Castilla, and F. Moreno. 1995. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J. Bacteriol. 177:7131-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes, F. 2000. The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol. Microbiol. 37:528-541. [DOI] [PubMed] [Google Scholar]
- 20.Heuermann, D., and R. Haas. 1995. Genetic organization of a small cryptic plasmid of Helicobacter pylori. Gene 165:17-24. [DOI] [PubMed] [Google Scholar]
- 21.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257:519-528. [DOI] [PubMed] [Google Scholar]
- 22.Hofinger, C., H. Karch, and H. Schmidt. 1998. Structure and function of plasmid pColD157 of enterohemorrhagic Escherichia coli O157 and its distribution among strains from patients with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 36:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 24.Hofreuter, D., S. Odenbreit, G. Henke, and R. Haas. 1998. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol. Microbiol. 28:1027-1038. [DOI] [PubMed] [Google Scholar]
- 25.Ilver, D., A. Arnqvist, J. Ogren, I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Borén. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 26.Keenleyside, W. J., and C. Whitfield. 1995. Lateral transfer of rfb genes: a mobilizable ColE1-type plasmid carries the rfbO:54 (O:54 antigen biosynthesis) gene cluster from Salmonella enterica serovar Borreze. J. Bacteriol. 177:5247-5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleanthous, H., C. L. Clayton, and S. Tabaqchali. 1991. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in gram-positive bacteria. Mol. Microbiol. 5:2377-2389. [DOI] [PubMed] [Google Scholar]
- 28.Kuipers, E. J., D. A. Israel, J. G. Kusters, and M. J. Blaser. 1998. Evidence for a conjugation-like mechanism of DNA transfer in Helicobacter pylori. J. Bacteriol. 180:2901-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 30.Minnis, J. A., T. E. Taylor, J. E. Knesek, W. L. Peterson, and S. A. McIntire. 1995. Characterization of a 3.5-kbp plasmid from Helicobacter pylori. Plasmid 34:22-36. [DOI] [PubMed] [Google Scholar]
- 31.Morlon, J., M. Chartier, M. Bidaud, and C. Lazdunski. 1988. The complete nucleotide sequence of the colicinogenic plasmid ColA. High extent of homology with ColE1. Mol. Gen. Genet. 211:231-243. [DOI] [PubMed] [Google Scholar]
- 32.Nedenskov-Sorensen, P., G. Bukholm, and K. Bovre. 1990. Natural competence for genetic transformation in Campylobacter pylori. J. Infect. Dis. 161:365-366. [DOI] [PubMed] [Google Scholar]
- 33.Pansegrau, W., W. Schroder, and E. Lanka. 1994. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem. 269:2782-2789. [PubMed] [Google Scholar]
- 34.Peck, B., M. Ortkamp, K. D. Diehl, E. Hundt, and B. Knapp. 1999. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 27:3325-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penfold, S. S., A. J. Lastovica, and B. G. Elisha. 1988. Demonstration of plasmids in Campylobacter pylori. J. Infect. Dis. 157:850-851. [DOI] [PubMed] [Google Scholar]
- 36.Rain, J. C., L. Selig, H. De Reuse, V. Battaglia, C. Reverdy, S. Simon, G. Lenzen, F. Petel, J. Wojcik, V. Schachter, Y. Chemama, A. Labigne, and P. Legrain. 2001. The protein-protein interaction map of Helicobacter pylori. Nature 409:211-215. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Saunders, N. J., J. F. Peden, D. W. Hood, and E. R. Moxon. 1998. Simple sequence repeats in the Helicobacter pylori genome. Mol. Microbiol. 27:1091-1098. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt, W., S. Odenbreit, D. Heuermann, and R. Haas. 1995. Cloning of the Helicobacter pylori recA gene and functional characterization of its product. Mol. Gen. Genet. 248:563-572. [DOI] [PubMed] [Google Scholar]
- 40.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPbeta plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 42.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 43.Wang, J., N. B. Shoemaker, G. R. Wang, and A. A. Salyers. 2000. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J. Bacteriol. 182:3559-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warren, J. R., and B. Marshall. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet i:1273-1275. [PubMed]