Abstract
Replicate populations of the social bacterium Myxococcus xanthus underwent extensive evolutionary adaptation to an asocial selective environment (liquid batch culture). All 12 populations showed partial or complete loss of their social (S) motility function after 1,000 generations of evolution. Mutations in the pil gene cluster (responsible for type IV pilus biogenesis and function) were found to be at least partially responsible for the loss of S motility in the majority of evolved lines. Restoration (partial or complete) of S motility in the evolved lines by genetic complementation with wild-type pil genes positively affected their fruiting body development and sporulation while negatively affecting their competitive fitness in the asocial regime. This genetic tradeoff indicates that mutations in the pil region were adaptive in the asocial selective environment. This finding was confirmed by experiments showing that defined deletions of pil gene regions conferred a competitive advantage under asocial conditions. Moreover, an amino acid substitution in an evolved genotype was located in a region predicted by genetic complementation analysis to bear an adaptive mutation.
Social interactions among individuals of the same species are integral to the life cycles of many animals (9, 10, 25, 26, 43). It is now known that many bacteria are also inherently social (32). Their relative simplicity makes bacteria attractive for studying numerous evolutionary questions (22-24, 27, 31, 35, 37, 39), including those pertaining to social interactions (1, 38, 40). The myxobacteria are of particular interest because of their diverse social behaviors, which include social motility, cooperative predation, and multicellular development (6, 11, 15, 21, 28, 34).
As predators, myxobacteria swarm together over solid surfaces in search of prey, which they kill and lyse by secreting multiple antibiotics and proteolytic enzymes (30). Their growth efficiency is density dependent because larger groups of bacteria produce higher concentrations of degradative enzymes per unit area than do smaller groups (29). The swarming of Myxococcus xanthus involves two genetically distinct yet interacting motility systems (12, 13): adventurous (A) and social (S). The A motility system allows individual cells to move independently of other cells, whereas S motility requires cell-cell proximity (15, 44). A motility has been shown to function primarily on hard agar surfaces, whereas S motility works primarily on soft agar (33), perhaps indicating that the two systems evolved to allow effective predation in different environments. Both systems are genetically complex, involving as many as 100 genes each (P. Youderian, personal communication).
Motility is also necessary for M. xanthus to form multicellular fruiting bodies, which they do under conditions of starvation. At least one of the two motility systems must be functional for development to occur (19). Upon exhausting available growth substrates, ∼105 M. xanthus cells swarm toward aggregation points in response to a quorum-sensing signal (18). These aggregates then develop into fruiting bodies, within which a fraction of the cells differentiate from rod-shaped vegetative cells into spherical spores that are resistant to heat, sonication, and detergent (17). The whole developmental process involves at least five stage-specific intercellular signals that mediate the expression of numerous developmentally regulated genes (3, 5, 8, 16, 34). Recent mutational analyses suggest there are ∼300 genes involved in development, representing a sizable portion of the 9.45-megabase M. xanthus genome (2; P. Youderian, personal communication).
In a previous study (38), we investigated the fate of M. xanthus social traits during evolution in an asocial selective environment, where neither motility nor development was necessary for growth or survival. After M. xanthus was grown in a nutrient-rich liquid regime for 1,000 generations, we observed major losses in social capacity in all 12 replicate lineages. In particular, mutants that were completely defective in S motility either became fixed or increased to majority type in every line, which suggested that these losses were adaptive in the asocial regime. Most but not all lines also lost the ability to form fruiting bodies and develop spores. Taken together, these results led us to hypothesize that the maintenance of M. xanthus social traits in general is costly to fitness in a physically unstructured environment with abundant resources. In this study, we test this hypothesis by genetically rescuing the evolved losses of S motility and then quantifying the effects of this restored function on competitive fitness in the asocial regime.
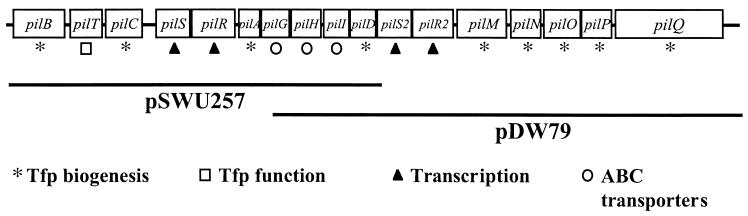
In previous screens for S-motility genes, a large number of knockout mutations have mapped to the pil region of M. xanthus (Fig. 1), a cluster of 17 loci involved in the biogenesis and function of type IV pili (12, 41, 42, 44-46). In our study, plasmids bearing ancestral wild-type pil genes were integrated into evolved clones that had become defective in S motility, and the resulting merodiploids were screened for restoration of S motility. Clones from four evolved lines that were rescued by the ancestral pil genes were then characterized in more detail. The extent of S-motility rescue of these clones was quantified, as were the effects of rescue on hard-agar motility, fruiting body morphology, spore production, and most importantly, competitive fitness in the asocial regime. The fitness effects of rescuing S motility in the evolved clones were compared to those of rescuing S motility in defined pil mutants. Finally, we searched for specific mutational changes in a defined pil segment of an evolved clone within which complementation analysis predicted the presence of at least one such mutation. One mutation that caused an amino acid substitution was identified.
FIG. 1.
pil region of the M. xanthus genome. The 17 genes in the pil cluster are shown along with the corresponding regions on the two plasmids used in this study (pSWU257 and pDW79). The symbol below each gene refers to its known or inferred function. Mutations in all genes except three are known to abolish S motility; mutations in pilS and pilS2 (both of which are homologous to sensor histidine kinases) do not appear to affect S motility, and pilR2 has not been analyzed. Tfp, type IV pili (41).
MATERIALS AND METHODS
Strains.
Ancestral strain S is a clonal reisolate of the fully motile (A+ S+) M. xanthus strain DK1622 (14). Ancestral strain R is a spontaneous rifampin-resistant mutant of strain S, and it provides a selectable marker in competition experiments. These two clones were used to initiate 12 independent lineages, S1 to S6 (derived from ancestor S) and R1 to R6 (derived from ancestor R), that evolved for 1,000 generations in liquid batch culture (38). At generation 1,000, three clones from each line (36 total) were isolated and stored at −80°C in 20% glycerol. Defined mutants DK10410 and DK10416 are derivatives of DK1622 that have markerless in-frame deletions in the pilA and pilB gene, respectively (45). Transformants of evolved clones and defined mutants are denoted by the lineage or strain name, respectively, followed by the transforming plasmid, with a plus sign indicating rescued S motility and a minus sign indicating lack of rescue (e.g., S2/pDW79/+, R6/pSWU257/−, and DK10410/pSWU257/+).
Transformations.
The two ancestors and 36 evolved clonal isolates were each transformed with plasmids pSWU257 (44) and pDW79 (42), which between them carry the ancestral (wild-type) versions of all known genes in the pil region (Fig. 1). The ΔpilA and ΔpilB mutants were transformed with pSWU257 only. Both plasmids carry a gene that confers resistance to kanamycin, and neither can replicate autonomously in M. xanthus. Upon transformation, these plasmids integrate into the chromosome via homologous recombination, resulting in a merodiploid state for the pil region originally contained on the plasmid. The two copies (one ancestral and one evolved) of the pil region therefore differ only by whatever mutations accumulated in that region during the 1,000 generations of experimental evolution.
To prepare competent cells of each strain, 1.5 ml of a mid-exponential-phase culture (∼108 cells/ml) were spun at 12,000 rpm for 2 min, then washed in 1 ml of sterile deionized water, and pelleted three times. After the final wash, the supernatant was removed, and the pellet was resuspended to a volume of ∼100 μl, to which 2 μl of the miniprep plasmid DNA of interest was added. Samples were electroporated at 400 Ω, 25 μF, and 650 V. One milliliter of CTT liquid medium (14) was added immediately after electroporation, and the sample was transferred to a flask containing 1.5 ml of CTT and shaken at 300 rpm for 4 to 8 h at 32°C. Samples were mixed with CTT soft agar (0.7%) at 50°C and plated on CTT hard agar (1.5%) with 40 μg of kanamycin per ml. Transformants were grown in CTT-kanamycin liquid and stored at −80°C in 20% glycerol.
Phenotypic screen of transformant motility.
Six transformants from each combination of evolved clone and plasmid were scored qualitatively for their ability to swarm on CTT soft agar (0.5%) plates. A+ S− genotypes do not swarm on soft agar (only on hard, 1.5% agar), so any swarming activity indicated at least partially functional S motility. Evolved clones that yielded S-motility-rescued transformants (S+) by a particular plasmid also produced some S− but kanamycin-resistant transformants by the same plasmid. These unrescued transformants may arise from gene conversion or from polar effects of pil mutations, e.g., if one end of the integrated plasmid DNA lies within an operon (7). Because these S− transformants nonetheless underwent vector integration and were resistant to kanamycin, they served as excellent controls for detecting any effects of transformation per se on fitness and other traits that are distinct from the effects specific to S-motility rescue.
Motility rate assay.
Clonal freezer stocks were inoculated into 5 ml of CTT liquid, grown to high density at 300 rpm and 32°C, and diluted to ∼2 × 107 to 3 × 107 cells ml−1. Cultures were then grown overnight, diluted, transferred in triplicate to total culture volumes of 11 ml, and grown overnight again. During this final growth cycle, cell densities were estimated during log-phase growth using a Coulter ZM particle counter, 10 ml of each culture was spun at 4,900 × g for 15 min, and pellets were resuspended to ∼5 × 109 cells ml−1 in TPM liquid (20). After resuspension in TPM liquid, 5-μl samples of each strain were placed at the center of three 0.5% agar CTT plates and three 1.5% agar CTT plates and allowed to dry, and the plates were incubated upside down at 32°C. Swarm perimeters were marked after 1 and 4 days of incubation.
To estimate motility rates, two perpendicular transects were made through the center of each swarm at random orientation. Along the four resulting radial vectors, the distance traveled by the swarm perimeter between days 1 and 4 was measured. The motility rate for each replicate was calculated as the mean of these four measurements. The motility rate reported for each strain represents the grand mean of 12 vector measurements across three replicate plates. The three replicate-plate means (not the 12 vector measurements) were used as independent observations for statistical comparisons. The Student-Newman-Keuls multiple-comparison test was used to compare the motility rates of evolved clones, their corresponding derived transformants, and their respective ancestor (S or R).
Sporulation assay.
Liquid cultures used to initiate sporulation assays were prepared using the same protocol as for the motility rate assays. After resuspension in TPM liquid, 100-μl aliquots were spotted in the center of TPM agar plates and allowed to dry. The plates were incubated for 72 h at 32°C, at which time developmental spots were harvested with a scalpel blade and suspended in 500 μl of TPM liquid. These developmental samples were sonicated to disperse spores and then heated in a 50°C water bath for 1 h to kill any nonspores that may have survived sonication. The samples were then plated in 10 ml of CTT soft agar (0.5%) at multiple dilutions, and colonies produced from germinated spores were counted after 6 or 7 days at 32°C. The reported counts indicate the number of cells that became viable spores per 100-μl spot on the TPM plate; each value reported is the mean of three replicate spots, and data were log10 transformed.
We also performed development experiments with mixed cultures of ancestor S and transformants of evolved clones S2 and R2a. The protocol above was followed except that, after resuspension of pellets, strain S was mixed at 99:1, 9:1, and 1:1 ratios with S− (unrescued) kanamycin-resistant derivatives of S2 and R2a and their S+ (rescued) counterparts. After development, sonication, and heating, these mixed samples were plated in 10 ml of CTT-kanamycin (40 μg/ml) soft agar as well as in nonselective CTT soft agar (0.5%). Spore counts for the kanamycin-sensitive ancestor, S, were calculated as the difference between total counts in nonselective and selective agar. As controls, development of pure cultures of the ancestor and each of the evolved variants was carried out in the same experimental block as the mixed cultures.
Assays of competitive fitness in the asocial regime.
Competitors were inoculated independently into 5 ml of CTT liquid and grown for 2 days at 300 rpm to high density. Each culture was then diluted to ∼2 × 107 to 5 × 107 cells ml−1 and grown for 24 h at 120 rpm before competing strains were mixed, diluted, and allowed to undergo a final preconditioning cycle together (also at 120 rpm). For all competitions, one strain resistant to either kanamycin or rifampin was mixed at an initial frequency of 0.1 with another strain sensitive to the corresponding antibiotic to give a total density of ∼2 × 107 cells/ml. Replication was introduced at this stage so that each pair of competitors was mixed and preconditioned in three to six (depending on the experiment) independent culture flasks.
After the 24-h preconditioning cycle, the mixed cultures were diluted 100-fold (0.1 ml into 9.9 ml) into fresh medium, at which time samples were diluted and plated in both selective and nonselective CTT soft agar (0.5% agar, 10 ml per plate) to determine the initial densities of each competitor. The mixed cultures were then propagated for either four or seven cycles with 100-fold daily dilution, after which samples were plated again to obtain final densities of each competitor. Competitions between pairs of evolved genotypes lasted 7 days, which allowed us to detect small fitness differences, whereas competitions between evolved and ancestral types lasted only 4 days, owing to much larger fitness differences. (In our original experimental design, all competitions ran for 7 days. In the competitions between ancestral and evolved genotypes, however, the fitness differences were so large that final plate counts of the ancestral strains were too small to allow accurate estimation of relative fitness. These competitions were therefore repeated in a second experiment, with appropriate internal controls, that lasted 4 days.)
All competitions involving a defined mutant ran for 7 days. Genotypes with functional S motility tend to form a ring of cells and organic matter at the glass-liquid-air interface under these culture conditions. As was done during the evolution experiment proper (38), this ring was scraped off with a sterile stick and mixed into the liquid prior to the daily transfers and prior to plating samples from any competition that involved an S+ strain.
The fitness (W) of one genotype (i) relative to another (j) is calculated as the ratio of their net rates of increase (23): Wij ∼ mi/mj. The net rate of increase (m) for genotype i (adjusted for the daily dilution into fresh medium) is mi = (1/t) ln [Dt × Ni(t)/Ni(0)], where D is the dilution factor (here, 100), t is the number of transfer cycles, and Ni(0) and Ni(t) are initial and final population densities (at the same stage of the growth cycle), respectively.
Sequencing.
Almost the entire ∼3.4-kb pil region common to both pSWU257 and pDW79 was sequenced from clone R6 and its direct evolutionary ancestor, strain R. Genomic DNA was prepared using an MBI Fermentas genomic DNA purification kit. The region of interest was PCR amplified from the genomic DNA samples with three pairs of 5′→3′ and 3′→5′ primers, the products of which have overlapping ends and span the entire 3.4-kb region. PCRs included 10 μl of 5× PCR buffer, 10 μl of 5× GC-Melt, and 5 U of Clontech Advantage GC Genomic Taq polymerase mix (all from a Clontech Advantage GC genomic polymerase kit), 2 μg of genomic DNA template, 100 pmol of each primer, 5 μl of 2 mM deoxynucleoside triphosphates (dNTPs), and high-pressure liquid chromatography (HPLC)-grade H2O to 50 μl. PCR conditions were 180 s at 95°C, 30 cycles (with 1 cycle consisting of 45 s at 95°C, 45 s at 55°C, and 120 s at 68°C), 180 s at 68°C, and storage at 4°C. Then 45 μl of the first-round PCR product sample was run on 1% TAE-agarose gels at 80 V, and bands of expected molecular weight were purified with an MBI Fermentas DNA extraction kit.
A second round of PCR was then performed under identical conditions to the first round except that 1 μl of each first-round, gel-purified fragment sample was used as the template. Second-round PCR samples were cleaned after amplification with Qiagen QIAquick purification columns and diluted in H2O to 6.5 ng/μl. Sequence cycling reactions were performed with an ABI Prism BigDye terminator cycle reaction kit and included 80 ng of the second-round PCR product as the template. Sequencing samples were processed by the Max-Planck Institute for Developmental Biology Sequencing Center. Resulting sequences were analyzed with SeqMan II software.
To avoid false-positive sequence differences introduced during PCR, the two-round amplification process was performed twice independently for each primer pair-by-strain combination. Both independently derived PCR products from each combination were sequenced with each sequencing primer. In total, 14 sequencing primers (some 5′→3′ and some 3′→5′) were used to sequence the entire 3.4-kb region. PCR and sequencing primer sequences, positions, and orientations are available upon request.
RESULTS
Qualitative screen for S-motility rescue.
The 36 evolved clones (three each from 12 lines) that were transformed with plasmids carrying wild-type pil genes were examined for their ability to swarm on soft agar, which is a screen for the presence of S motility (Table 1). Some or all clones from 4 of the 12 evolved lines (S5a, R1a, R2b, and R3a) showed rescue of S motility by pSWU257 alone, and only plasmid pDW79 rescued clones from four lines (S2, S3a, S4, and R5a). Certain clones from four lines (S5b, R1b, R2a, and R6) were rescued to some degree by both plasmids. Note that two distinct clones from the same line sometimes showed different patterns of rescue (e.g., R2a and R2b). None of the clones tested for S1, S6, or R4 were rescued by either plasmid.
TABLE 1.
Rescue of S motility by ancestral pil genes in S− evolved lines
| Evolved line | S motilitya with transforming plasmid:
|
|
|---|---|---|
| pSWU257 | pDW79 | |
| S1 | − | − |
| S2 | − | + |
| S3a | − | + |
| S3b | − | − |
| S4 | − | + |
| S5a | + | − |
| S5b | + | + |
| S6 | − | − |
| R1a | + | − |
| R1b | + | + |
| R2a | + | + |
| R2b | + | − |
| R3a | + | − |
| R3b | − | − |
| R4 | − | − |
| R5a | − | + |
| R5b | − | − |
| R6 | + | + |
Minus signs indicate no discernible S motility, and plus signs indicate detectable S motility. When different clones from the same evolved population exhibited distinct rescue patterns with respect to the two plasmids, these patterns are distinguished by the suffixes a and b.
Three clones each from lines S2, S4, R2, and R6 were compared to their rescued counterparts for motility rate on soft agar and sporulation frequency. Transformants of the three S2 clones (one transformant from each clone) all showed similar increases in motility relative to their unrescued parents (data not shown). These three transformants varied widely, however, in their sporulation levels (data not shown), indicating that genetic variation among the three clones influenced the extent to which rescue of S motility enhanced sporulation efficiency. Such variation in the effects of S-motility rescue on sporulation level was also seen, although to a lesser degree, among the clones from lines S4, R2, and R6.
One clone from each of these four evolved lines was chosen for further characterization. The focal clone from line R2 exhibited the R2a rescue pattern shown in Table 1; the other focal clones are from lines that exhibited only one pattern. The focal clone from line R2 is hereafter referred to as R2a, and the other three focal clones are simply designated by the evolved line from which they were isolated.
Quantification of motility rates.
Mutations that knock out S motility cause a complete inability to swarm on soft agar, but only partially hinder swarming on hard agar (33). The extent of S-motility rescue was therefore quantified by measuring the motility of four evolved clones, their rescued variants, and their ancestors on both soft (0.5%) and hard (1.5%) agar plates (Table 2). On soft agar, the rescued variant of evolved clone R6 was fully restored to its ancestral motility rate, whereas the other three evolved clones were only partially restored. In S2 and R2a, rescued clones swarmed at approximately one-third the ancestor's rate, whereas S4 was restored to about two-thirds the ancestral level. Control transformants of the evolved clones that were not rescued (see Materials and Methods) showed no significant differences in motility on either soft or hard agar from the parental clones (data not shown). This result indicates that restoration of S motility resulted from complementation by ancestral pil genes that were integrated into the chromosome of the rescued transformants rather than some confounding effect of transformation.
TABLE 2.
Motility rates of S+ ancestral, S− evolved, and S+ rescued genotypesa
| Strain | Soft agar
|
Hard agar
|
||||||
|---|---|---|---|---|---|---|---|---|
| Motility rate (mm/day) | EC | Ancestor | % Rescue | Motility rate (mm/day) | EC | Ancestor | % Rescue | |
| S ancestor | 4.00 | 2.58 | ||||||
| S2 | 0.33 | *** | 0.97 | *** | ||||
| S2/pDW79/+ | 1.31 | *** | *** | 27 | 2.25 | *** | ** | 80 |
| S4 | 0.36 | *** | 0.69 | *** | ||||
| S4/pDW79/+ | 2.67 | *** | *** | 63 | 2.78 | *** | * | 112 |
| R ancestor | 4.03 | 2.61 | ||||||
| R2a | 0.33 | *** | 1.38 | *** | ||||
| R2a/pSWU257/+ | 1.36 | ** | *** | 28 | 2.59 | *** | ns | 98 |
| R6 | 0.34 | *** | 1.63 | *** | ||||
| R6/pSWU257/+ | 3.98 | *** | ns | 99 | 3.45 | *** | *** | 186 |
Columns labeled EC indicate statistical significance of the difference in motility rate between rescued (S+) and unrescued (S−) evolved clones. Columns labeled ancestor indicate significance of differences between all evolved genotypes (rescued and unrescued) and their ancestor. Significance levels are based on two-tailed t tests. ***, P < 0.001. **, P < 0.01. *, P < 0.05. ns, P > 0.05. The percent rescue columns show the extent of motility rate rescue as a percentage of the difference between the rates of unrescued evolved clones and their respective ancestors.
Complementation with the wild-type pil genes tended to restore motility on hard agar to an even greater degree than on soft agar (Table 2). All four evolved clones retained some hard-agar motility but at reduced rates compared to the ancestors. Of the two evolved clones showing one-third restoration of soft-agar motility, S2 motility was restored 80% on hard agar, while R2a was fully restored. Clones S4 and R6, which showed two-thirds and full restoration of soft-agar motility, respectively, both achieved motility rates on hard agar that were significantly higher than the ancestral rate. These results imply that deficits in hard-agar motility of the evolved lines (38) were caused directly by losses of S motility rather than by mutations in A-motility genes.
Fruiting ability and sporulation levels.
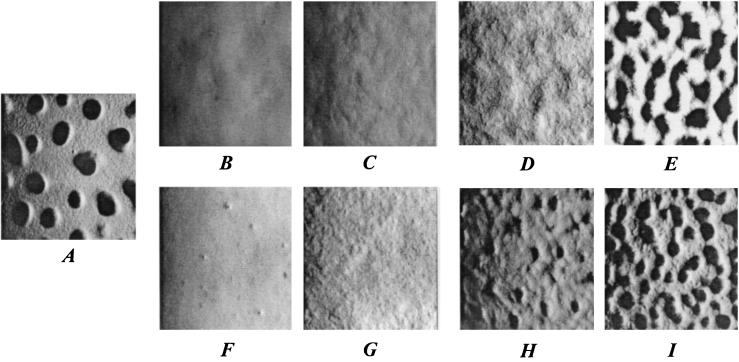
The effects of rescuing S motility on fruiting body formation varied significantly among the four evolved clones (Fig. 2). The partial rescue of social motility in S2 (Fig. 2B and C) and R2a (Fig. 2F and G) had little effect on their aggregation ability. Neither these evolved clones nor their transformed counterparts formed distinct fruiting bodies within the spots of starving cells. The transformants did, however, show greater surface variegation during starvation than did their unrescued parents, perhaps indicating some tendency toward aggregation.
FIG. 2.
Developmental phenotypes of the ancestor and evolved clones before and after restoration of S motility. (A) Ancestor S. (B) Evolved clone S2. (C) Rescued evolved clone S2/pDW79/+. (D) Evolved clone S4. (E) Rescued evolved clone S4/pDW79/+. (F) Evolved clone R2a. (G) Rescued evolved clone R2a/pSWU257/+. (H) Evolved clone R6. (I) Rescued evolved clone R6/pSWU257/+. Magnification, ×50.
The greater extent of genetic rescue of S motility in S4 and R6 produced more dramatic improvements in their fruiting body formation. S4 did not develop visible aggregates, whereas S4/pDW79/+ developed fruiting bodies only slightly smaller and more closely spaced than the ancestral fruiting bodies (Fig. 2A, D, and E). R6 retained the ability to form small fruiting bodies, but its rescued counterpart, R6/pSWU257/+, formed even more fruiting bodies than the ancestor, although they were slightly smaller than the ancestral mounds (Fig. 2A, H, and I).
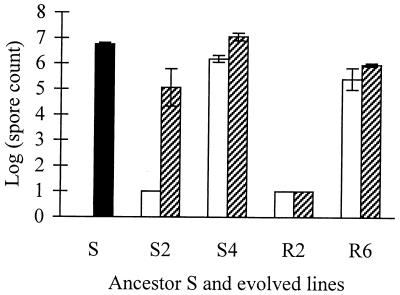
While S-motility rescue of S2 did not restore the ability to make fruiting bodies, it did restore the sporulation level to about four orders of magnitude above that in the unrescued state, although this restored level was still only a few percent of that of the ancestor (Fig. 3). By contrast, neither R2a nor its rescued counterpart, R2a/pSWU257/+, produced any spores. The unrescued clones of S4 and R6 were both somewhat defective at sporulating (spore counts, ∼10 and 3% of those for the ancestor, respectively). S4/pDW79/+ sporulation was raised to a level slightly higher than that of the ancestor, whereas sporulation of R6/pSWU257/+ was only partially restored.
FIG. 3.
Sporulation levels of the ancestor and S− and S+ variants of evolved clones. The solid column shows the ancestor, open columns are untransformed evolved clones defective for S motility (S−), and hatched columns are transformed derivatives of each evolved clone with rescued S motility (S+). Error bars indicate 95% confidence intervals. Short columns with no error bars indicate a sporulation level below the level of detection, which is shown by the column height.
Competitive fitness in the asocial regime.
Unrescued S− transformants of the evolved clones (e.g., R6/pSWU257/−) were used as controls to detect any effect of transformation on fitness other than the intended effect of S-motility complementation. Such unintended effects might be caused by either vector integration or expression of kanamycin resistance. However, in competitions between the four evolved clones and their respective S− transformants, there were no significant differences in fitness in the asocial (liquid batch) culture regime (data not shown). Thus, transformation per se had no effect on competitive fitness.
Transformants of the ancestors were also obtained in order to use kanamycin resistance as a marker in competitions between ancestral and evolved clones. We tested for possible fitness effects of each plasmid's transformation of the R ancestor in the asocial regime. Such effects might be caused by the kanamycin resistance marker, vector integration, or the merodiploid state (S+ S+) of the transformants for the vector-borne pil genes. These controls were performed by separately competing strains R, R/pSWU257, and R/pDW79 (all of which are rifampin resistant) against the rifampin-sensitive evolved clone S2. We observed no significant fitness differences among these three ancestral variants (analysis of variance, F = 1.671, 2 and 6 degrees of freedom, P = 0.265), indicating that any marker or transformation effect is small relative to the fitness difference between evolved clones and their marked ancestors (Table 3).
TABLE 3.
Rescue effects on relative fitness of evolved clones in the asocial batch regimea
| Evolved clone | Relative fitness
|
|
|---|---|---|
| Rescued (S+) vs. evolved (S−) | Ancestor (S+) vs. evolved (S−) | |
| S2 | 0.963*** | 0.741*** |
| S4 | 0.960** | 0.680*** |
| R2a | 0.992ns | 0.793* |
| R6 | 0.942** | 0.720*** |
Asterisks indicate statistical significance of the fitness differences between rescued (S+) and unrescued (S−) evolved clones and between the ancestor and unrescued evolved clones. The null hypothesis (equal relative fitness) is 1. Significance levels are based on one-tailed t tests. ***, P < 0.001. **, P < 0.01. *, P < 0.05. ns, P > 0.05.
Among the comparisons of primary interest for this study, we observed significant negative effects (4 to 6%) on asocial fitness of rescuing S motility in three of the four evolved clones tested (S2, S4, and R6 [Table 3]). We measured much greater differences in asocial fitness between the unrescued evolved clones and their ancestors (Table 3, right column), which means that the rescued pil genes account for only a portion of the fitness gains that accrued during 1,000 generations in the asocial regime.
Of the four focal clones, the S motility of R6 alone was fully restored to its ancestral level, whereas S2, S4, and R2a were restored only partially (Table 2). It is possible that complete restoration of S motility in these latter clones (via identification and rescue of additional S-motility mutations) would result in even greater losses of asocial fitness than those observed here. The observation that the complete rescue of S motility in R6 produced the largest decrement in asocial fitness is consistent with this possibility.
In principle, the fitness effects of rescuing S− mutations in the pil region of M. xanthus may depend on genetic background, the specific mutations being complemented, or both. We therefore examined the fitness effects of rescuing S motility, using pSWU257, in two defined S− mutants in which parts of either pilA or pilB had been deleted in frame (45). We then competed the S+ parent strain (DK1622) and the two S+ rescued strains (DK10410/pSWU257/+ and DK10416/pSWU257/+) against their respective S− ΔpilA and ΔpilB mutants. As summarized in Table 4, the S+ parent DK1622 had similarly low fitness in competition with each mutant (0.834 and 0.848, respectively, for ΔpilA and ΔpilB). And as expected, both S+ rescued strains had low fitness values, similar to the S+ parent, when they competed against the S− mutants. In competitions between the parent and each of the two S+ rescued Δpil strains, fitness differences were very small (1 to 2%) relative to differences between S− and S+ genotypes (14 to 21%). These results clearly indicate that functional S motility directly harms fitness in the asocial regime.
TABLE 4.
Rescue effects on relative fitness of defined Δpil mutants in the asocial regimea
| Δpil mutant | Relative fitness
|
||
|---|---|---|---|
| Rescued Δpil strain (S+) vs. Δpil mutant (S−) | Parent (DK1622, S+) vs. Δpil mutant (S−) | Parent (DK1622, S+) vs. rescued Δpil strain (S+) | |
| ΔpilA | 0.791*** | 0.834** | 1.024*** |
| ΔpilB | 0.856*** | 0.848*** | 1.013ns |
Asterisks indicate statistical significance of the fitness differences between rescued (S+) and unrescued (S−) Δpil mutants and between the parent and unrescued Δpil mutants. The null hypothesis (equal relative fitness) is 1. Significance levels are based on one-tailed t tests in the left and middle fitness columns and two-tailed t tests in the right column. ***, P < 0.001. **, P < 0.01. *, P < 0.05. ns, P > 0.05.
The average fitness difference between the two defined Δpil mutants and their rescued counterparts was 0.177 (Table 4), whereas the mean difference between rescued and unrescued forms of the evolved clones was only 0.036 (Table 3); this contrast is highly significant (two-tailed t test, t = 5.293, 4 degrees of freedom, P = 0.0061). It is evident that factors other than restoration of wild-type pil genes into the evolved clones influenced the fitness effects of the rescue events. These factors may include additional mutations in the evolved genomes that interact with the pil loci, variation in the properties of the particular pil mutations that were rescued, or both. Clear evidence for additional beneficial mutations in the evolved clones is seen by comparing the mean fitness difference from their DK1622 ancestor (0.266, Table 3) with the mean fitness difference between the defined pil mutants and DK1622 (0.159, Table 4).
Developmental cheating.
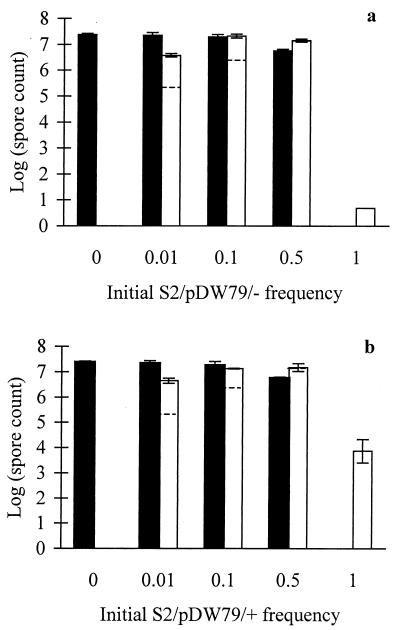
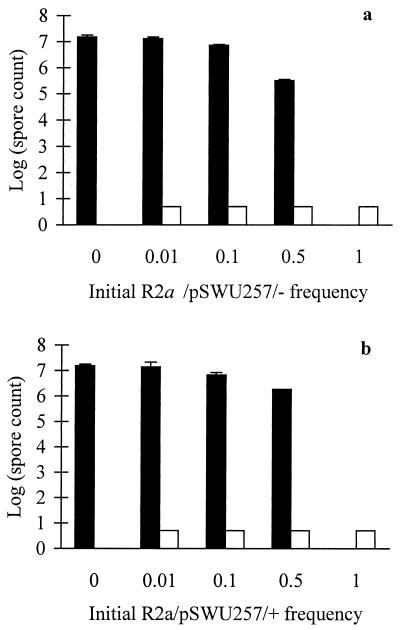
We screened two developmentally defective evolved clones, S2 and R2a, and their rescued counterparts for their tendency to cheat during development when mixed with their developmentally proficient ancestor. In this social system, developmental cheating is defined as the tendency for a defective genotype to outperform a proficient genotype during development of a mixed culture (40). Such cheating goes beyond mere extracellular complementation of a developmental defect (whereby a defective strain is restored to wild-type sporulation efficiency when mixed with the wild type). That is, the cheater actually performs better in a mixture than does a neutrally marked wild-type strain mixed at the same frequency with the wild type. Figure 4 shows that both the rescued and unrescued transformants of S2 exhibited similar cheating behavior at low frequencies (0.01 and 0.1). Both S2 variants also depressed the sporulation of the wild-type strain when mixed at a 1:1 ratio (0.5 frequency). Thus, although restoration of S motility in clone S2 had a dramatic effect on sporulation in pure culture (Fig. 3), it had no discernible effect on cheating behavior.
FIG. 4.
Effect of S-motility rescue on developmental cheating in mixed culture. Solid columns indicate spore counts for the ancestor, and open columns give the counts for unrescued S2/pDW79/− (a) and rescued S2/pDW79/+ (b). Dashed lines in the open columns are the expected sporulation levels for a neutrally marked wild-type strain at each initial frequency. Spore counts for evolved clone variants that exceed the expected level indicate developmental cheating (40). Error bars indicate 95% confidence intervals. The short column with no error bars indicates a sporulation level below the level of detection, which is shown by the column height.
By contrast, neither the S− nor S+ transformants of R2a showed even partial extracellular complementation of their sporulation defects by the ancestor (Fig. 5). However, at an initial frequency of 0.5, the unrescued S− transformant reduced the ancestor's sporulation to a greater extent than did the rescued S+ transformant (Fig. 5). This observation suggests that functional S motility in a strain that cannot itself sporulate nonetheless may facilitate the sporulation of a developmentally proficient S+ strain with which it is mixed. Thus, S2 and R2a respond very differently to mixing with the wild-type progenitor (Fig. 4 and 5), even though they show similar losses of S motility and their motility can be rescued to a similar degree (Table 2). Evidently, other loci must be involved in the developmental cheating exhibited by S2.
FIG. 5.
Effect of S-motility rescue on sporulation of the ancestor in mixed culture. Solid columns indicate spore counts for the ancestor, and open columns give the counts for unrescued R2a/pSWU257/− (a) and rescued R2a/pSWU257/+ (b). Error bars show 95% confidence intervals. Short columns with no error bars indicate a sporulation level below the level of detection, which is shown by the column height.
Identification of a specific evolutionary mutation.
The S motility of some evolved clones was rescued at least partially by both of the plasmids used in this study. These plasmids share ∼3.4 kb of overlapping DNA that includes part of pilG and the complete coding regions of pilH, pilI, and pilD (Fig. 1). Mutants with S− genotypes rescued to S+ by both plasmids are expected to bear a mutation in this overlapping region that is responsible for at least the minimum degree of S-motility loss that is rescued by either of the plasmids. This prediction assumes the straightforward explanation of phenotypic rescue being caused by direct genetic complementation. (Theoretically, the rescue could be due to a gene dosage effect or an undefined epistatic interaction.)
The S motility of the R6 clone was rescued by both plasmids (Table 1), and therefore this clone was chosen for sequencing of the pil region overlapped by pSWU257 and pDW79 along with the same region in its evolutionary ancestor, strain R. One nucleotide substitution was found in R6, where a guanine residue was found to have replaced a thymine residue at position 612 of the pilH coding region (46), resulting in an amino acid replacement of a cysteine residue by a tryptophan residue. The identification of this nonsynonymous substitution in the appropriate pil region thus confirms the prediction that was based upon phenotypic rescue by direct genetic complementation. The pilH gene codes for an ABC transporter homologue that is necessary for production of type IV pili and therefore also for S motility.
DISCUSSION
We previously demonstrated that replicate evolving lines of M. xanthus consistently lost social motility function during 1,000 generations of adaptation to a nutrient-rich and physically unstructured regime (38). In this study, we have used a candidate gene approach to identify a set of 17 pil loci (Fig. 1) that are responsible, at least partially, for the losses of S motility in many lines that evolved under this asocial regime (Tables 1 and 2). To test the hypothesis that mutations in one or more pil genes were causally involved in adaptive changes, we utilized genetic manipulations in both directions, i.e., restoring a functional pil cluster to the derived lines and deleting pil genes from the ancestral strain. Both manipulations confirmed an effect of these genes on competitiveness and in the directions predicted if the loss of pil-encoded functions were adaptive in the asocial regime. The actual magnitude of the fitness effects in the two directions differed, but this variation may arise because different alleles were used in the two backgrounds or because the two backgrounds had diverged at other loci that influence fitness. In support of the latter possibility, the fitness benefit attributable to the loss of S motility accounts for only a fraction of the overall fitness gains in the evolved lines (Table 3). All these findings together clearly indicate that the S− mutations that were substituted in the evolving lines provided a significant benefit under the asocial regime.
We have also identified by sequencing a nonsynonymous mutation in the pilH gene of an evolved clone. It will be interesting to examine the effects of this particular mutation on S motility and asocial fitness, in both the evolved and ancestral backgrounds. We also seek to identify and examine other mutations in the pil region in order to understand the types of mutations that can confer losses of S motility and concomitant gains in asocial fitness. For example, are most adaptive changes in this region caused by point mutations, or are deletions and insertions important as well?
The intent of performing evolution experiments in the laboratory is not to replicate nature in all its bewildering complexity, but rather to explore more rigorously the realm of possibilities under well-defined conditions. In particular, we have studied populations of M. xanthus as they evolve and genetically adapt to a simple environment in order to understand the costs as well as the benefits of their complex social and developmental processes. We have now identified two distinct situations in which defective genotypes actually have an advantage relative to their socially and developmentally proficient counterparts.
First, certain defective genotypes have a selective advantage—but only when mixed with proficient genotypes—during fruiting body development and sporulation (40) (see also Fig. 4). This advantage in a mixed population is termed cheating because it demonstrably harms the overall group function while enhancing the relative reproductive success of the cheaters (40). Second, we have shown here that mutations in the pil cluster which cause social and developmental defects are directly advantageous in a nutrient-rich and physically unstructured regime. These manipulations support the hypothesis that social and developmental functions were lost under the asocial regime because they were costly to maintain, not simply because they decayed in the absence of positive selection for them (4, 38). Therefore, social and developmental functions in M. xanthus incur costs as well as benefits, and the degree of their expression may vary in response to local selective conditions.
Although the intent of our evolution experiments was not to reproduce nature in all its complexity, the demonstration that social motility is costly to maintain has implications for an understanding of natural populations. Myxobacteria in the soil are presumably subject to diverse selective factors, including varying resource availability as well as different physical surfaces on which they move. Maintaining S motility may be detrimental in environments where it is unnecessary for survival and growth. Such costs might reflect allocation of energy to synthesizing pili and running the motility motor (36), or the physical cohesiveness of piliated cells might interfere with efficient extraction of diffuse resources.
S motility might also be detrimental to M. xanthus living in habitats with relatively hard surfaces. A motility is more effective than S motility for swarming on hard surfaces, whereas S motility is more effective on soft surfaces (33). Shi and Zusman (33) hypothesized that these two motility systems evolved to allow effective predatory swarming over different surfaces. Following this reasoning, natural populations that experience primarily hard surfaces would depend less on S motility to search for prey and might even benefit by the loss of S motility given its costs. However, Shi and Zusman (33) and we have observed that the A- and S-motility systems are, in fact, positively synergistic on surfaces ranging from 0.3 to 1.5% agar. Thus, eliminating S motility inhibits motility even on hard agar, where A motility is most effective. This synergism indicates that both motility systems contribute to swarming over a range of agar surfaces, but it does not exclude the possibility that loss of one or the other system might be beneficial in some natural environments.
Even if there are natural environments in which the loss of S motility is sometimes favored, most M. xanthus populations presumably experience intermittent bouts of starvation, during which they undergo fruiting body development and sporulation. Evolutionary losses of S motility tend to disrupt this process, as we have shown by rescuing S motility and, in most cases, observing enhanced development, sporulation, or both (Fig. 2 and 3). Any benefit of losing S motility would thus be offset in nature by impaired development, to the extent that development is an important component of overall fitness and to the extent that deficient genotypes must exist independently (i.e., excluding their advantage as developmental cheaters). Considering both the synergism between the two motility systems and the positive contribution of S motility to fruiting body development, we expect that complete losses of S motility are harmful to the overall fitness of M. xanthus in most natural habitats, despite the costs of maintaining S motility that we have demonstrated here. Nonetheless, it is important that anyone intent on isolating a diverse collection of myxobacteria from nature consider the possibility that some genotypes, or even entire species, may have lost their social motility and ability to produce fruiting bodies.
Acknowledgments
We thank Dale Kaiser for helpful discussions and sharing the plasmids used in this study, Phil Youderian for sharing unpublished data, Iris Dinkelacker for technical help, and two anonymous reviewers for helpful recommendations.
This work was supported by grants from Michigan State University and the National Science Foundation and funding from the Max- Planck Institute for Developmental Biology.
REFERENCES
- 1.Chao, L., and B. R. Levin. 1981. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl. Acad. Sci. USA 78:6324-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, H. W., I. M. Keseler, and L. J. Shimkets. 1990. Genome size of Myxococcus xanthus determined by pulsed-field gel electrophoresis. J. Bacteriol. 172:4206-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, Y., and D. Kaiser. 1989. dsg, a gene required for cell-cell interaction early in Myxococcus development. J. Bacteriol. 171:3719-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper, V. S., and R. E. Lenski. 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407:736-739. [DOI] [PubMed] [Google Scholar]
- 5.Downard, J., S. V. Ramaswamy, and K. Kil. 1993. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J. Bacteriol. 175:7762-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dworkin, M., and D. Kaiser (ed.). 1993. Myxobacteria II . American Society for Microbiology, Washington, D.C.
- 7.Gill, R., and L. J. Shimkets. 1993. Genetic approaches for analysis of myxobacterial behavior, p. 129-156. In M. Dworkin and D. Kaiser (ed.), Myxobacteria II. American Society for Microbiology: Washington, D.C.
- 8.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton, W. D. 1972. Altruism and related phenomena, mainly in social insects. Annu. Rev. Ecol. Syst. 3:193-232. [Google Scholar]
- 10.Heinsohn, R., and C. Packer. 1995. Complex cooperative strategies in group-territorial African lions. Science 269:1260-1262. [DOI] [PubMed] [Google Scholar]
- 11.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of motility in nonmotile mutants of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176. [Google Scholar]
- 13.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 14.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser, D., and C. Crosby. 1983. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 3:227-245. [Google Scholar]
- 16.Kaiser, D., and L. Kroos. 1993. Intercellular signaling, p. 257-284. In M. Dworkin and D. Kaiser (ed.), Myxobacteria II. American Society for Microbiology, Washington, D.C.
- 17.Kaiser, D., and R. Losick. 1993. How and why bacteria talk to each other. Cell 73:873-885. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, H. B., and L. Plamann. 1996. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol. Lett. 139:89-95. [DOI] [PubMed] [Google Scholar]
- 19.Kim, S. K., and D. Kaiser. 1990. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 4:896-905. [DOI] [PubMed] [Google Scholar]
- 20.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 21.Kuspa, A., L. Kroos, and D. Kaiser. 1986. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev. Biol. 117:267-276. [DOI] [PubMed] [Google Scholar]
- 22.Lenski, R. E., and B. R. Levin. 1985. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and the predictions for natural communities. Am. Nat. 125:585-602. [Google Scholar]
- 23.Lenski, R. E., M. R. Rose, S. C. Simpson, and S. C. Tadler. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315-1341. [Google Scholar]
- 24.Levin, B. R. 1988. Frequency-dependent selection in bacterial populations. Philos. Trans. R. Soc. B 319:459-472. [DOI] [PubMed] [Google Scholar]
- 25.Martin, J. 1979. Miss Manners' guide to excruciatingly correct behavior. Warner, New York, N.Y.
- 26.Maynard Smith, J., and G. R. Price. 1973. The logic of animal conflict. Nature 246:15-18. [Google Scholar]
- 27.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 28.Reichenbach, H. 1999. The ecology of the myxobacteria. Environ. Microbiol. 1:15-21. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg, E., K. H. Keller, and M. Dworkin. 1977. Cell density-dependent growth of Myxococcus xanthus on casein. J. Bacteriol. 129:770-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg, E., and M. Varon. 1984. Antibiotics and lytic enzymes, p. 109-125. In E. Rosenberg (ed.), Myxobacteria: development and cell interactions. Springer-Verlag, New York, N.Y.
- 31.Rosenzweig, R. F., R. R. Sharp, D. S. Treves, and J. Adams. 1994. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137:903-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro, J. A., and M. Dworkin (ed.). 1997. Bacteria as multicellular organisms. Oxford University Press, New York, N.Y.
- 33.Shi, W., and D. R. Zusman. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. USA 90:3378-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 35.Sniegowski, P. D., P. J. Gerrish, and R. E. Lenski. 1997. Evolution of high mutation rates in experimental populations of Escherichia coli. Nature 387:703-705. [DOI] [PubMed] [Google Scholar]
- 36.Spormann, A. M. 1999. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol. Mol. Biol. Rev. 63:621-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travisano, M., J. A. Mongold, A. F. Bennett, and R. E. Lenski. 1994. Experimental tests of the roles of adaptation, chance, and history in evolution. Science 267:87-90. [DOI] [PubMed] [Google Scholar]
- 38.Velicer, G. J., L. Kroos, and R. E. Lenski. 1998. Loss of social behaviors by Myxococcus xanthus during evolution in an unstructured habitat. Proc. Natl. Acad. Sci. USA 95:12376-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velicer, G. J., and R. E. Lenski. 1999. Evolutionary trade-offs under conditions of resource abundance and scarcity: experiments with bacteria. Ecology 80:1168-1179. [Google Scholar]
- 40.Velicer, G. J., L. Kroos, and R. E. Lenski. 2000. Developmental cheating in the social bacterium Myxococcus xanthus. Nature 404:598-601. [DOI] [PubMed] [Google Scholar]
- 41.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 42.Wall, D., P. E. Kolenbrander, and D. Kaiser. 1999. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J. Bacteriol. 181:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, E. O. 1975. Sociobiology: the new synthesis. Harvard University Press, Cambridge, Mass.
- 44.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 45.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109-121. [DOI] [PubMed] [Google Scholar]
- 46.Wu, S. S., J. Wu, Y. L. Cheng, and D. Kaiser. 1998. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol. Microbiol. 29:1249-1261. [DOI] [PubMed] [Google Scholar]