Abstract
Salmonella enterica serovar Pullorum is a fowl-adapted bacterial pathogen that causes dysentery (pullorum disease). Host adaptation and special pathogenesis make S. enterica serovar Pullorum an exceptionally good system for studies of bacterial evolution and speciation, especially regarding pathogen-host interactions and the acquisition of pathogenicity. We constructed a genome map of S. enterica serovar Pullorum RKS5078, using I-CeuI, XbaI, AvrII, and SpeI and Tn10 insertions. Pulsed-field gel electrophoresis was employed to separate the large DNA fragments generated by the endonucleases. The genome is 4,930 kb, which is similar to most salmonellas . However, the genome of S. enterica serovar Pullorum RKS5078 is organized very differently from the majority of salmonellas, with three major inversions and one translocation. This extraordinary genome structure was seen in most S. enterica serovar Pullorum strains examined, with different structures in a minority of S. enterica serovar Pullorum strains. We describe the coexistence of different genome structures among the same bacteria as genomic plasticity. Through comparisons with S. enterica serovar Typhimurium, we resolved seven putative insertions and eight deletions ranging in size from 12 to 157 kb. The genomic plasticity seen among S. enterica serovar Pullorum strains supported our hypothesis about its association with bacterial evolution: a large genomic insertion (157 kb in this case) disrupted the genomic balance, and rebalancing by independent recombination events in individual lineages resulted in diverse genome structures. As far as the structural plasticity exists, the S. enterica serovar Pullorum genome will continue evolving to reach a further streamlined and balanced structure.
The bacterial genus Salmonella consists of more than 2,400 documented species, many of which are important pathogens in humans or animals (4, 14, 16, 41, 42, 44, 45). Selander and colleagues have assembled three sets of reference strains for representative Salmonella species (5, 7, 8). The Salmonella species are very closely related to one another, as judged by genomic DNA reassociation rates (9, 15), which could be higher than 90%. Despite such close relatedness, different Salmonella species may have drastically different biological properties, especially in host range and the nature of diseases they cause. For example, S. enterica serovar Typhimurium infects many host species, including humans, mice, and fowl, but S. enterica serovar Typhi, a close relative of S. enterica serovar Typhimurium , is strictly limited to humans. S. enterica serovar Pullorum and S. enterica serovar Gallinarum, on the other hand, are both specific to fowl, but they cause distinct diseases, with S. enterica serovar Pullorum causing dysentery (pullorum disease) and S. enterica serovar Gallinarum causing typhoid fever (45, 46). Our long-term goal is, through genomic comparisons among representative Salmonella species, to explore the mechanisms of genomic divergence and evolution that have brought about the genetic differences and made each of the Salmonella species unique.
Representative strains of two Salmonella serovars have been completely sequenced, S. enterica serovar Typhimurium LT2 (33) and S. enterica serovar Typhi CT18 (39). Base-to-base comparisons of these sequenced Salmonella genomes would surely resolve all the genetic differences between them and provide new insights into the mechanisms of phylogenetic divergence and evolution of these bacteria. However, there are more than 2,400 Salmonella serovars, each of which is unique. Sequencing all of them is out of the question. Based on the great genetic similarity among all Salmonella species, we suggest and are testing a complementary approach to comparative genomics of salmonellas: determining the overall genome structure and locating genomic differences by physical mapping. This can be done on a very large number of selected Salmonella species within a relatively short time. Once insertions are located, they can be further analyzed by sequencing and functional studies. Deletions can be defined through comparisons with the genomic sequence of S. enterica serovar Typhimurium. Inversions and translocations can be very efficiently resolved by physical mapping. Previously, we have mapped large-scale genomic insertions, inversions, and translocations in a number of Salmonella species (18, 20, 22-24). We have now optimized the techniques to greater accuracy and higher efficiency for systematic comparisons among Salmonella spp.
S. enterica serovar Pullorum is highly adapted to fowl, although S. enterica serovar Pullorum infections in primates have been reported (36). However, because of the high specificity of S. enterica serovar Pullorum for fowl, S. enterica serovar Pullorum infections in mammals are extremely rare and therefore have not been a serious public health issue.
As a fowl-specific pathogen, on the other hand, S. enterica serovar Pullorum continues to cause economic losses worldwide. The increasing problems of antibiotic resistance, long persistence of the bacteria in chickens after infection (49), and poor knowledge of the immunology of Salmonella infection in poultry (50) call for a better understanding of the genetics of S. enterica serovar Pullorum and pathogenesis of S. enterica serovar Pullorum infection in order to develop alternative measures for the control of this pathogen. As the first step toward this, we constructed a genome map of S. enterica serovar Pullorum on strain RKS5078. We located numerous insertions and deletions, ranging from 12 to 157 kb, as well as several major genomic rearrangements on the genome. These findings suggest possible roles of these genomic changes in the phylogenetic divergence and evolution of S. enterica serovar Pullorum and lead us to conclude that the genome of S. enterica serovar Pullorum is still evolving to reestablish a genomic balance and eventually to complete the process of speciation.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
S. enterica serovar Pullorum strains RKS5078, maintained at the Salmonella Genetic Stock Center (SGSC, www.ucalgary.ca/≈kesander) as SGSC2294, and RKS2242 (SGSC2295) were obtained from R. K. Selander, and strains 490 (SGSC2737), 498 (SGSC2738), 499 (SGSC2739), 501 (SGSC2740), 504 (SGSC2741), 505 (SGSC2742), 507 (SGSC2743), 509 (SGSC2745), 510 (SGSC2746), 512 (SGSC2747), 513 (SGSC2748), 514 (SGSC2749), 515 (SGSC2750), and R278 (SGSC2751) were obtained from C. Poppe. Previously, we have made I-CeuI maps on two S. enterica serovar Pullorum strains, RKS2266 (SGSC2508) and RKS2246 (SGSC2509) (28), so these two strains were not included in this study. RKS2266 and RKS2246, however, will be compared with RKS5078 in this study.
All S. enterica serovar Pullorum strains were identified serologically (1, 9,12:—:—) and distinguished from S. enterica serovar Gallinarum (also 1,9,12:—:—) by the ornithine test (S. enterica serovar Pullorum produces rapid decarboxylation of ornithine, whereas S. enterica serovar Gallinarum does not). Tn10 insertion mutants of S. enterica serovar Typhimurium LT2 were obtained from numerous sources and have been described before (2, 43). The bacteria were routinely grown in Luria-Bertani (LB) broth or on LB plates (22). Tetracycline was used at 20 μg/ml for Tn10 insertion mutants. The bacteria were maintained in 15% glycerol at −70°C; a single colony was picked prior to use.
Transfer of Tn10 insertions through bacteriophage P22-mediated transduction.
A large number of Tn10 insertions into genes with known functions have been mapped on the genome of S. enterica serovar Typhimurium LT2 (21). We transferred Tn10 insertions from S. enterica serovar Typhimurium LT2 to S. enterica serovar Pullorum RKS5078 by bacteriophage P22-mediated transduction to locate the same genes through homologous recombination. We made P22 lysates from a selected set of Tn10 insertion mutants of S. enterica serovar Typhimurium LT2 by growing a 3-ml overnight culture of these selected Tn10 mutants in LB broth and inoculating these cultures with P22 at a multiplicity of infection of 1:100, followed by coincubation for 6 h. After removal of the cell debris by centrifugation, the lysates, 1011 PFU/ml, were ready for use in transduction.
For transferring the Tn10 insertions to S. enterica serovar Pullorum RKS5078, we spread 100 μl of an overnight culture of S. enterica serovar Pullorum RKS5078 and 20 μl of lysate onto an LB plate containing tetracycline. A colony was picked up and restreaked on another tetracycline plate for single-colony isolation. One colony from the second tetracycline plate was used for phenotype tests and mapping.
Enzymes and chemicals.
I-CeuI, AvrII, and SpeI were purchased from New England Biolabs; XbaI and proteinase K were from Boehringer Mannheim. [32P]dCTP was from New England Nuclear. Most other chemicals were from Sigma Chemical Co.
PFGE methods and genomic mapping.
Preparation of intact genomic DNA, endonuclease cleavage of DNA in agarose blocks, and separation of the DNA fragments by pulsed-field gel electrophoresis (PFGE) were done as described previously (22, 29). PFGE was performed with the Bio-Rad CHEF Mapper or Bio-Rad CHEF DRII electrophoresis system. For PFGE, we normally use three cycles of conditions: the first for general separation at 30 s of ramping to 90 s for 16 h at full voltage and a buffer temperature of 12°C; the second for zooming in on crowded areas of small bands at 3 s of ramping to 6 s; and the third for zooming in on crowded areas of larger bands at pulsing times based on the sizes of the bands. The total run times for the second and third cycles are usually 6 to 12 h, depending on the extent of the separation. Most runs were carried out at a 120o angle. For very crowded areas of bands, a 150o angle was used.
For determining the sizes of DNA fragments on the PFGE gel, we most often used only a λ ladder (New England Biolabs) as size markers, but in many cases we also used bacterial genomic DNA cleaved with an endonuclease as markers. Among the ones we often used were S. enterica serovar Typhimurium LT2 and S. enterica serovar Typhi Ty2 DNA cleaved with XbaI, AvrII, or SpeI; the sizes of these fragments had been determined previously (21, 24). These markers significantly improved the precision of the size estimation. Genomic mapping methods with I-CeuI have been described previously (19) and further optimized (25). The technique of double cleavage and end labeling was also described previously (21).
RESULTS
I-CeuI cleavage of S. enterica serovar Pullorum RKS5078 genomic DNA.
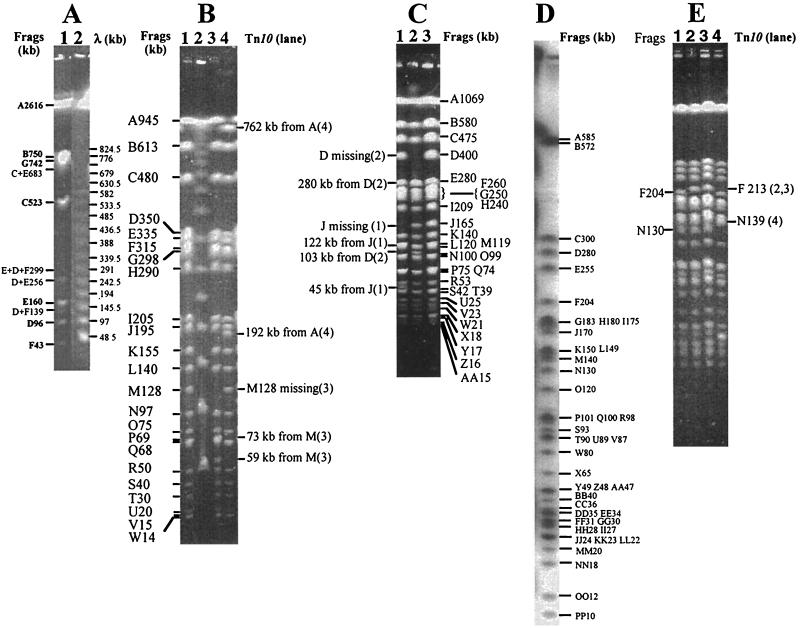
I-CeuI is an intron-encoded endonuclease (10, 31, 32), which cleaves DNA within bacterial rrl genes and thus determines the copy number and genomic distributions of rrl genes (19). Cleavage of genomic DNA by this endonuclease generated seven fragments in S. enterica serovar Pullorum RKS5078 with sizes similar to those of S. enterica serovar Typhimurium (21). As our previous work has demonstrated that I-CeuI fragments of similar sizes are homologous among all Salmonella species, we designated the seven I-CeuI fragments of S. enterica serovar Pullorum with the same letters as in S. enterica serovar Typhimurium based on their sizes. The order of the seven I-CeuI fragments in S. enterica serovar Typhimurium is ABCDEFG (21). However, the order in S. enterica serovar Pullorum RKS5078 was different (Fig. 1A). As shown in Fig. 1A, the partial I-CeuI cleavage products in S. enterica serovar Pullorum RKS5078, C+E, D+E, D+F, etc., determined the order FDEC; results from several experiments eventually determined the order as ABFDECG (see the I-CeuI map in Fig. 2).
FIG. 1.
Endonuclease cleavages of genomic DNA of S. enterica serovar Pullorum RKS5078. (A) I-CeuI cleavage pattern. Lanes: 1, S. enterica serovar Pullorum RKS5078; 2, λ DNA concatemer as DNA size markers. (B) XbaI cleavage patterns of S. enterica serovar Pullorum RKS5078 and representative Tn10 insertion mutants. Lanes: 1, S. enterica serovar Pullorum RKS5078; 2, λ DNA concatemer; 3, an anonymous Tn10 insertion in fragment (Frag) M (128 kb); 4, Tn10 insertion in dadB. (C) AvrII cleavage patterns of S. enterica serovar Pullorum RKS5078 and representative Tn10 insertion mutants. Lanes: 1, an anonymous Tn10 insertion in J (165 kb); 2, Tn10 insertion in ompC; 3, S. enterica serovar Pullorum RKS5078. (D) Radioautograph of end-labeled SpeI cleavage products of S. enterica serovar Pullorum RKS5078. (E) SpeI-cleaved genomic DNA of S. enterica serovar Pullorum RKS5078 and representative Tn10 insertion mutants. Lanes: 1, S. enterica serovar Pullorum RKS5078; 2, Tn10 insertion in nadA; 3, Tn10 insertion in bio; 4, Tn10 insertion in aspC. See the map locations of these genes in Fig. 2.
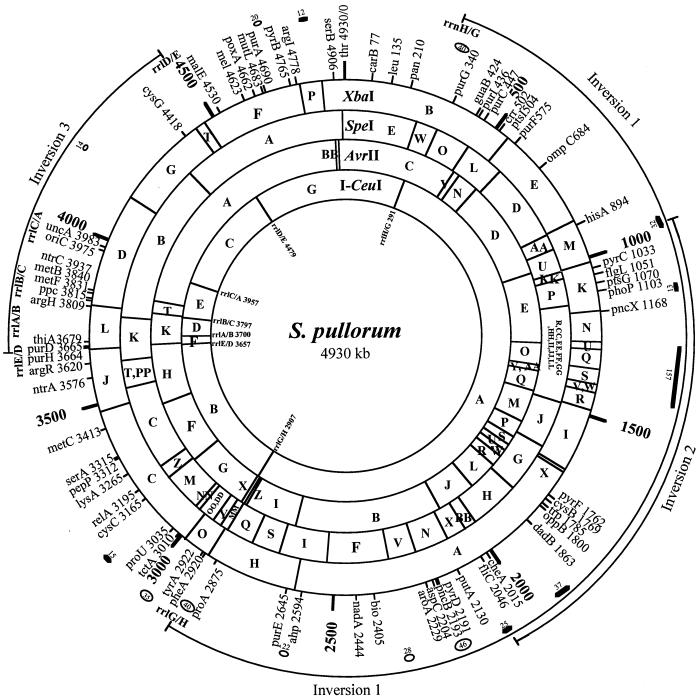
FIG. 2.
Genome map of S. enterica serovar Pullorum RKS5078. The outermost lines define the ranges of the inversions relative to S. typhimurium LT2. Thick short lines show the location and size of insertions (in kilobases). Ovals show the deletions (in kilobases). Some fragments were mapped to the same genomic regions, but their order was not determined.
Cleavages of S. enterica serovar Pullorum RKS5078 genomic DNA with XbaI, AvrII, and SpeI.
Taking advantage of the XbaI and AvrII cleavage sites within the Tn10 DNA sequence, we located the genes in which Tn10 had inserted by cleavage with XbaI and AvrII and PFGE separation. Most of the Tn10 insertions transferred from S. enterica serovar Typhimurium were inserted at homologous sites in the genome of S. enterica serovar Pullorum RKS5078, as confirmed by phenotype tests, such as auxotropism (18) and loss of motility (17). Most transductants tested had the expected phenotype. A small number of Tn10 insertions were not inserted into homologous sites; we called them anonymous Tn10 insertions. These anonymous Tn10 insertions were also useful, however, for determining the neighboring relationships of some fragments from XbaI, AvrII, and SpeI cleavages. Fig.ures 1B and 1C show XbaI and AvrII cleavages, respectively, of S. enterica serovar Pullorum RKS5078 and representative Tn10 insertion mutants.
Unlike XbaI or AvrII, SpeI does not cut the Tn10 sequence. However, the SpeI fragments that have Tn10 insertions can be recognized by a size increase (9.3 kb). This feature is sometimes very advantageous for reliably assigning a Tn10 to a certain genomic location by the increased size of an SpeI fragment, whereas in the case of XbaI and AvrII, a Tn10 inserted within a few kilobases of the end of a fragment is often very difficult to locate on the genome. SpeI cleaved genomic DNA of S. enterica serovar Pullorum RKS5078 into 42 fragments (Fig. 1D). Fig.ure 1E shows examples of Tn10 insertion mutants cleaved with SpeI.
Genome map of S. enterica serovar Pullorum RKS5078.
A genome map was then constructed after all cleavage and Tn10 results were summarized (Fig. 2). This map starts with thr as 0 kb and goes clockwise. On the map, we present genomic locations for 72 genes; locations for 30 additional genes are not shown for a cleaner presentation. The location for oriC (at 3,975 kb from thr) was inferred from the Escherichia coli K-12 map by its distances from uncA (atpA) and rrlC. The first four genes, thr, carB, leu, and pan, had the same order and spacing among them as in S. enterica serovar Typhimurium LT2. The next gene clockwise on the map is rrlH/G; it is a hybrid between rrlH and rrlG created by homologous recombination, which resulted in the inversion of I-CeuI A. All genes between rrlH/G and rrlG/H, covering 2,616 kb (kb 291 to 2907) were in the reverse order (inversion 1) relative to those in S. enterica serovar Typhimurium LT2 except for the genes between hisA and putA (not inclusive), where there was another inversion of about 1,100 kb (inversion 2). A third inversion (inversion 3) was found between rrlD and rrlE, resulting in another pair of hybrid rrl genes, rrlE/D and rrlD/E. Within this third inversion, there was a translocation: I-CeuI D, which is between I-CeuI C and E in S. enterica serovar Typhimurium LT2 and most other Salmonella genomes, is now between I-CeuI E and F. This translocation results in three hybrid rrl genes: when I-CeuI D “left” the original location, the flanking rrlC and rrlA would join to become rrlC/A; when it was inserted to the current position, i.e., between I-CeuI E and F, two hybrid rrl genes, rrlB/C and rrlA/B, would be generated.
Genomic comparisons between S. enterica serovar Pullorum and S. enterica serovar Typhimurium (21), whose map was greatly refined recently by adding more Tn10 insertions and the SpeI data to improve the resolution (S.-L. Liu, unpublished data), revealed seven areas with increased physical distances between genes, which we assume to be insertions, including 32 kb between hisA and pyrC, 13 kb between phoP and pncX, 157 kb between pncX and pyrF, 37 kb between dadB and cheA, 25 kb between fliC and putA, 15 kb between proU and cysC, and 12 kb between argI and serB. The sizes are net increases in genomic DNA. At this stage, we do not rule out the possibilities of simultaneous deletions in the vicinity of the “insertion” area (so the actual insertion could be larger), nor do we have evidence that these increased sizes did not result from translocations of genomic segments from other places of the genome. Some or all of the seven areas of increase might contain several smaller insertions. Interestingly, two insertions, one between genes hisA and pyrC (32 kb) and one between fliC and putA (25 kb), were found in about the same places as the crossover leading to inversion 2, suggesting the possibility that the two insertions have a high level of sequence similarity and are the actual sites of homologous recombination leading to inversion 2.
Genomic comparisons between S. enterica serovar Pullorum and S. enterica serovar Typhimurium also revealed eight areas with reduced distances between genes, which we suppose to be genomic deletions, including 40 kb between rrlH/G and purG, 46 kb between pyrD and pncB, 28 kb between aroA and bio, 22 kb between ahp and purE, 40 kb between proA and rrlG/H, 35 kb between tyrA and tctA, 14 kb between uncA and cysG, and 20 kb between purA and pyrB. Again, these sizes are net decreases in genomic DNA and we do not rule out any other genomic events (e.g., insertions) that might have occurred in the vicinity of these areas.
Plastic genome structure of S. enterica serovar Pullorum.
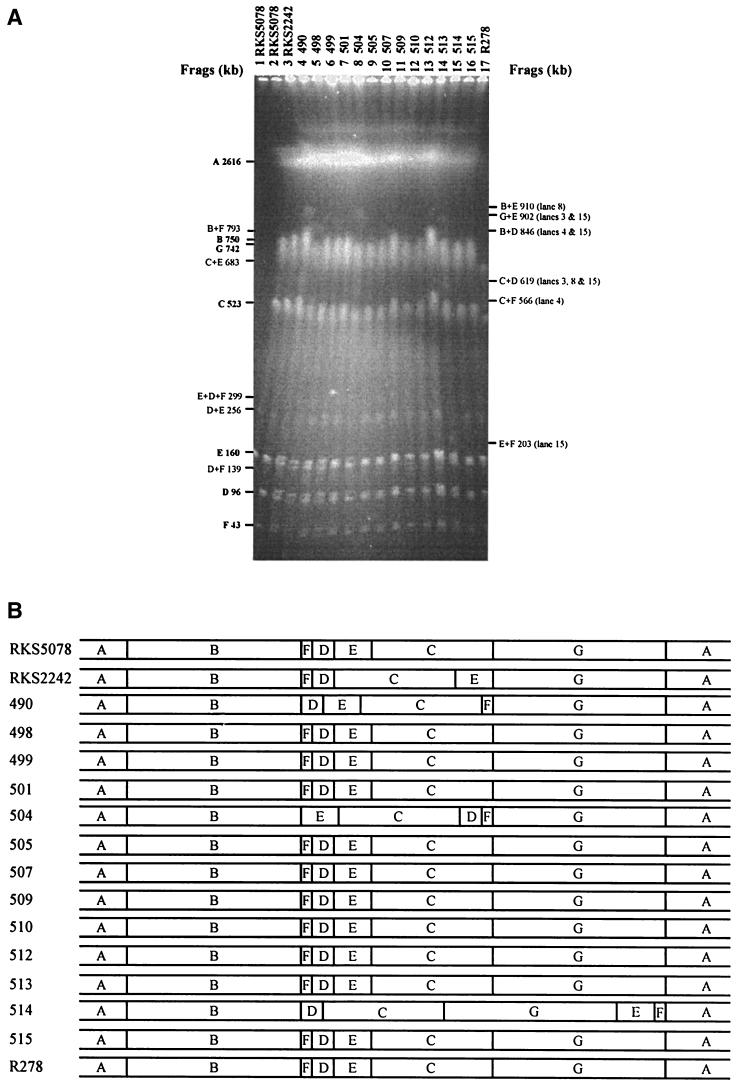
Having seen the extraordinary genome structure in S. enterica serovar Pullorum RKS5078 in addition to the other two S. enterica serovar Pullorum strains that we previously mapped with I-CeuI (28), we wondered whether different S. enterica serovar Pullorum strains would all have different genome structures, a phenomenon we call plastic genome structure, as in the case of S. enterica serovar Typhi (27). We carried out I-CeuI analysis on the strains listed in the Materials and Methods section and found other different genome structures among them, although they all had indistinguishable genome sizes. RKS5078 represented the dominant genome type: 12 strains had the same genome structure as RKS5078, with the seven I-CeuI fragments arranged in the order ABFDECG, including RKS5078, 498, 499, 501, 505, 507, 509, 510, 512, 513, 515, and R278. The remaining four strains each represented a distinct genome type, with strain RKS2242 being ABFDCEG, 490 being ABDECFG, 504 being ABECDFG, and 514 being ABDCGEF (Fig. 3).
FIG. 3.
Population genome structure of S. enterica serovar Pullorum. (A) I-CeuI cleavage patterns of wild-type strains of S. enterica serovar Pullorum. (B) I-CeuI maps based on data in panel A.
The previously mapped S. enterica serovar Pullorum strains had other genome structures that were not found here, with RKS2266 being ABDCFEG and RKS2246 being ABEDCFG (28). Most cleavage fragments with XbaI, AvrII, or SpeI were identical in size among all 16 strains, indicating that they were a tight phylogenetic group of bacteria (i.e., they were all “S. enterica serovar Pullorum”), though with significant variations in size of a small number of cleavage fragments, presumably a result of different genomic inversions or translocations in different strains.
DISCUSSION
We have constructed a genome map of S. enterica serovar Pullorum in strain RKS5078 with an average resolution of about 15 kb. On the map, we determined the size and basic structure of the genome, located over 100 genes, revealed three inversions and one translocation, and resolved seven insertions and eight deletions ranging from 12 to 157 kb, relative to S. enterica serovar Typhimurium. In many ways, this genome was similar to those of most Salmonella species that we have mapped to date, e.g., the size of the genome and the number of rrl genes, two features of phylogenetic significance (30). Especially, the lengths of the seven I-CeuI fragments were typical of salmonellas (19, 28). Even gene order was similar: if the seven I-CeuI fragments were aligned to the genome of S. enterica serovar Typhimurium, there would be perfect colinearity (except for the inversion within I-CeuI A) with indistinguishable spacing between genes except for the seven insertions and eight deletions. However, several features did clearly distinguish S. enterica serovar Pullorum from other salmonellas, the most striking of which was the extraordinary arrangement of the seven I-CeuI fragments. This special arrangement of the seven I-CeuI fragments seen in RKS5078 represented the dominant genome type of S. enterica serovar Pullorum. Why did most S. enterica serovar Pullorum strains take this particular genome structure?
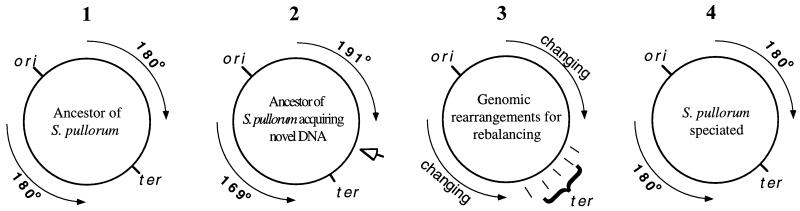
The simplest explanation is rebalancing of the genome by recombination between homologous sites, such as the rrn operons, as a compensation mechanism after the genomic balance is disrupted be the 157-kb insertion. This rebalancing, occurring independently in individual S. enterica serovar Pullorum cells, creates different genome structures. Cells with better balanced genomes will have larger population sizes. Currently, there are several coexisting genome types of S. enterica serovar Pullorum, so we hypothesize that the genome of S. enterica serovar Pullorum is still evolving to reach a precise balance through further refinements. The plastic genome stage will last until a cell with a nearly balanced genome appears. This cell will replicate most efficiently, develop lager populations, and eventually replace all other populations. This hypothesized genomic evolution and speciation of S. enterica serovar Pullorum is illustrated by a model which we call the adopt-adapt model (Fig. 4).
FIG. 4.
Hypothesized genomic evolution and speciation of S. enterica serovar Pullorum—the adopt-adapt model. Panel 1, Ancestor of S. enterica serovar Pullorum having a balanced genome; panel 2, the ancestor having acquiring (adopting) exogenous DNA; panel 3, the genome undergoing adaptive genomic reorganization to reestablish a balance (different genome structures coexist—the plastic genome stage); and panel 4, S. enterica serovar Pullorum with a balanced and streamlined genome.
S. enterica serovar Pullorum has two outstanding biological features, being adapted to fowl and causing only dysentery. Based on the close relatedness and the vast genetic similarity between S. enterica serovar Pullorum and other salmonellas such as S. enterica serovar Gallinarum, it is reasonable to assume that much of the ability of S. enterica serovar Pullorum to cause dysentery might be encoded by the inserted DNA. On the other hand, its host adaptation to fowl might be the result of the genomic deletions: the lost DNA might be present in most other Salmonella species that have a broad host range. We are in the process of cloning the seven insertion areas for sequencing and functional analyses. We are also mapping the genome of S. enterica serovar Gallinarum, aiming at finding any insertions and deletions. We anticipate finding the set of deletions that are common to S. enterica serovar Pullorum and S. enterica serovar Gallinarum that may account for both Salmonella serovars' becoming adapted to fowl.
The contributions of genomic insertions and deletions to bacterial evolution and speciation are continually being documented (11, 12, 26, 34, 35). Having focused on mainly genomic insertions (18, 23, 24) and inversions and translocations (20, 27), we have for the first time mapped genomic deletions on the genome of S. enterica serovar Pullorum. Sequence comparisons of DNA segments that are present in S. enterica serovar Typhimurium but absent from S. enterica serovar Pullorum with those absent from other host-adapted Salmonella species, such as S. enterica serovar Typhi, will help in elucidating the mechanisms of host adaptation of salmonellas and in providing new understanding about pathogen-host interaction.
The recent sequence comparisons of pairs of closely related bacteria have provided further insights into and great details about the mechanisms of bacterial divergence, such as Helicobacter pylori 26695 and J99 (1, 48), E. coli K-12 and O157:H7 (6, 13, 40), Neisseria meningitidis z2491 and MC58 (38, 47), and two Rickettsia strains (3, 37). Many lines of evidence from these studies have indicated additions of genes as the main cause of divergence. Genomes also lose genes during adaptation to new niches. In addition to deletions, genes decay gradually and lose functions eventually (37). However, genomic balance and rebalancing have not been given due attention as an important factor in genomic evolution and bacterial speciation.
The genome of S. enterica serovar Pullorum may have provided a snapshot of salmonellas in the midst of evolution: acquiring new genes, losing no-longer-needed ancestral genes, and rearranging the genome for rebalancing. Further analyses of the insertions and deletions of S. enterica serovar Pullorum may bring about new discoveries on pathogenesis and host-pathogen interactions of salmonellas, potentially leading to novel strategies of control and utilization of salmonellas and other bacteria. More attention focused on genomic rearrangement, especially the phenomenon of genomic plasticity, may substantially update our understanding of bacterial genomic evolution.
Acknowledgments
We thank R. K. Selander and C. Poppe for the strains used in this study and Danni Qi, Ning Qi, and Lili Lei for technical assistance.
This study was supported by an operating grant from the Medical Research Council of Canada (MRC, grant GOP-38106) to S.-L.L, an operating grant from the Natural Sciences and Engineering Research Council of Canada (NSERC, grant 216912-00) to S.-L.L, an operating grant from the Canadian Institutes for Health Research (CIHR, grant MOP-47817) to S.-L.L, an NSERC operating grant to K.E.S., NIH grant RO1AI34829 from the National Institute of Allergy and Infectious Diseases to K.E.S., and an MRC operating grant to R.N.J. W.-Q. Liu is a summer student of K.E.S. supported by the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomicsequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Altman, E., J. R. Roth, A. Hessel, and K. E. Sanderson. 1996. Transposons in current use in genetic analysis in salmonellas, p. 2613-2626. In F. Neidhard, R. Curtiss III, C. A. Gross, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 3.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 4.Beltran, P., J. M. Musser, R. Helmuth, J. J. I. Farmer, W. M. Frerichs, I. K. Wachsmuth, K. Ferris, A. C. McWhorter, J. G. Wells, A. Cravioto, and R. K. Selander. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltran, P., S. A. Plock, N. H. Smith, T. S. Whittam, D. C. Old, and R. K. Selander. 1991. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J. Gen. Microbiol. 137:601-606. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett, I. I. I., C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, E. F., F. S. Wand, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, E. F., F.-S. Want, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationships among the salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosa, J. H., D. J. Brenner, W. H. Ewing, and S. Falkow. 1973. Molecular relationships among the salmonellae. J. Bacteriol. 115:307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauthier, A., M. Turmel, and C. Lemieux. 1991. A group I intron in the chloroplast large subunit rRNA gene of Chlamydomonas eugametos encodes a double-strand endonuclease that cleaves the homing site of this intron. Curr. Genet. 19:43-47. [DOI] [PubMed] [Google Scholar]
- 11.Groisman, E. A., M. H. Saier, Jr., and H. Ochman. 1992. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 11:1309-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groisman, E. A., M. A. Sturmoski, F. R. Solomon, R. Lin, and H. Ochman. 1993. Molecular, functional, and evolutionary analysis of sequences specific to Salmonella. Proc. Natl. Acad. Sci. USA 90:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 14.Le Minor, L. 1984. Salmonella Lignieres 1900: p. 427-458. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, Md.
- 15.Le Minor, L. 1988. Typing of Salmonella species. Eur. J. Clin. Microbiol. Infect. Dis. 7:214-218. [DOI] [PubMed] [Google Scholar]
- 16.Le Minor, L., and M. Y. Popoff. 1987. Designation of Salmonella enterica sp. nov. as the type and only species of the genus Salmonella. Int. J. Syst. Bacteriol. 37:465-468. [Google Scholar]
- 17.Liu, S.-L., T. Ezaki, H. Miura, K. Matsui, and E. Yabuuchi. 1988. Intact motility as a Salmonella invasion-related factor. Infect. Immun. 56:1967-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, S.-L., A. Hessel, H.-Y. M. Cheng, and K. E. Sanderson. 1994. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella paratyphi B. J. Bacteriol. 176:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-CeuI, an intron-encoded endonuclease, specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella enteritidis shows an inversion relative to Salmonella typhimurium LT2. Mol. Microbiol. 10:655-664. [DOI] [PubMed] [Google Scholar]
- 21.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end-labeling, and pulsed-field gel electrophoresis. J. Bacteriol. 175:4104-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, S.-L., and K. E. Sanderson. 1992. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J. Bacteriol. 174(5):1662-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, S.-L., and K. E. Sanderson. 1995. The chromosome of Salmonella paratyphi A is inverted by recombination between rrnH and rrnG. J. Bacteriol. 177:6585-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, S.-L., and K. E. Sanderson. 1995. The genomic cleavage map of Salmonella typhi Ty2. J. Bacteriol. 177:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, S.-L., and K. E. Sanderson. 1995. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J. Bacteriol. 177:3355-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, S.-L., and K. E. Sanderson. 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl. Acad. Sci. USA 92:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, S.-L., and K. E. Sanderson. 1996. Highly plastic chromosomal organization in Salmonella typhi. Proc. Natl. Acad. Sci. USA 93:10303-10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S.-L., and K. E. Sanderson. 1998. Homologous recombination between rrn operons rearranges the chromosome in host-specialized species of Salmonella. FEMS Microbiol. Lett. 164:275-281. [DOI] [PubMed] [Google Scholar]
- 29.Liu, S.-L., and K. E. Sanderson. 1998. Physical analysis of the Salmonella typhimurium genome, p. 371-381. In P. H. Williams, J. Ketley, and G. Salmond (ed.), Methods in microbiology. Academic Press, New York, N.Y.
- 30.Liu, S.-L., A. B. Schryvers, K. E. Sanderson, and R. N. Johnston. 1999. Bacterial phylogenetic clusters revealed by genome structure. J. Bacteriol. 181:6747-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall, P., T. B. Davis, and C. Lemieux. 1994. The I-CeuI endonuclease: purification and potential role in the evolution of Chlamydomonas group I introns. Eur. J. Bacteriol. 220:855-859. [DOI] [PubMed] [Google Scholar]
- 32.Marshall, P., and C. Lemieux. 1991. Cleavage pattern of the homing endonuclease encoded by the fifth intron in the chloroplast large subunit rRNA-encoding gene of Chlamydomonas eugametos. Gene 104:241-245. [DOI] [PubMed] [Google Scholar]
- 33.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 34.Morrow, B. J., J. E. Graham, and R. Curtiss III. 1999. Genomic subtractive hybridization and selective capture of transcribed sequences identify a novel Salmonella typhimurium fimbrial operon and putative transcriptional regulator that are absent from the Salmonella typhi genome. Infect. Immun. 67:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochman, H., and U. Bergthorsson. 1998. Rates and patterns of chromosome evolution in enteric bacteria. Curr. Opin. Microbiol. 1:580-583. [DOI] [PubMed] [Google Scholar]
- 36.Ocholi, R. A., L. U. Enurah, and P. S. Odeyemi. 1987. Fatal case of salmonellosis (Salmonella pullorum) in a chimpanzee (Pan troglodytes) in the Jos Zoo. J. Wildl. Dis. 23:669-670. [DOI] [PubMed] [Google Scholar]
- 37.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 38.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 39.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 40.Perna, N. T., G. I. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 41.Popoff, M. Y., J. Bockemuhl, and F. W. Brenner. 2000. Supplement 1999 (no. 43) to the Kauffmann-White scheme. Res. Microbiol. 151:893-896. [DOI] [PubMed] [Google Scholar]
- 42.Reeves, M. W., G. M. Evins, A. A. Heiba, B. D. Plikaytis, and J. J. Farmer III. 1989. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 27:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanderson, K. E., A. Hessel, S.-L. Liu, and K. E. Rudd. 1996. The genetic map of Salmonella typhimurium, edition VIII, p. 1903-1999. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 44.Selander, R. K., P. Beltran, N. H. Smith, R. Helmuth, F. A. Rubin, D. J. Kopecko, K. Ferris, B. D. Tall, A. Cravioto, and J. M. Musser. 1990. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 58:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selander, R. K., J. Li, and K. Nelson. 1996. Evolutionary genetics of Salmonella enterica, p. 2691-2707. In F. C. Neidhardt et al.(ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 46.Shivaprasad, H. L. 2000. Fowl typhoid and pullorum disease. Rev. Sci. Technol. 19:405-424. [DOI] [PubMed] [Google Scholar]
- 47.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 48.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 49.Wigley, P., A. Berchieri, Jr., K. L. Page, A. L. Smith, and P. A. Barrow. 2001. Salmonella enterica serovar Pullorum persists in splenic macrophages and in the reproductive tract during persistent, disease-free carriage in chickens. Infect. Immun. 69:7873-7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang-Barber, L., A. K. Turner, and P. A. Barrow. 1999. Vaccination for control of Salmonella in poultry. Vaccine 17:2538-2545. [DOI] [PubMed] [Google Scholar]