Abstract
The putative regulatory CcaR protein, which is encoded in the β-lactam supercluster of Streptomyces clavuligerus, has been partially purified by ammonium sulfate precipitation and heparin affinity chromatography. In addition, it was expressed in Escherichia coli, purified as a His-tagged recombinant protein (rCcaR), and used to raise anti-rCcaR antibodies. The partially purified CcaR protein from S. clavuligerus was able to bind DNA fragments containing the promoter regions of the ccaR gene itself and the bidirectional cefD-cmcI promoter region. In contrast, CcaR did not bind to DNA fragments with the promoter regions of other genes of the cephamycin-clavulanic acid supercluster including lat, blp, claR, car-cyp, and the unlinked argR gene. The DNA shifts obtained with CcaR were prevented by anti-rCcaR immunoglobulin G (IgG) antibodies but not by anti-rabbit IgG antibodies. ccaR and the bidirectional cefD-cmcI promoter region were fused to the xylE reporter gene and expressed in Streptomyces lividans and S. clavuligerus. These constructs produced low catechol dioxygenase activity in the absence of CcaR; activity was increased 1.7- to 4.6-fold in cultures expressing CcaR. Amplification of the ccaR promoter region lacking its coding sequence in a high-copy-number plasmid in S. clavuligerus ATCC 27064 resulted in a reduced production of cephamycin C and clavulanic acid, by 12 to 20% and 40 to 60%, respectively, due to titration of the CcaR regulator. These findings confirm that CcaR is a positively acting autoregulatory protein able to bind to its own promoter as well as to the cefD-cmcI bidirectional promoter region.
Secondary metabolites play different roles in the producer strains and usually are formed in nature at very low levels, indicating the existence of tight control mechanisms for their biosynthesis (5, 20, 21). Streptomyces clavuligerus produces β-lactam antibiotic cephamycin C (7-methoxy-3′-carbamoyl-deacetylcephalosporin C) (17) and β-lactamase inhibitor clavulanic acid (reviewed in references 11 and 18). This strain also produces a β-lactamase that is sensitive to clavulanic acid (25), a β-lactamase inhibitory protein (BLIP) (8), and a BLIP-homologous protein (BLP) (27).
The genes encoding cephamycin C and clavulanic acid biosynthesis are clustered in the genome forming the so-called β-lactam supercluster (37). Genes for cephamycin C biosynthesis include lat and pcd, involved in the formation of the α-aminoadipic precursor of the antibiotic, as well as structural genes involved in the early steps of the pathway (pcbAB and pcbC), resulting in the formation of isopenicillin N, the middle steps of the pathway (cefD and cefE), forming deacetylcephalosporin C, and the late specific C-7 methoxylation (cmcI and cmcJ) and carbamoylation steps (cmcH) of cephamycin biosynthesis (17). We described a few years ago a regulatory gene, ccaR, located in the cephamycin gene cluster that appears to control both cephamycin C and clavulanic acid biosynthesis (27, 36). Disruption of ccaR prevents synthesis of cephamycin and clavulanic acid, whereas complementation of a disrupted mutant with the ccaR gene restores the production of both antibiotics to normal levels (27). In addition, this mutant did not express the claR gene, which encodes a regulatory protein required for clavulanic acid biosynthesis (23, 29).
The regulation of expression of genes for cephamycin C and clavulanic acid biosynthesis is still poorly understood. The pcbC gene, encoding isopenicillin N synthase, is transcribed as a small monocistronic messenger (31) and as part of a polycistronic transcript together with the lat and pcbAB genes, both of them encoding enzymes for the early steps of the pathway (1). The cefD and cefE genes, encoding enzymes for the middle steps of the pathway, are cotranscribed (15), forming a polycistronic transcript with early gene pcd (26, 28). Northern analysis of ccaR indicates that this gene is transcribed as a monocistronic mRNA of 0.9 kb (27). Other transcriptional units in the cephamycin C-clavulanic acid supercluster that have been described (23, 24, 30) are indicated in Fig. 1.
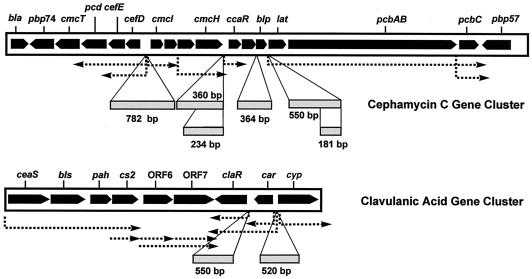
FIG. 1.
Organization of the cephamycin C-clavulanic acid gene cluster. Dotted arrows, transcriptional units reported by several authors; boxes, DNA fragments used in mobility shift experiments (sizes are indicated below).
Recently a report concluded that the CcaR regulatory protein binds the promoter of the lat gene (16), but presumably it might also bind the promoters of other structural genes encoding key enzymes in cephamycin biosynthesis. CcaR affects also clavulanic acid by an unknown mechanism, which might be mediated by the expression of the LysR-type regulatory protein encoded by claR. It was, therefore, of interest to study the role of ccaR by purifying the CcaR protein and performing in vitro interaction studies. We report in this article that CcaR is an autoregulatory activator that interacts with the cefD-cmcI bidirectional promoter and also with its own promoter.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. Escherichia coli strains were grown at 37°C in TY medium or in 2× TY medium (20 g of tryptone/liter and 10 g of yeast extract/liter, pH 7.2) supplemented with ampicillin (100 μg/ml) when required. S. clavuligerus ATCC 27064 and the strains derived from it were grown in TSB medium (30 g of Trypticaseine soy broth [Pronadisa, Madrid, Spain]/liter) for 36 h at 220 rpm and 28°C. Five milliliters of this culture was used to inoculate 100 ml of TSB, and the culture was grown in the same conditions for 36 h. Streptomyces lividans 1326 was grown in YEME medium (12) supplemented with MgCl2 (5 mM) and glycine (0.5%). Cultures of S. clavuligerus or S. lividans transformants were supplemented with thiostrepton (5 μg/ml) or neomycin (1 μg/ml) when required.
TABLE 1.
Strains and plasmids used in this work
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdr17 supE44 relA1 lac[F′ proAb lac1qZΔM15Tn10(TetI)] | 3 |
| S. lividans 1326 | Host in transformation experiments | 12 |
| S. clavuligerus 27064 | Producer of cephamycin C and clavulanic acid | ATCCa |
| S. clavuligerus(pB17B) | Transformant carrying ccaR in multicopy | 27 |
| S. clavuligerus ccaR::aph | ccaR-disrupted mutant | 27 |
| pBluescript I KS(+), pBluescript II SK(+) | E. coli general cloning vectors; Ampr | Stratagene |
| pQE30 | Expression vector to purify histidine-tagged proteins; Ampr | Qiagen |
| pQE30-ccaR | pQE30 containing ccaR | This work |
| pIJ4083 | Promoter-probe plasmid using xylE as reporter | 6 |
| pIJ6021 | Used to get a 550-bp BamHI-KpnI fragment carrying the tipA promoter and the to terminator | 7 |
| pIJ699 | Bifunctional E. coli/Streptomyces positive selection vector; 9.5 kb; Kanr Vphr Tsrr | 12 |
| pULVK99 | Bifunctional E. coli/Streptomyces positive selection vector; 7.8 kb; Kanr Tsrr | 4 |
| pIK | Bifunctional plasmid obtained by ligation of pBluescript II SK(+) and pIJ4083 | This work |
| pKTK | Intermediate vector; 4.9 kb; contains in pIJ699 sequentially the aph gene, the PtipA promoter, and the to terminator | This work |
| pKTCK | Intermediate vector; 5.75 kb; contains in pIJ699 sequentially the aph gene, the to terminator, the PtipA promoter, and the ccaR gene | This work |
| pCX | Intermediate vector; 4.8 kb; contains in pIJ699 sequentially the PccaR promoter, the xylE gene, and the to terminator | This work |
| pIKT, pCXT, pCXTccaR, pBXT-D and -I, pBXTccaR-D and -I | pIJ699-derived plasmids containing the inserts shown in Fig. 3 | This work |
| PccaR-234 | pULVK99-derived plasmid; 8 kb; carries in the EcoRI site a 234-nt DNA fragment containing the ccaR promoter | This work |
ATCC, American Type Culture Collection.
DNA manipulations.
Restriction endonuclease digestions of DNA were carried out according to the manufacturer's recommendations, and the DNA fragments were purified from agarose gels as described by Polman and Larkin (32). DNA ligation, plasmid isolation, and E. coli and Streptomyces transformations were performed by standard procedures (12, 34). PCR mixtures (50 μl) contained 20 ng of template DNA, Taq polymerase (1 U), 0.5 μM (each) primer, and deoxynucleoside triphosphate as follows: 35 μM dGTP and dCTP and 15 μM dTTP and dATP. The following oligonucleotides were used as primers: ccaR-1 (5′-AAGGATCCGTGAGGATCCGGCTCCTGG) and ccaR-2 (5′-TCCCCGCCGTTGTGAGAAGA), ccaR-3 (5′-GTGGACATGGCTTCGGCGTAAT) and the SK reverse primer, ccaR-4 (5′-GGGGGTAGGGAGGGGAGAGT) and ccaR-3, ccaR-5 (5′-GGAGGGAGCATATGAACACCTGGAATGATGTG), DI-1 (5′-GCTACCGCCATGTCAACG) and DI-2 (5′-CATTGCCCTCTTCCTTGA), blp-1 (5′-GCGCGAAAGCCCTGAATGAC) and blp-2 (5′-GCGGACCTCCATGTCTTCTTC), lat-1 (5′-CGGGCTTCGGGAGAAACAC) and lat-2 (5′-TCGCCCATGGGTGAGAACTC), lat-3 (5′-CCATTCAGGGCAGTTCACAAAGA), and argR-1 (5′-GCTGATTCCGCCGCTGGTC) and argR-2 (5′-CGGCTGGCGGTTGAGGAT). The underlined region in the ccaR-1 sequence corresponds to a BamHI site, and that in the ccaR-5 sequence corresponds to an NdeI site.
The following DNA fragments used for mobility shift experiments (Fig. 1) were obtained by PCR: blp-364, obtained with oligonucleotides blp-1 and blp-2, DI-782, obtained with oligonucleotides DI-1 and DI-2, ccaR-360, obtained with oligonucleotides ccaR-1 and ccaR-2, ccaR-234, obtained with oligonucleotides ccaR-3 and ccaR-4, lat-550, obtained with oligonucleotides lat-1 and lat-2, and lat-181, obtained with oligonucleotides lat-3 and lat-2. Probe argR-221, containing the promoter region of the argR gene and used as negative control (33), was obtained by PCR with argR-1 and argR-2. The ccaR gene was obtained (i) by PCR with oligonucleotides ccaR-1 and ccaR-2 as an 855-bp DNA fragment lacking the ATG start codon, which was inserted in plasmid pQE30, and (ii) by PCR using oligonucleotides ccaR-2 and ccaR-5, resulting in a 884-bp DNA fragment containing the complete ccaR gene, which was inserted in plasmids pCXT ccaR, pBXT ccaR-I, and pBXT ccaR-D. In addition, a 550-bp NcoI-PstI DNA fragment containing the claR promoter and a 520-bp SacI-SalI DNA fragment containing the bidirectional promoter car-cyp were prepared.
To prepare labeled DNA for binding assays, the DNA fragments containing the promoters were subcloned into the EcoRV site of pBluescript II SK(+) and rescued by digestion with XbaI-SalI. The DNA fragments were labeled at both ends with [α-32P]dCTP (Amersham) and Klenow DNA polymerase (34). The labeled probes were purified by filtration through the Wizard DNA clean-up system (Promega).
The sequence of the lat-181 probe was determined by the Sanger method. The nucleotide sequence of this probe was as follows: 5′-CCATTCAGGGCAGTTCACAAAGAGCCATCGAGAGGCGTCCGAGAGAGCTGGAAGAGGGGTCCAAGAGCATGGTGGGTCATTATTGTGATCCTAAAATGTCCAGTTCACCGCCATGACAGCAGAGGCTGGAAAGTCCCCCATAATTCAGCCTGATCCCCCAGGAGTTCTCACCCATGGGCGA-3′, in which the putative nucleotides reported to be the target for CcaR (16) are underlined.
Plasmids constructions. (i)Plasmid pIKT.
Plasmids pBluescript II SK(+) and pIJ4083 (6) were linearized with HindIII and ligated to form 10.6-kb bifunctional vector pIK. Then, a 550-bp BamHI/SstI fragment from pIJ6021 (12) containing the tipA promoter and the to terminator of lambda phage was inserted into a BamHI/SstI fragment of linearized pIK to give 11.1-kb pIKT.
(ii) Plasmids pCXT, pCXTccaR, pBXT-D, pBXTccaR-D, pBXT-I, and pBXTccaR-I
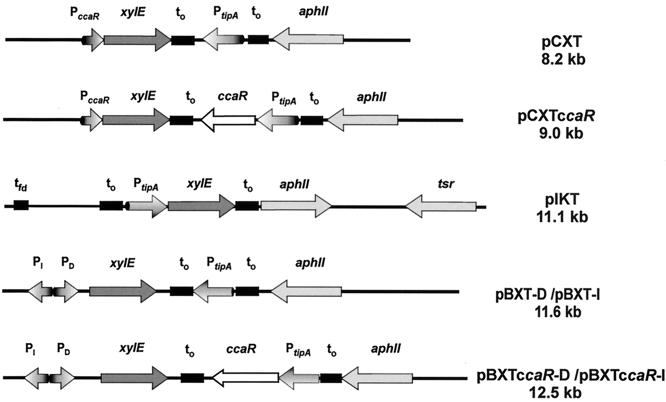
These plasmids were constructed in E. coli, and the fragments listed in Table 1 were subcloned in Streptomyces basal vector pIJ699 with adequate cohesive or blunt ends. The final constructions are shown in Fig. 7.
FIG. 7.
Plasmid constructions used in this work to test expression of xylE under the control of the ccaR promoter. xylE, reporter gene encoding catechol oxygenase; PccaR, promoter of the S. clavuligerus ccaR gene; PtipA, promoter of the thiostrepton-induced tipA gene; PDI, bidirectional cefD-cmcI promoter of S. clavuligerus; aphII, aminoglycoside phosphotransferase (kanamycin) resistance gene; to, lambda phage terminator. Note that constructs pCXT ccaR, pBXT ccaR-D, and pBXT ccaR-I contain the complete ccaR gene in addition to the xylE reporter construct to study the effect of increasing CcaR protein concentration on reporter gene expression.
Overproduction and purification of recombinant rCcaR from E. coli.
An 855-bp DNA fragment obtained by PCR using oligonucleotides ccaR-1 and -2 as primers was digested with BamHI and SalI and fused in frame to BamHI- and SalI-digested pQE30 to form pQE30-ccaR. The correct fusion was confirmed by DNA sequencing, and this plasmid was transformed into E. coli XL1-Blue to form E. coli(pQE30-ccaR). This transformant was grown in 2× TY medium at 37°C for 10 h, and the culture was used to seed (1% inoculum) a 2× TY culture that was grown at 25°C to an optical density at 600 nm of 0.15 to 0.2, induced with IPTG (isopropyl-β-d-thiogalactopyranoside; 0.5 mM), and incubated for an additional 17 h. Cells were harvested, washed with binding buffer (6 M urea, 0.1 M sodium phosphate, 0.01 M Tris-HCl, pH 8.0), and disrupted by sonication. The cell lysate was centrifuged at 18,100 × g for 15 min at 4°C, and the supernatant was loaded into a Ni2+-nitrilotriacetic acid resin (Qiagen). His-tagged CcaR (rCcaR) was eluted with 0.5 M imidazole and dialyzed against 50 mM TES (N-tris[hydroxymethyl]methyl-2-aminoethanesulfonic acid) buffer to remove the urea. The purified rCcaR protein was confirmed by immunoblotting against anti-His antibodies (Sigma Co., St. Louis, Mo.). Most of the rCcaR formed by E. coli was present in inclusion bodies in all the conditions tested; therefore urea treatment was required to solubilize the protein and subsequent dialysis was required for refolding.
Preparation of anti-rCcaR protein antibody.
rCcaR was separated from small contaminant proteins by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE). The 29-kDa rCcaR band was excised from gels, homogenized in sterile water, and eluted with an Electro-Eluter 422 (Bio-Rad). The eluted protein was injected intradermically into New Zealand White rabbits with Freund's complete adjuvant. The rabbits were injected with a booster every 2 weeks until 2.5 mg of rCcaR protein was used. The antiserum was precipitated with solid ammonium sulfate (100%), and the antibodies were resuspended in phosphate-buffered saline buffer (50 mM Na2HPO4, 300 mM NaCl, pH 7.6) and desalted through a PD10 column (Amersham Pharmacia). Additional purification of the immunoglobulin Gs (IgGs) was achieved in a protein A-Sepharose column (Amersham Pharmacia) equilibrated with buffer D (3 M NaCl, 1.5 M glycine, pH 8.9), and the IgGs were eluted with 0.1 M citric acid, pH 4.5. The antibodies were equilibrated in 500 mM Tris-HCl, pH 8.9, and stored at −80°C. Western blots of proteins were made in Immobilon-P transfer membranes (Millipore Corp.) in accordance with the manufacturer protocol.
Purification of CcaR from S. clavuligerus.
The mycelium from a 36-h culture of S. clavuligerus(pB17B) was washed with and resuspended in buffer A (100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 5% glycerol, 20 mM Tris-HCl, pH 7.5). Cells were disrupted by sonication, and the cell debris was removed by centrifugation at 13,200 × g and 4°C for 30 min. The cell extract was fractionated with ammonium sulfate (0 to 40%, 40 to 60%, and 60 to 80%), and the precipitated proteins were resuspended in buffer B (20 mM HEPES, 1.5 mM MgCl2, 50 mM KCl, 1 mM EDTA, 1 mM DTT, pH 7.9) and desalted through a Sephadex G-25 column equilibrated with buffer C (0.01 M sodium phosphate, pH 7.0). The CcaR protein was found in the 40-to-60% fraction by immunodetection (Western blotting) after SDS-12% PAGE and by gel retardation assays.
CcaR-containing fractions were applied to a 5-ml HiTrap heparin-agarose column (Amersham Pharmacia) equilibrated with 0.01 M phosphate buffer at a flow rate of 0.3 ml/min. The column was washed with 40 ml of 0.01 M phosphate buffer at a flow rate of 0.5 ml/min, and the proteins were eluted in the same buffer with a two-step gradient: 0 to 1 M NaCl in 25 ml and 1 to 2 M NaCl in 25 ml at a flow rate of 5 ml/min. Fractions were suspended (1:1) in 50% glycerol and stored at −80°C until required.
Enzyme activities.
The catechol-2,3-dioxygenase activity of the xylE reporter gene was measured as described by Kieser et al. (12) in dialyzed cell extracts of 48-h cultures.
DNA-protein binding assays.
DNA-binding tests were performed by the electrophoretic mobility shift assay (EMSA) in a final volume of 20 μl containing 80 mM HEPES, 200 mM KCl, 20 mM MgCl2, 0.5 mM MnCl2, 40% glycerol, 16 mM Tris-HCl, pH 7.5, poly[(dI-dC)] (1 μg), and 0.1 to 0.2 μg of CcaR preparation. End-labeled DNA (1 to 3 ng) was then added, and the reaction mixture was incubated for 30 min at 25°C.
Nondenaturing polyacrylamide gels were made in TBE buffer (89 mM Tris-HCl, 89 mM boric acid, 2 mM EDTA) (34) and prerun for 90 min prior to application of the samples. Each EMSA was performed at the optimal conditions for the specific promoter interaction being studied. Assay mixtures made with the ccaR promoter were applied to a 10% polyacrylamide (60:1) gel, and the electrophoresis was developed for 20 h at 110 V. To test binding of the DI bidirectional promoter, the assay mixture was applied to a 9% polyacrylamide (29:1) gel, while mobility assays with the argR promoter needed 9% polyacrylamide (40:1) gels. In the last two cases the electrophoresis was developed for 5 h at 100 V. The gels were dried and exposed to Kodak X-ray films at −80°C.
RESULTS
Purification of CcaR: identification of S. clavuligerus proteins giving positive reaction with anti-rCcaR antibodies.
To determine if the CcaR protein regulates cephamycin C biosynthesis by binding to promoters of the cephamycin C gene cluster, ammonium sulfate precipitate fractions (0 to 40%, 40 to 60%, and 60 to 80%) obtained from cell extracts of S. clavuligerus(pB17B) (36-h culture) were tested by immunoblotting using anti-rCcaR antibodies. A positive immunoblotting signal was found in the 40-to-60% ammonium sulfate precipitate fraction. The protein giving the immunoreaction was further purified from the 40-to-60% ammonium sulfate precipitate by HiTrap heparin-agarose chromatography, and the eluted fractions were tested by immunoblotting with antibodies against rCcaR. Fractions 24 to 29 from the column showed a positive immunoblotting signal, suggesting that these fractions contain CcaR.
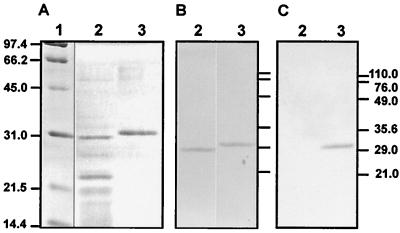
Analysis by SDS-PAGE showed that fractions 24 to 29 contained two major protein bands of about 30 and 23 kDa (Fig. 2A, lane 2). The 30-kDa protein (expected size of CcaR according to the amino acid sequence is 28.3 kDa) reacted with anti-rCcaR antibodies (Fig. 2B, lane 2). The same antibodies reacted clearly with a positive control of recombinant rCcaR obtained in E. coli (Fig. 2B, lane 3), as did commercial antihistidine antibodies (Fig. 2C, lane 3). The rCcaR protein showed a molecular mass 1 kDa higher than that of endogenous S. clavuligerus CcaR; this correlated well with the increase in molecular mass due to the six histidine residues (about 1 kDa).
FIG. 2.
(A) SDS-PAGE of CcaR preparations. (B) Immunoblotting using antibodies against rCcaR. (C) Immunoblotting using antihistidine antibodies. Lane 1, molecular weight markers; lanes 2, fraction 26 from a heparin-agarose column (2 μg of protein); lanes 3, rCcaR purified from E. coli pQE30-CcaR (6 μg).
Identification of proteins with promoter-binding activity.
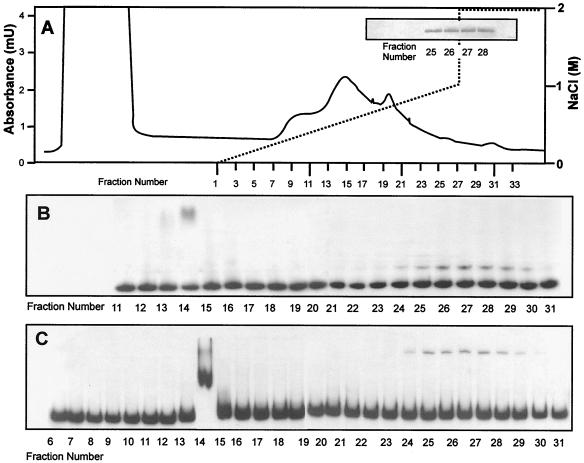
Heparin-agarose-eluted fractions were then tested by EMSA using labeled DNA fragment DI-782 containing the bidirectional DI promoter region (Fig. 3B) and the ccaR-360 DNA fragment containing the ccaR promoter region (Fig. 3C). Fractions 24 to 29, showing a positive immunoblotting signal with anti-rCcaR antibodies (Fig. 3A, inset), gave a clear mobility shift for both promoter-containing fragments.
FIG. 3.
Purification of the CcaR protein from S. clavuligerus(pB17B) by HiTrap heparin-agarose. (A) The ammonium sulfate (40 to 60%) precipitate obtained from the cell extract was suspended in 10 mM phosphate buffer, pH 7.0, and 10.5 ml (105 mg of protein) of the suspension was applied to a heparin-agarose column. The proteins were eluted with a 0-to-2 M NaCl gradient. Solid line, protein; dashed line, NaCl. (Inset) Fractions giving positive signals in immunoblotting assays with anti-rCcaR antibodies. (B) Gel mobility shift of the ccaR-360 DNA fragment containing the ccaR promoter with fractions eluted from the heparin-agarose column. (C) Mobility shift of the DI-782 DNA fragment containing the cefD-cmcI bidirectional promoter with the same fractions eluted from the heparin-agarose column.
Cell extracts from S. clavuligerus ccaR::aph, a ccaR-disrupted mutant (27), were precipitated with ammonium sulfate (40 to 60%) and purified through the same heparin-agarose column, and fractions 24 to 29, equivalent to those containing CcaR in S. clavuligerus(pB17B), were tested by EMSA with the DI-782 and ccaR-360 DNA fragments. No alteration of the electrophoretic mobility of any of the promoter regions was found with these fractions from the ccaR-disrupted mutant.
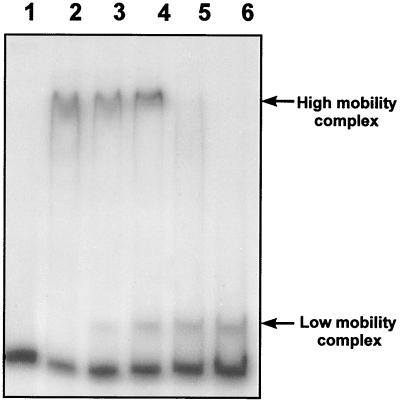
In addition, fraction 14, eluted from the heparin-agarose column, gave a retardation of the electrophoretic mobilities of both promoters (Fig. 3B and C). However, proteins present in fraction 14 gave a high mobility shift (Fig. 4, lane 2), which was different from the low mobility shift exerted by the proteins present in fractions 24 to 29 (Fig. 4, lane 6). There was no interaction between the proteins present in both fractions, as shown in EMSAs in which high-mobility shift- and low-mobility shift-producing proteins were mixed (Fig. 4, lanes 3 to 5). Moreover the proteins present in fraction 14 gave mobility shift on all the promoters tested, including the argR promoter. Proteins forming the high-mobility complex were also found in the heparin-purified extracts of S. clavuligerus ccaR::aph (lacking CcaR), indicating that their DNA-binding ability is not due to CcaR. None of the proteins present in faction 14 showed a molecular mass close to 30 kDa. It is likely that the mobility shift produced by fraction 14 is due to the RNA polymerase complex that might elute in these fractions.
FIG. 4.
Lack of interaction in the mobility shift of the ccaR-234 DNA probe by proteins present in different fractions eluted from heparin-agarose. The lanes contain 2 ng of labeled probe and the following amounts of protein: lane 1, none; lane 2, 0.2 μg from fraction 14; lane 3, 0.15 μg from fraction 14 and 0.05 μg from fraction 26; lane 4, 0.1 μg from fraction 14 and 0.1 μg from fraction 26; lane 5, 0.05 μg from fraction 14 and 0.15 μg from fraction 26; lane 6, 0.2 μg from fraction 26.
Specificity of the binding of CcaR to different promoter regions of the cephamycin C-clavulanic acid supercluster.
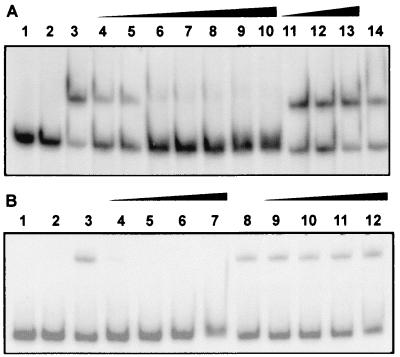
The binding observed with the DI and ccaR promoters was due to the CcaR protein, as deduced from the lack of retardation in EMSA experiments made with fractions eluted from heparin of cell extracts obtained from disrupted mutant S. clavuligerus ccaR::aph. To confirm the specificity of the binding, mobility shift assays were performed in the presence of antibodies against rCcaR. The mobility shift of the fragments containing the bidirectional DI (Fig. 5B) or ccaR (Fig. 5A) promoters was prevented when antibodies against CcaR were added to the preparation (Fig. 5A, lanes 4 to 10, and B, lanes 4 to 7). This neutralizing effect is specific for the antibodies against rCcaR since no neutralization was observed when anti-rabbit IgG antibodies from goats were added to the assay mixture (Fig. 5A, lanes 11 to 13, and B, lanes 10 to 12).
FIG. 5.
(A) Mobility shift of the 360-nt DNA fragment (3 ng) containing the ccaR promoter. Lanes 1 and 2, free probe; lanes 3 and 14, complete mobility shift reaction mixture; lanes 4 to 10, complete mobility shift reaction mixture supplemented, respectively, with 10, 15, 20, 25, 30, 40, or 50 μl of anti-rCcaR antibodies at 1/5,000 dilution; lanes 11 to 13, complete mobility shift reaction mixture supplemented with 3 μl of undiluted anti-rabbit IgG from goats (Sigma Co.). (B) Mobility shift of the 782-nt DNA fragment containing the bidirectional DI promoter. Lanes 1 and 2, free probe; lanes 3 and 8, complete mobility shift reaction mixture; lanes 4 to 7, complete mobility shift assay mixture supplemented with 5, 10, 15, or 30 μl, respectively, of a 1/5,000 dilution of anti-rCcaR antibodies from goats; lanes 9 to 12, complete mobility shift reaction mixture supplemented with 3 μl of undiluted goat anti-rabbit antibodies.
To test whether CcaR was able to bind additional promoters, DNA fragments corresponding to the blp and lat promoter regions of the cephamycin cluster (Fig. 1) were obtained by PCR. In addition NcoI-PstI DNA probe claR-550 and SacI-SalI probe car-cyp-520 (both regions are located in the clavulanic acid gene cluster [Fig. 1]) were also tested in mobility shift experiments using pooled fractions 24 to 29 from heparin-agarose.
The heparin-purified CcaR protein did not bind to DNA fragments containing cephamycin promoters lat or blp, clavulanic acid promoter claR or car-cyp, or the argR promoter under several binding conditions. To confirm the specificity of the binding, assay mixtures were made by using labeled probes and increasing the amounts of unlabeled probes. When the ccaR-234 probe was used, a 15-fold excess of unlabeled probe (7.5 ng) was enough to completely prevent the gel shift. For the cefDI-782 probe the gel shift was prevented in the presence of a 30-fold excess of unlabeled probe (15 ng). The specificity of the binding was confirmed by using the unlabeled claR-550 probe as negative control for ccaR-234 gel shifts. A gel shift with ccaR-234 was always obtained, even in the presence of 100 ng of claR-550.
The results of gel shift studies using the soluble rCcaR obtained in E. coli were less clear than the results obtained with the purified native CcaR protein from S. clavuligerus, probably due to the urea solubilization and subsequent refolding treatment of the protein.
Lack of gel shift of the lat promoter by S. clavuligerus purified CcaR.
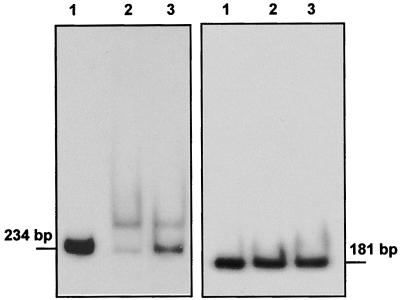
Since gel shift for the lat promoter had been previously described and since the probe used in our work was 550 bp, we obtained by PCR an additional probe of the lat promoter of only 181 bp, lat-181, which contained the sequence CGTCCGAGAGAGCTGGAAGAGGG, reported to be the target for CcaR binding (16). This probe was sequenced to confirm that no errors in the sequence were produced in the PCR procedure. No gel shift of the lat-181 DNA probe was obtained (Fig. 6, right) under conditions in which the ccaR-234 probe was shifted (Fig. 6, left). Additionally unlabeled lat-181 or lat-550 (at 50 ng of DNA per reaction mixture) did not compete with the mobility shift of the ccaR-234 DNA fragment, even though the proportion of labeled to unlabeled probe was 1:100.
FIG. 6.
(Left) Mobility shift of the ccaR-234 probe. Lane 1, free probe; lane 2, assay mixture with 1 μg of purified CcaR; lane 3, assay mixture with 1.5 μg of purified CcaR. (Right) Mobility shift of the lat-181 probe. Lane 1, free probe; lane 2, assay mixture with 1 μg of purified CcaR; lane 3, assay mixture with 1.5 μg of purified CcaR.
CcaR stimulates expression of the reporter xylE gene coupled to ccaR or the DI bidirectional promoters.
The xylE gene, encoding the catechol oxygenase of Pseudomonas aeruginosa was expressed in S. lividans by using the promoter of the ccaR gene (PccaR) or the bidirectional promoter PDI (in both orientations) in plasmid constructs (pCXT and pBXT-D and -I). Other constructs derived from these vectors, carrying, additionally, a copy the ccaR gene (pCXTccaR and pBXT ccaR-D and -I), were also used (Fig. 7). As a positive control, expression of xylE from the tipA promoter in the construct in S. lividans(pIKT) was studied. Two negative controls were used: (i) transformant S. lividans(pIKT) in the absence of thiostrepton (inducer of PtipA) and (ii) S. lividans(pIJ4083), containing the xylE gene without a promoter. By comparing the expression of xylE in S. lividans(pIKT) with that in S. lividans(pCXT) (Table 2), it was concluded that the ccaR promoter (PccaR) has about 20% of the strength of the induced tipA promoter (PtipA) in S. lividans. However, when the expression in S. lividans(pCXTccaR) (which carries in addition the ccaR gene) was studied, it was found that expression from PccaR increased 4.4-fold. These results confirmed that CcaR is a positive-acting autoregulatory protein since overexpression of CcaR leads to increased XylE activity expressed from the ccaR promoter.
TABLE 2.
Effect of CcaR on the catechol dioxygenase activity expressed from the tipA, ccaR, or DI promoter
| Experiment | Strain | Catechol dioxygenase activity
|
|
|---|---|---|---|
| mU/min/mg of protein | % | ||
| I | S. lividans(pIKT) | 101.0 +/− 0.85 | 100.0 |
| S. lividans(pCXT) | 19.5 +/− 0.75 | 19.3 | |
| S. lividans(pCXTccaR) | 87.0 +/− 1.00 | 86.1 | |
| II | S. lividans(pIKT) | 95.5 +/− 0.85 | 100.0 |
| S. lividans(pBXT-D) | 50.0 +/− 1.00 | 52.3 | |
| S. lividans(pBXTccaR-D) | 83.5 +/− 2.50 | 87.4 | |
| S. lividans(pBXT-I) | 48.0 +/− 0.40 | 50.2 | |
| S. lividans(pBXTccaR-I) | 83.0 +/− 1.00 | 86.9 | |
| III | S. lividans(pIKT) | 95.5 +/− 0.85 | |
| S. clavuligerus(pIKT) | 77.5 +/− 0.05 | ||
| S. lividans(pCXT) | 15.5 +/− 0.75 | 100.0 | |
| S. clavuligerus(pCXT) | 51.5 +/− 0.05 | 332.0 | |
The bidirectional PDI promoter expressed xylE in S. lividans(pBXT-D) and S. lividans(pBXT-I) with about 50% of the intensity of xylE expressed from the induced PtipA. The levels of expression in both orientations of the bidirectional DI promoter were similar. Additionally, introduction of ccaR in constructions S. lividans(pBXT ccaR-D) and S. lividans(pBXT ccaR-I) resulted in an increase of xylE expression of 1.7-fold independently of the bidirectional promoter orientation.
The expression of the ccaR promoter in S. lividans(pCXT) is weaker (33%) than that in S. clavuligerus(pCXT), indicating that the endogenous CcaR protein present in S. clavuligerus indeed exerts a positive effect.
Titration of CcaR by the ccaR promoter region results in lower production of cephamycin C and clavulanic acid.
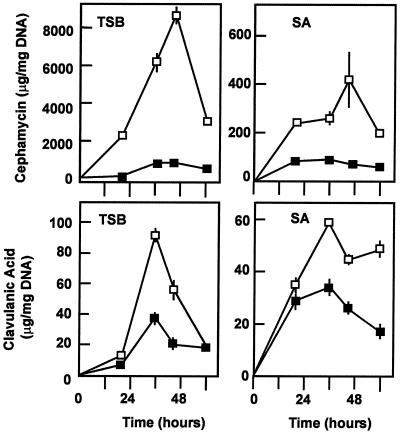
To confirm that the binding of CcaR is required for increasing the expression from both the ccaR and the DI promoters, the ccaR-234 DNA fragment containing the ccaR promoter (Fig. 1) was inserted into EcoRI-digested and end-filled pULVK99 and then transformed into S. clavuligerus. Cultures of S. clavuligerus(pULVK99) and transformant S. clavuligerus(Pc-234) carrying multiple copies of the ccaR promoter were grown in TSB and SA (starch-asparagine) culture media (24), and the production of cephamycin C and clavulanic acid was measured (Fig. 8). A clear reduction in the production of clavulanic acid and cephamycin in transformant S. clavuligerus(Pc-234) in both media was observed throughout the fermentation. This effect can be explained by the titration of CcaR by the ccaR-234 DNA fragment. The effect on cephamycin production was more drastic (the cephamycin level was 10 to 20% of that produced by the control strain in TSB medium), but clavulanic acid production was also affected (40 to 60% production in relation to that produced by the control strain in TSB medium). Therefore, we concluded that the CcaR protein is an activator controlling the expression of its own ccaR gene; additionally it activates expression of the cefD gene, which encodes the epimerase required for the middle steps of the pathway, and of the cmcI gene, encoding the late methoxylation reaction of the cephamycin pathway.
FIG. 8.
Titration effect of multiple copies of the ccaR-234 promoter region. Cephamycin C (top) and clavulanic acid (bottom) production by S. clavuligerus(pULVK99) (□) and S. clavuligerus(Pc-234) (▪) in TSB medium (left) and SA medium (right).
DISCUSSION
Genes encoding ActII-ORF4-like transcriptional regulatory proteins essential for antibiotic biosynthesis have been found in actinorhodin (9), daunorubicin (19), undecylprodigiosin (22), cephamycin (27), and many other gene clusters. These proteins are pathway-specific transcriptional activators of antibiotic structural genes (38). DnrI binds bidirectional promoters dnrG-dpsE, encoding early genes (dnrG-dspABCD-dspEF) of the daunorubicin pathway, and dnrC-dnrD, encoding late genes in the pathway (dnrDKPQS) (35). The Streptomyces coelicolor actII-ORF4-encoded protein activates transcription from the actIII-actI and the actVI-ORF1-ORFA intergenic regions, which contain divergently arranged promoters for the early and late steps of actinorhodin biosynthesis, respectively (2).
As shown in this article, CcaR binds the bidirectional promoter of cefD-cmcI of the cephamycin gene cluster of S. clavuligerus. Therefore it controls expression of the middle (cefDE) and late (cmcI) steps of the cephamycin biosynthesis pathway. When S. clavuligerus is grown in TSB medium, the cefD promoter expresses transcripts of 2.6 and 4.1 kb carrying the cefDE and the cefDE-pcd genes, respectively (28). In addition, a large transcript starting at the cefD promoter region was reported by Kovacevic et al. (15). The pcd gene is an early gene of the cephamycin biosynthetic pathway involved in the formation of α-aminoadipic acid, a cephamycin C precursor. Therefore, by controlling pcd expression from the cefD-cmcI promoter, CcaR controls simultaneously early, middle, and late genes of the pathway. However, the regulatory activity of CcaR might extend to other nontested promoters of the cephamycin pathway and is very likely to control formation of proteins required for the expression of clavulanic acid regulatory protein ClaR.
Binding of E. coli crude recombinant CcaR to the lat promoter has been recently reported (16). In our hands, the 550-bp DNA fragment located upstream of and proximal to the lat gene is not shifted by CcaR. Additionally a 181-bp DNA fragment upstream of the lat gene, which contains the sequence described as the target for CcaR binding, is not shifted either (Fig. 6). However in our assays we used a purified native preparation of CcaR from S. clavuligerus, and therefore the results reported by these authors may be due to another protein present in their extracts.
CcaR also binds to its own ccaR promoter. Negative autoregulation of the transcriptional activators BarA and FurA in Streptomyces virginiae and Streptomyces lavendulae has been reported (13, 14). BarA binds its own promoter in the absence of butyrolactones but not in the presence of butyrolactones. Autoregulation of ActII-ORF4 and DnrI has not been reported. CcaR is a regulatory activator protein. In the absence of CcaR, transformant S. lividans(pCXT) showed 20% of the XylE activity of transformant S. lividans(pCXT ccaR), carrying ccaR. The higher XylE activity in S. clavuligerus(pCXT) than in S. lividans(pCXT) might be due to the presence of CcaR in the former strain, which carries the endogenous ccaR gene in the chromosome. The possibility that low-molecular-weight inducers are involved in S. clavuligerus CcaR autoregulation cannot be excluded in spite of the fact that butyrolactones have not been found yet in S. clavuligerus (10).
CcaR belongs to the family of SARP proteins (Streptomyces antibiotic regulatory proteins). It has been proposed that these proteins recognize specific heptameric sequences that sometimes overlap with the −35 regions of structural genes (38). SARP boxes similar to those reported for the ActII-ORF4- (2) and dnrI (35)-controlled genes occur in the bidirectional (723-nucleotide [nt]) region between the ATG start codons of cefD and cmcI (Fig. 9). A triple palindromic SARP box, separated by 13 and 6 nt, is present 43 nt upstream of the start codon of cefD. In the complementary strand in the region corresponding to the cmcI promoter two SARP boxes are separated by only 15 nt at a position located 224 nt upstream of the cmcI start codon. In addition, two SARP boxes separated by 16 nt are present 395 nt upstream of the cmcI ATG start codon. The significance of these sequences will remain unclear until footprinting experiments confirm their binding to CcaR.
FIG. 9.
SARP-like sequences found in the cefD-cmcI bidirectional promoter (nucleotide accession no. SCCEFDA and AFO73896) in comparison to the sequences present in act and dnr genes. SARP box distances for cefD, cmcI, lat, and ccaR are in relation to the ATG start codon, while the distances for the act and dnr genes are in relation to the transcription initiation site.
Acknowledgments
This work was supported by the Spanish Ministry of Science and Technology (FD97-1419-CO2-O2). I. Santamarta received a fellowship from the University of León.
We are grateful to A. de la Fuente, F. J. Enguita, and C. de Torre for their interest and helpful discussions, to A. Jiménez for revising the manuscript, and to M. Mediavilla for technical assistance.
REFERENCES
- 1.Alexander, D. C., M. J. Brumlik, L. Lee, and S. E. Jensen. 2000. Early cephamycin biosynthesis genes are expressed from a polycistronic transcript in Streptomyces clavuligerus. J. Bacteriol. 182:348-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, P., M. A. Fernández-Moreno, and F. Malpartida. 1999. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J. Bacteriol. 181:6958-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullock, W. O., J. M. Fernández, and J. M. Short. 1987. XL1-blue: a high efficiency plasmid transforming recA Escherichia coli strains. BioTechniques 5:376-379. [Google Scholar]
- 4.Chary, V. K., J. L. de la Fuente, P. Liras, and J. F. Martín. 1997. amy as a reporter gene for promoter activity in Nocardia lactamdurans: comparison of promoters of the cephamycin cluster. Appl. Environ. Microbiol. 63:2977-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. [DOI] [PubMed] [Google Scholar]
- 6.Clayton, T. M., and M. J. Bibb. 1990. Streptomyces promoter-probe plasmids that utilise the xylE gene of Pseudomonas putida. Nucleic Acids Res. 18:1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Fuente, A., E. Cisneros, and A. Talavera. 1994. Restriction end-converting vectors with repeated multiple cloning sites. Gene 139:83-86 [DOI] [PubMed] [Google Scholar]
- 8.Doran, J. L., B. K. Leskiw, S. Aippersbach, and S. E. Jensen. 1990. Isolation and characterization of a β-lactamase-inhibitory protein from Streptomyces clavuligerus and cloning and analysis of the corresponding gene. J. Bacteriol. 172:4909-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Moreno, M. A., J. L. Caballero, D. A. Hopwood, and F. Malpartida. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66:769-780. [DOI] [PubMed] [Google Scholar]
- 10.Horinouchi, S., and T. Beppu. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859-864. [DOI] [PubMed] [Google Scholar]
- 11.Jensen, S. E., and A. S. Paradkar. 1999. Biosynthesis and molecular genetics of clavulanic acid. Antonie Leeuwenhoek 75:125-133. [DOI] [PubMed] [Google Scholar]
- 12.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 13.Kinoshita, H., T. Tsuji, H. Ipposhi, T. Nihira, and Y. Yamada. 1999. Characterization of binding sequences for butyrolactone autoregulator receptors in streptomycetes. J. Bacteriol. 181:5075-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitani, S., H. Kinoshita, T. Nihira, and Y. Yamada. 1999. In vitro analysis of the butyrolactone autoregulator receptor protein (FarA) of Streptomyces lavendulae FRI-5 reveals that FarA acts as a DNA-binding transcriptional regulator that controls its own synthesis. J. Bacteriol. 181:5081-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovacevic, S., M. B. Tobin, and J. R. Miller. 1990. The β-lactam biosynthesis genes for isopenicillin N-epimerase and deacetoxycephalosporin C synthetase are expressed from a single transcript in Streptomyces clavuligerus. J. Bacteriol. 172:3952-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyung, Y.-S., W.-S. Hu, and D. H. Sherman. 2001. Analysis of temporal and spatial expression of the CcaR regulatory element in the cephamycin C biosynthetic pathway using green fluorescent protein. Mol. Microbiol. 40:530-541. [DOI] [PubMed] [Google Scholar]
- 17.Liras, P. 1999. Biosynthesis and molecular genetics of cephamycins. Antonie Leeuwenhoek 75:109-124. [DOI] [PubMed] [Google Scholar]
- 18.Liras, P., and A. Rodríguez-García. 2000. Clavulanic acid, a β-lactamase inhibitor: biosynthesis and molecular genetics. Appl. Microbiol. Biotechnol. 54:467-475. [DOI] [PubMed] [Google Scholar]
- 19.Madduri, K., and C. R. Hutchinson. 1995. Functional characterization and transcriptional analysis of the dnrR1 locus, which controls daunorubicin biosynthesis in Streptomyces peucetius. J. Bacteriol. 177:1208-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martín, J. F., and A. L. Demain. 1980. Control of antibiotic synthesis. Microbiol. Rev. 44:230-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martín, J. F., S. Gutiérrez, and J. F. Aparicio. 2000. Secondary metabolites, p. 213-237. In Encyclopedia of microbiology, vol. 4. Academic Press, San Diego, Calif.
- 22.Narva, K. E., and J. S. Feitelson. 1990. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2). J. Bacteriol. 172:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paradkar, A. S., K. A. Aidoo, and S. E. Jensen. 1998. A pathway-specific transcriptional activator regulates late steps of clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol. Microbiol. 27:831-843. [DOI] [PubMed] [Google Scholar]
- 24.Paradkar, A. S., and S. E. Jensen. 1995. Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. J. Bacteriol. 177:1307-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez-Llarena, F., J. F. Martín, J. J. R. Coque, J. L. de la Fuente, M. Galleni, J.-M. Frère, and P. Liras. 1997. The bla gene of the cephamycin cluster of Streptomyces clavuligerus encodes a class A β-lactamase of low enzymatic activity. J. Bacteriol. 179:6035-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Llarena, F. 1997. Caracterización de la agrupación de genes de cefamicina C en Streptomyces clavuligerus. Ph.D. thesis. University of León, León, Spain.
- 27.Pérez-Llarena, F. J., P. Liras, A. Rodríguez-García, and J. F. Martín. 1997. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both β-lactam compounds. J. Bacteriol. 179:2053-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Llarena, F. J., A. Rodríguez-García, F. J. Enguita, J. F. Martín, and P. Liras. 1998. The pcd gene encoding piperideine-6-carboxylate dehydrogenase involved in biosynthesis of α-aminoadipic acid is located in the cephamycin cluster of Streptomyces clavuligerus. J. Bacteriol. 180:4753-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Redondo, R., A. Rodríguez-García, J. F. Martín, and P. Liras. 1998. The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene 211:311-321. [DOI] [PubMed] [Google Scholar]
- 30.Pérez-Redondo, R. 2000. Genética de la producción de ácido clavulánico en Streptomyces clavuligerus. Ph.D. thesis. University of León, León, Spain.
- 31.Petrich, A. K., B. K. Leskiw, A. S. Paradkar, and S. E. Jensen. 1994. Transcriptional mapping of the genes encoding the early enzymes of the cephamycin biosynthetic pathway of Streptomyces clavuligerus. Gene 142:41-48. [DOI] [PubMed] [Google Scholar]
- 32.Polman, J. K., and J. M. Larkin. 1989. Purification of DNA from agarose gels. Biotechnol. Tech. 3:329-332. [Google Scholar]
- 33.Rodríguez-García, A., M. Ludovice, J. F. Martín, and P. Liras. 1997. Arginine boxes and the argR gene in Streptomyces clavuligerus: evidence for a clear regulation of the arginine pathway. Mol. Microbiol. 25:219-228. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Tang, L., A. Grimm, Y.-X. Zhang, and R. Hutchinson. 1995. Purification and characterization of the DNA-binding protein DnRI, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol. Microbiol. 22:801-813. [DOI] [PubMed] [Google Scholar]
- 36.Walter, N. J., B. Barton, and A. J. Earl. 18August1994. Novel compounds. International patent WO 94/18326-A1.
- 37.Ward, J. M., and J. E. Hodgson. 1993. The biosynthetic genes for clavulanic acid and cephamycin production occur as a “super-cluster” in three Streptomyces. FEMS Microbiol. Lett. 110:239-242. [DOI] [PubMed] [Google Scholar]
- 38.Wietzorreck, A., and M. Bibb. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA binding fold. Mol. Microbiol. 25:1181-1184. [DOI] [PubMed] [Google Scholar]