Abstract
Biogenesis of the light-driven proton pump bacteriorhodopsin in the archaeon Halobacterium salinarum requires coordinate synthesis of the bacterioopsin apoprotein and carotenoid precursors of retinal, which serves as a covalently bound cofactor. As a step towards elucidating the mechanism and regulation of carotenoid metabolism during bacteriorhodopsin biogenesis, we have identified an H. salinarum gene required for conversion of lycopene to β-carotene, a retinal precursor. The gene, designated crtY, is predicted to encode an integral membrane protein homologous to lycopene β-cyclases identified in bacteria and fungi. To test crtY function, we constructed H. salinarum strains with in-frame deletions in the gene. In the deletion strains, bacteriorhodopsin, retinal, and β-carotene were undetectable, whereas lycopene accumulated to high levels (≈1.3 nmol/mg of total cell protein). Heterologous expression of H. salinarum crtY in a lycopene-producing Escherichia coli strain resulted in β-carotene production. These results indicate that H. salinarum crtY encodes a functional lycopene β-cyclase required for bacteriorhodopsin biogenesis. Comparative sequence analysis yields a topological model of the protein and provides a plausible evolutionary connection between heterodimeric lycopene cyclases in bacteria and bifunctional lycopene cyclase-phytoene synthases in fungi.
Microbial rhodopsins consist of a covalent complex between an opsin, containing seven transmembrane α-helices, and retinal, a light-sensitive cofactor. Members of the microbial rhodopsin family were first identified in the archaea, but examples have been found recently in fungi and marine proteobacteria (35). Archaeal rhodopsins act as light-driven ion transporters and phototaxis receptors, and they share a common reaction mechanism in which the primary event is photoisomerization of retinal from an all-trans to a 13-cis configuration (27, 35). Fungal and bacterial rhodopsins are thought to function in the same way as archaeal rhodopsins, based on similarities in their photochemistry and ion transport properties (4, 5, 7, 35). Although the functional properties of microbial rhodopsins have been characterized extensively, relatively little is known about their biogenesis.
We have studied the biogenesis of a well-characterized member of the microbial rhodopsin family, bacteriorhodopsin (BR). BR is produced by the archaeon Halobacterium salinarum from bacterioopsin (BO) and carotenoids, which are synthesized by the organism and converted to retinal, which forms a covalent linkage with BO. BR is the most abundant of four rhodopsins produced by H. salinarum, which also include halorhodopsin and sensory rhodopsins I and II (27, 35). Studying the biogenesis of BR and other rhodopsins in H. salinarum is attractive because molecular genetic tools are available, including transformation (11), selectable markers (16, 24), and targeted gene replacement (29). Significantly, the genome sequence of a closely related organism, Halobacterium sp. strain NRC-1, has been determined (26), so functional genomic approaches are possible. BR production in H. salinarum has been used as a model for several steps in membrane protein biogenesis, such as protein insertion into the membrane (12, 13), biosynthesis and assembly of membrane-bound cofactors (30), and assembly of protein-lipid complexes (17, 18, 20).
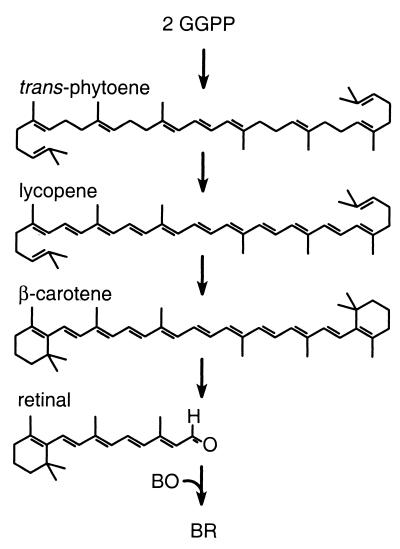
In studies of cofactor assembly during BR biogenesis, we have focused on identifying cellular factors that participate in the biosynthesis and metabolism of carotenoids and are required for BR production. A pathway for retinal biosynthesis in archaeal halophiles closely related to H. salinarum has been proposed based on chemical inhibition studies, the in vitro conversion of isoprenoid precursors to the C40 carotenoid β-carotene, and the carotenoid biosynthetic pathways of other organisms (23, 37). In this pathway (Fig. 1), a series of condensation reactions among isoprenoids of various lengths leads to formation of the C40 carotenoid, phytoene. Phytoene then undergoes four desaturation reactions to yield lycopene. Both ends of lycopene are then cyclized to form β-carotene, which is thought to serve as the immediate precursor of retinal.
FIG. 1.
Proposed carotenoid metabolic pathway leading to BR formation. Geranylgeranyl pyrophosphate (GGPP) is a C20 isoprenoid precursor of phytoene. Partially desaturated intermediates in the conversion of trans-phytoene to lycopene are not shown. The reactions that generate BR from β-carotene and BO are unknown but are proposed to include a symmetric oxidative cleavage followed by binding of retinal to BO.
We previously identified two genes, brp and blh, that are required for generating BR from BO and carotenoid in H. salinarum (30). The brp gene was discovered in earlier studies of spontaneous insertion mutations that abolished BR production (6), but a precise role of the gene could not be ascertained because of transcriptional polar effects of insertions in brp (30). The blh gene was identified as a paralog of brp by sequence analysis of the Halobacterium sp. NRC-1 genome (30). Deletion of both brp and blh abolished BR production and led to the accumulation of β-carotene, suggesting that these genes encode proteins that catalyze the conversion of β-carotene to retinal (30). brp and blh were the first genes found to be required for the assembly of a microbial rhodopsin from carotenoids and a microbial opsin.
In the work reported here, we have identified a third carotenoid metabolic gene required for BR biogenesis. We searched for a lycopene cyclase gene because the conversion of lycopene to β-carotene is thought to be a key regulatory step in BR biogenesis (15). We identified a gene in H. salinarum, designated crtY, that encodes a protein homologous to a recently discovered class of lycopene cyclases found in both bacteria (22, 41) and fungi (2, 39, 40). Deletion studies of crtY in H. salinarum and heterologous expression studies in Escherichia coli indicate that the gene encodes a lycopene β-cyclase required for synthesizing β-carotene during BR production. The implications of these findings for BR biogenesis in H. salinarum and the evolution of lycopene cyclases are discussed.
MATERIALS AND METHODS
Materials.
Oligonucleotides were obtained from Operon (Alameda, Calif.), Taq polymerase was obtained from Promega (Madison, Wis.), and restriction endonucleases and ligase were obtained from New England Biolabs (Beverly, Mass.). Lycopene, β-carotene, and retinal were obtained from Sigma.
Plasmid construction.
Plasmids used in this study are listed in Table 1. Plasmids were propagated in the E. coli strain DH5α except where noted. A plasmid suitable for general gene replacement was constructed by combining the 5.1-kb KpnI-EcoRI fragment of pMPK85 (17) with the 1.4-kb KpnI-EcoRI fragment from pMPK408 (29). The resulting plasmid, pMPK428, contains a mevinolin resistance determinant, the ura3 cassette, a polylinker site, and features to allow propagation in E. coli. Two-step PCR was used to construct the ΔcrtY plasmids. As a first step, reactions were carried out with the primer pairs 1 (TCTAGATCTAGATGAACCGTGATCTGAACAGC) and 2 (GTGCTCGATCTCGATCTCGACAGCCAGGAACGTGAGGT) or 3 (GAGATCGAGATCGAGCAGCAGTACACCACCGGAATCAC) and 4 (AAGCTTAAGCTTACCCACAACAGGAGGGTTTC), using MPK1 (19) genomic DNA as template. The PCR products were used as template in the second PCR step with the primers 1 and 4, yielding a 1.5-kb product with two XbaI sites at the 5′ end and two HindIII sites at the 3′ end of the fragment (restriction sites underlined in the primer sequences). The sites were duplicated to increase restriction efficiency. The PCR product was fully digested with XbaI and HindIII and combined with the 6.5-kb XbaI-HindIII fragment from pMPK428 to yield pMPK430. This plasmid harbors a 1.5-kb section of DNA homologous to the H. salinarum chromosome with an in-frame crtY deletion of 555 bp and an 18-bp insertion at the deletion site. pMPK431, a similar plasmid that harbors an in-frame crtY deletion of 228 bp with an 18-bp insertion at the deletion site, was constructed by the same strategy except that the primers 1 and 5 (GTGCTCGATCTCGATCTCCTGCGTGATGACGAACAGAT) and 3 and 4 were used in the first PCR step. To create a complementation strain containing crtY at the ura3 locus, PCR was carried out with the primers 1 and 6 (AGATCTAGATCTAAAAGCCGCGCCGGTTCAACGCCACCGGGCTAGCA) by using MPK1 genomic DNA as template. The resulting product contained the crtY gene and flanking DNA with XbaI and BglII sites introduced in the primers. The PCR product was digested with XbaI and BglII and ligated into the XbaI-BglII fragment of pMPK424, which contains the mevinolin resistance determinant, the ura3 cassette, and features to allow propagation in E. coli (30). pMPK424 was prepared from a methylation-deficient strain (CSH26 dam). The resulting plasmid, pMPK432, contains the crtY open reading frame, 605 bp of upstream flanking sequence to allow normal transcription, and a 21-bp sequence derived from the bop transcription terminator (14) to ensure the transcription termination of crtY.
TABLE 1.
Microbial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| MPK1 | H. salinarum Rub− Vac− | 19 |
| MPK407 | MPK1 Δura3 | 29 |
| MPK426 | MPK407 ΔcrtY (codons 45 to 229) | This study |
| MPK427 | MPK407 ΔcrtY (codons 121 to 229) | This study |
| MPK428 | MPK426 ura3::crtY | This study |
| Plasmids | ||
| pACCAR16 ΔcrtX | pACYC-based plasmid containing E. uredovora carotenogenic genes | 25 |
| pBAD TOPO TA | pUC-based expression vector with arabinose-inducible promoter | Invitrogen |
| pMPK85 | pAT153-based plasmid containing mevinolin resistance determinant (Mevr) | 17 |
| pMPK408 | Litmus 28 (NEB)-based plasmid containing ura3 | 29 |
| pMPK424 | pAT153-based plasmid containing Mevrura3, and unique XbaI, BglII sites flanked by 2.0-kb ura3 flanking sequence | 30 |
| pMPK428 | 1.4-kbp KpnI-EcoRI ura3 fragment of pMPK408 combined with 5.1-kbp KpnI-EcoRIpAT153, Mevr fragment of pMPK85 | This study |
| pMPK430 | ΔcrtY (codons 45 to 229) PCR fragment in pMPK428 | This study |
| pMPK431 | ΔcrtY (codons 121 to 229) PCR fragment in pMPK428 | This study |
| pMPK432 | 1.4-kbp XbaI-BglII crtY PCR fragment in pMPK424 | This study |
| pMPK434 | Partial BstXI digest of pACCAR16ΔcrtX, recircularized | This study |
| pMPK438 | crtY open reading frame in pBAD TOPO TA vector | This study |
| pMPK454 | pBAD TOPO TA vector, circularized | This study |
To create a lycopene-producing strain of E. coli, the plasmid pACCAR16ΔcrtX (25) (provided by J. von Lintig) was modified. This plasmid contains four functional Erwinia uredovora genes that result in β-carotene production. The plasmid was partially digested with BstXI, which cuts twice within the E. uredovora crtY gene, and self-ligated after trimming the 3′ overhangs with the large Klenow fragment of DNA polymerase I (New England Biolabs). The ligated sample was then transformed into E. coli JM109, and pink colonies were obtained at a frequency of ≈0.5%. The deletion junction in crtY in the resulting plasmid, pMPK434, was verified in one isolate. This plasmid was found by UV-visible spectroscopy to promote lycopene production without promoting β-carotene production. To construct an expression vector for H. salinarum crtY, the primers GTGGCGTGTCCGACTCGACA and TCAACGCCACCGGGCTAGCA were used in a PCR with MPK1 genomic DNA as template. The PCR product was cloned directly into the pBAD TOPO TA expression vector (Invitrogen, Carlsbad, Calif.), and the resulting plasmid, pMPK438, was transformed into E. coli JM109 with pMPK434. To obtain a control strain containing only the expression vector, the pBAD TOPO TA vector was self-ligated to make pMPK454 and introduced into pMPK434-containing E. coli JM109.
H. salinarum strain construction.
H. salinarum strains (Table 1) were obtained by using a two-step ura3-based gene replacement method as described previously (29). Briefly, strains were transformed as described elsewhere (19) with selection for mevinolin resistance and then replated on medium containing 5-fluoroorotic acid to select for recombinants. Colonies resistant to 5-fluoroorotic acid were screened by PCR to identify the desired recombinants. The ΔcrtY strain lacking codons 45 to 229, MPK426, was isolated by transforming pMPK430 into MPK407 (29). The ΔcrtY strain lacking codons 121 to 229, MPK427, was isolated by transforming pMPK431 into MPK407. The complementation strain with crtY integrated at the ura3 locus, MPK428, was isolated by transforming pMPK432 into MPK426. For all strains, recombinants resistant to 5-fluoroorotic acid were obtained at a frequency of ≈1:100, of which 1 to 20% had the desired gene replacement. The structures of the bop, brp, blh, crtY, and ura3 loci were confirmed in all strains by PCR and Southern blot analysis as described previously (19). The sequences of the entire deleted crtY open reading frame of MPK426 and the deletion junction within the crtY open reading frame in MPK427 were confirmed by ABI PRISM Big Dye Primer Cycle sequencing (Applied Biosystems, Inc., Foster City, Calif.) by using various primers. Sequencing reactions were analyzed with a 337XL automated DNA sequencer (Applied Biosystems, Inc.) at the University of Wisconsin Biotechnology Center.
Quantitation of BR, BO, and carotenoids in H. salinarum.
H. salinarum strains were grown under conditions to induce BR synthesis, cell lysates were prepared, and BR and BO levels were measured as described elsewhere (30), except that cultures were incubated for 86 to 90 h. Total carotenoid was extracted from cell lysates as described previously (15), except that lysates were illuminated for 3 min with a >520-nm light prior to extraction to convert all retinal to the all-trans isomer.
To identify the major carotenoid in ΔcrtY strains, the extracts were fractionated by high-performance liquid chromatography (HPLC) on an HPLX solvent delivery system (Rainin Instrument Co., Inc., Emeryville, Calif.) coupled with a reverse-phase Alltech Altima C18 column (250 by 4.6 mm; 5 μm particle size) (Alltech Associates, Inc., Deerfield, Ill.) and an Alltech Econosphere C18 5-μm guard column. The mobile phase was a gradient of solvent A (85% acetonitrile, 15% methanol) and solvent B (dichloromethane) eluting at 1 ml/min. The solvent change over a 25-min sample run was programmed as follows: elution with 100% solvent A, 1 min; gradient to 32% solvent B, 1.5 min; isocratic elution with solvent B, 5.5 min; gradient to 65% solvent B, 0.5 min; isocratic elution with solvent B, 6 min; gradient to 100% solvent A, 0.5 min; and reequilibration with 100% solvent A, 4 min. The eluate was monitored at 380 nm for the first 8 min and at 474 nm for the remainder of each run with a Dynamax UV-1 variable wavelength UV-visible absorbance detector (Rainin Instrument Co., Inc.). HPLC fractions eluting at ≈11 min were collected and evaporated under nitrogen gas. The UV-visible spectrum was obtained by dissolving the sample in hexane and scanning on a Perkin-Elmer λ2 Spectrophotometer (Applied Biosystems, Inc.). For mass spectrometry analysis, an aliquot of the sample was evaporated to dryness and resuspended in acetone to a lycopene concentration of 10 μM. Mass spectrometry was performed at the University of Wisconsin Biotechnology Center on a Bruker Biflex III matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) instrument (Bruker Analytical Systems, Billerica, Mass.) using 2,5-dihydrobenzoic acid as a matrix.
Simultaneous measurement of retinal, β-carotene, and lycopene was carried out using the HPLC conditions described above. Standard curves were generated with commercial all-trans retinal, β-carotene, and lycopene. The efficiency of extraction of retinal was approximately 55% as judged by parallel extractions of purified BR (data not shown). The difference in retinal yields between this study and our earlier results (30) is likely due to a more accurate standard curve generated by using freshly obtained retinal.
Extraction and analysis of carotenoids in E. coli.
To analyze carotenoid production in E. coli, Luria-Bertani medium (50 ml in a 125-ml Erlenmeyer flask) containing chloramphenicol (170 μg/ml) and ampicillin (150 μg/ml) was inoculated with 1 ml of overnight culture grown in the same medium to saturation. Cultures were grown in the dark at 28°C with shaking at 250 rpm for approximately 4 h to an optical density at 600 nm (OD600) of ≈0.5. Cells were then induced by the addition of 0.5 ml of 20% arabinose to a final concentration of 0.2% arabinose. Cultures were removed at 0, 3, and 6 h after induction and cells were harvested by centrifuging at 6,000 × g for 20 min at 4°C. The cell pellet was resuspended in 50 ml of 50 mM Tris-Cl (pH 8.0) and centrifuged again at 6,000 × g for 20 min, followed by a brief centrifugation to remove all traces of the supernatant. The cell pellet was then resuspended in 3 ml of Tris-Cl buffer followed by vigorous vortexing to obtain a homogenous suspension. A 2.5-ml aliquot of the cell suspension was added to 22.5 ml of acetone and the mixture was stirred vigorously for 20 min. Twelve milliliters of hexane-water (4:1) was added to the mixture, which was then stirred for 2 min, poured into a 100-ml graduated cylinder, and allowed to settle for 5 min. The upper layer containing the carotenoids was then removed and evaporated to dryness under nitrogen. The remaining 0.5 ml of the cell suspension was solubilized in electrophoresis sample buffer, and total cell protein was measured by a bicinchoninic acid assay (Pierce, Rockford, Ill.).
The carotenoid extracts were analyzed and quantified using the same HPLC method described above, except that the eluate was monitored at 474 nm throughout the run. To confirm that β-carotene was synthesized in E. coli strains expressing H. salinarum crtY, fractions eluting at ≈13 min were collected, evaporated to dryness under nitrogen, resuspended in isopropanol, and scanned to obtain UV-visible spectra. Aliquots of the β-carotene sample were prepared, and mass spectrometry analysis was performed as described above.
RESULTS
Sequence identification of Halobacterium sp. NRC-1 crtY.
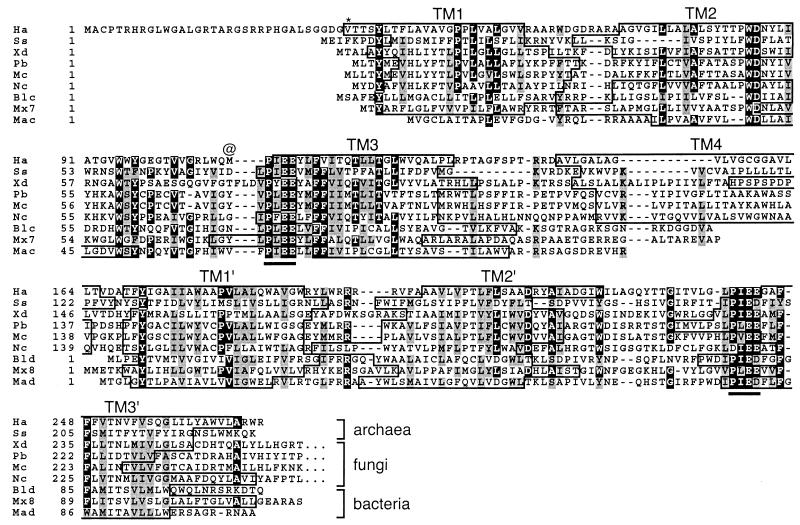
In the initial annotation of the Halobacterium sp. NRC-1 genome sequence (26), BLAST analyses (1) with the National Center for Biotechnology Information sequence database using known plant (LCYB-like or CrtL-like) and bacterial (CrtY-like) lycopene β-cyclases yielded no promising candidates. However, subsequent BLAST searches revealed a single gene encoding a putative membrane protein homologous to members of a newly discovered class of heterodimeric CrtYc and CrtYd lycopene β-cyclases in bacteria (22, 41) and bifunctional lycopene cyclase-phytoene synthases in fungi (2, 39, 40). This gene had been assigned as encoding a 162-amino-acid protein (Fig. 2, Ha, ampersand) corresponding to geranylgeranyl transferase (26), but this function now appears unlikely. Further analysis indicated that the encoded protein is longer at the N terminus than initially assigned, taking codon third-position GC bias into account (data not shown) and allowing for the use of GUG as a start codon. Two potential GUG codons are upstream of the assigned start, yielding a protein of 237 amino acids (Fig. 2, Ha, asterisk) or 270 amino acids (Fig. 2, full-length Ha protein).
FIG. 2.
Alignment of lycopene cyclases. Ha, Halobacterium sp. NRC-1 CrtY; Ss, the predicted CrtY ortholog from S. solfataricus (33); Xd, the CrtY portion of CrtYB from Xanthophyllomyces dendrorhous (40); Pb, the CarR portion of CarRA from P. blakesleeanus (2); Mc, the CarR portion of CarRP from M. circinelloides (39); Nc, the CrtY portion of the predicted CrtYB from N. crassa (32); Blc and Bld, the heterodimeric lycopene cyclase proteins CrtYc and crtYd from Brevibacterium linens (22); Mx7 and Mx8, the predicted proteins encoded by open reading frames 7 and 8 in the carotenogenic gene cluster of M. xanthus (9); Mac and Mad, CrtYc and CrtYd from M. aurum (41). Sequences of the heterodimeric proteins were artificially fused for alignment. Sequence alignment was obtained through the use of the software program ClustalW (38). Identical residues are highlighted in black and similar residues are shaded in gray. A possible alternate N terminus in Halobacterium sp. NRC-1 is denoted by an asterisk, and the initially assigned N terminus is denoted by the @ symbol. The conserved PxE(E/D) motif previously found to be required for lycopene cyclase activity (2) is underlined. The boundaries of transmembrane segments (TM) predicted by TMHMM (21) are boxed.
The predicted Halobacterium sp. NRC-1 crtY gene product aligns well with bacterial and fungal enzymes that possess lycopene cyclase activity (Fig. 2, Xd, Pb, Mc, Blc, Bld, Mac, and Mad). The similarity extends to orthologs of these proteins predicted from the nucleotide sequences of other carotenogenic organisms, including the archaeon Sulfolobus solfataricus, the fungus Neurospora crassa, and the bacterium Myxococcus xanthus (Fig. 2, Ss, Nc, Mx7, and Mx8). The bacterial proteins are thought to act as a heterodimer of two closely related monomers, CrtYc and CrtYd (22, 41). In the bifunctional CrtYB proteins of fungi, two domains homologous to the bacterial proteins are fused to form an N-terminal lycopene cyclase domain, which in turn is fused to a C-terminal phytoene synthase domain (2, 39, 40). Halobacterium sp. NRC-1 CrtY is similar to the N-terminal domain of CrtYB. The polypeptide includes two homologous domains (34% identity over 101 amino acids) related to the monomeric bacterial enzymes (Fig. 2). Each domain contains the highly conserved PXE(E/D) motif that is present in all members of this class of lycopene cyclases (Fig. 2) and is required for catalytic activity of the Phycomyces blakesleeanus enzyme (2).
The structural similarity of Halobacterium sp. NRC-1 CrtY to other lycopene cyclases is reinforced by comparing their membrane topology (Fig. 2) as computed by TMHMM (21). The bacterial CrtYcs and CrtYds are predicted to contain three transmembrane segments (Fig. 2, TM1 to -3 and TM1′ to -3′, respectively), which are also evident in the repeated domains of the fungal and Halobacterium sp. NRC-1 proteins. A fourth transmembrane helix links the repeated domains of the fungal and archaeal proteins (Fig. 2, TM4). (TM1 was not identified in the Mycobacterium aurum CrtYc sequence but was predicted when 14 additional N-terminal residues from an upstream GUG start were included [data not shown]. TM2 was not identified in the P. blakesleeanus and Mucor circinelloides proteins, but significant peaks in the TM2 region were apparent in hydropathy plots generated by TMHMM [21].)
The identification of Halobacterium sp. NRC-1 crtY is also supported by the proximity of the gene to blh, which appears to partially substitute for brp function in retinal biosynthesis during BR production (30). The crtY gene is directly upstream of blh, and their open reading frames are predicted to overlap by eight codons (data not shown), based on the putative start codon of blh (30). This arrangement raises the possibility that the genes are translationally coupled in an operon and that the gene products are involved in the same metabolic process. Thus, sequence similarity, predicted topology, and gene arrangement suggest that Halobacterium sp. NRC-1 crtY encodes a lycopene β-cyclase required for BR biogenesis.
Deletion of crtY eliminates BR expression.
Genetic analysis was required to determine whether the crtY gene encodes a functional product and whether the putative lycopene β-cyclase is required for the synthesis of the retinal cofactor of BR. We used H. salinarum as a strain for genetic analysis because all of our previous work on BR biogenesis has been performed in this strain. The strains used in our laboratory are derived from H. salinarum R1 (19), which was isolated from Halobacterium sp. NRC-1 (31). We have found very few differences between the two organisms among several genes sequenced in our laboratory, including brp, ura3, and secY, and among a subset of genes available from the two organisms at the National Center for Biotechnology Information (unpublished results). Thus, use of the Halobacterium sp. NRC-1 genome sequence for analysis of H. salinarum is valid.
To examine the role of crtY in H. salinarum, two ΔcrtY strains were constructed. MPK426 has a deletion of codons 45 to 229 of the proposed full-length open reading frame. MPK427 has a deletion of codons 121 to 229 that was designed to minimize possible polar effects on blh transcription. Such effects might occur if transcriptional signals near the 5′ end of the gene were deleted. Colonies from both ΔcrtY strains were pale red and distinct from the purple color of the wild type. This result suggested that BR production in the ΔcrtY strains was reduced due to the loss of crtY.
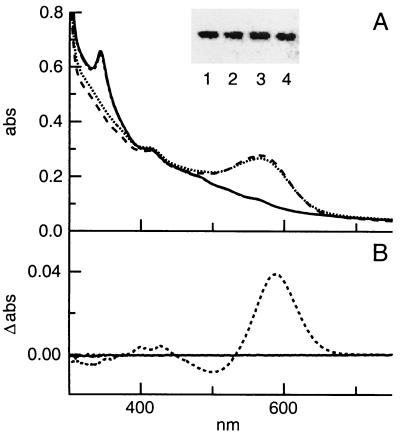
To accurately measure BR levels, cells were grown under microaerobic conditions known to induce BR synthesis (28). Cultures were grown in the dark to prevent differences in BR levels from affecting cell energy states. Cell lysates from the ΔcrtY strains lacked BR, as shown by the absence of the BR peak at 570 nm (Fig. 3A). As measured by light-dark difference spectroscopy (19), BR represented ≈4% of total cell protein in the wild-type strain but was undetectable in both ΔcrtY strains (Fig. 3B and 4). Levels of BO were only slightly reduced (Fig. 3A, insert, and 4), as determined by immunoblotting with a BO C-terminal antibody. The slight reduction in BO could not account for the complete elimination of BR synthesis in ΔcrtY strains. The absence of BR without a significant reduction in the levels of apoprotein BO suggests that the loss of crtY results in a defect in the synthesis of the BR retinal cofactor.
FIG. 3.
(A) UV-visible spectra of cell lysates from wild-type (dotted line), ΔcrtY (MPK426) (solid line), ΔcrtY (MPK427) (dashed and dotted line), and ΔcrtY ura3::crtY complementation (dashed line) strains. Cells were grown under microaerobic conditions to induce BR synthesis. Spectra of lysates were obtained at a total cell protein concentration of ≈3 mg/ml and normalized for slight differences in protein concentration. Inset, immunoblot of wild-type (lane 1), ΔcrtY (MPK426) (lane 2), ΔcrtY (MPK427) (lane 3), and ΔcrtY ura3::crtY complementation (lane 4) lysates. Total cell protein (4.0 μg) was electrophoresed and immunoblotted with the C-terminal BR antibody, BR-114. The blot was analyzed by incubating with a fluorescein-conjugated secondary antibody and scanning with a fluoroimager. (B) Light-dark difference spectroscopy of wild-type (dotted line) and ΔcrtY (MPK426) (solid line) lysates. Spectra were obtained from lysates dark-adapted for ≈12 h or light-adapted for 5 min. A light-dark difference spectrum was calculated, and the BR level was determined from the value at 587 nm. The estimated detection threshold is ≈1% of the wild-type level of BR.
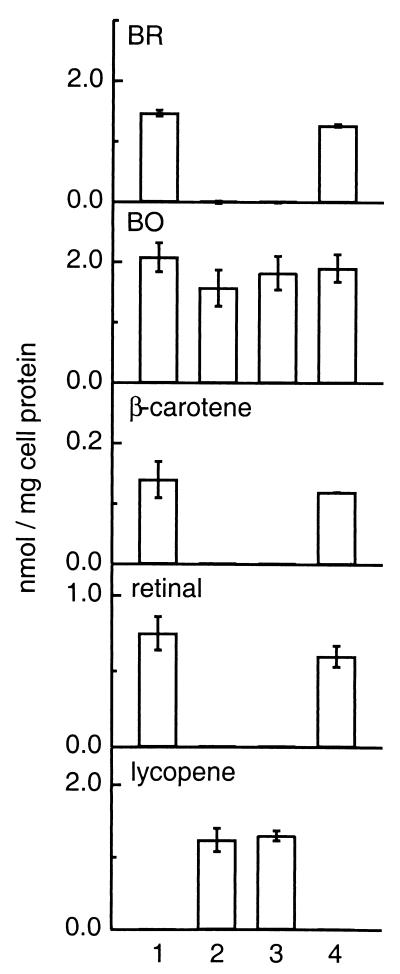
FIG. 4.
Comparison of BR, BO, β-carotene, retinal, and lycopene levels in wild-type (sample 1), ΔcrtY (MPK426) (sample 2), ΔcrtY (MPK427) (sample 3), and ΔcrtY ura3::crtY complementation (sample 4) strains. Cells were grown under microaerobic conditions to induce BR, and BR, BO, β-carotene, retinal, and lycopene levels were determined as described in Materials and Methods. All values are an average of three independent determinations and the error bars indicate 1 standard deviation.
ΔcrtY strains accumulate lycopene and do not synthesize β-carotene or retinal.
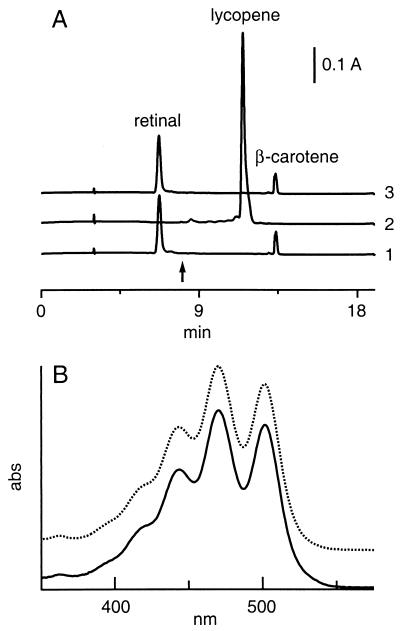
In addition to lacking a characteristic BR peak, spectra of ΔcrtY cell lysates exhibited a sharp peak at ≈340 nm that was absent from that of the wild type (Fig. 3A). A similar peak in cell lysate spectra from a Halobacterium sp. strain lacking synthesis of C50 carotenoids had been attributed to an accumulation of lycopene (37), which has an absorbance maximum near 340 nm in aqueous environments that is attributed to aggregation (10, 37). To examine the possibility that lycopene had accumulated in the ΔcrtY strains, ΔcrtY and wild-type lysates were extracted with acetone and hexane to recover carotenoids and retinal (15) and fractionated by HPLC to quantify lycopene, β-carotene, and retinal. The ΔcrtY extracts exhibited a major component eluting at 11.1 min that comigrated with commercial lycopene (Fig. 5A, trace 2). This peak was not detected in the wild-type extract (Fig. 5A, trace 1). To confirm that this major carotenoid species was lycopene, peak HPLC fractions from both ΔcrtY strains were collected and compared with commercial lycopene. The UV-visible spectra in hexane were similar, with both ΔcrtY extracts and commercial lycopene having absorption maxima at 470 nm (Fig. 5B). MALDI-TOF mass spectrometry yielded mass ion values of 536.42 and 536.43 for the ΔcrtY (MPK426) fraction and commercial lycopene, respectively. These results confirmed that lycopene is the major carotenoid accumulating in the ΔcrtY strains.
FIG. 5.
(A) Comparison of lycopene, β-carotene, and retinal accumulation in the wild-type (trace 1), ΔcrtY (MPK426) (trace 2) and ΔcrtY ura3::crtY (trace 3) strains. Carotenoids were extracted from cells as described elsewhere (15) and analyzed by reverse-phase HPLC as described in Materials and Methods. At the time indicated by the arrow, the detector was switched from 380 nm, the wavelength at which retinal was quantified, to 474 nm to quantify lycopene and β-carotene. Traces are shown normalized to total cell protein and offset on the vertical axis for clarity. (B) UV-visible spectra of lycopene obtained from ΔcrtY extract (solid line) and from a commercial source (dotted line). Total carotenoid was extracted from the ΔcrtY strain MPK426 as described previously (15). Both H. salinarum extract and commercial lycopene were purified by reverse-phase HPLC as described in Materials and Methods, dissolved in hexane, and scanned on a Perkin-Elmer λ2 spectrophotometer. The absorbance scale is arbitrary, and spectra are offset on the vertical axis for clarity.
The accumulation of lycopene in the ΔcrtY strains was accompanied by a decrease of both β-carotene and retinal to undetectable levels (Fig. 5A, trace 2, and Fig. 4, samples 2 and 3). The simplest interpretation of these results is that crtY encodes a lycopene β-cyclase that catalyzes the conversion of lycopene to β-carotene, the precursor of the retinal cofactor of BR.
ΔcrtY is complemented by an intact copy of crtY.
To demonstrate that the ΔcrtY phenotype was caused by the loss of crtY and not a polar effect on the downstream blh gene or a spurious mutation in an unknown gene, a complementation strain was constructed. The crtY open reading frame, flanked by 605 bp of upstream sequence to allow normal expression and regulation, was integrated at the ura3 locus. The resulting strain, ΔcrtY ura3::crtY, yielded purple colonies identical to those of the wild type. When grown under microaerobic conditions, BR, β-carotene, and retinal were restored to near wild-type levels and lycopene was undetectable (Fig. 3A; 4, sample 4; 5A, trace 3). As expected, BO levels were similar to those in the wild type (Fig. 3A, inset, and 4). The slight reduction in levels of BR, retinal, and β-carotene in the complementation strain may be due to a decrease in transcription or mRNA stability of crtY when expressed from a different locus. These results confirm that the ΔcrtY phenotype is due solely to the loss of crtY.
Expression of H. salinarum crtY in lycopene-producing E. coli is sufficient to yield β-carotene.
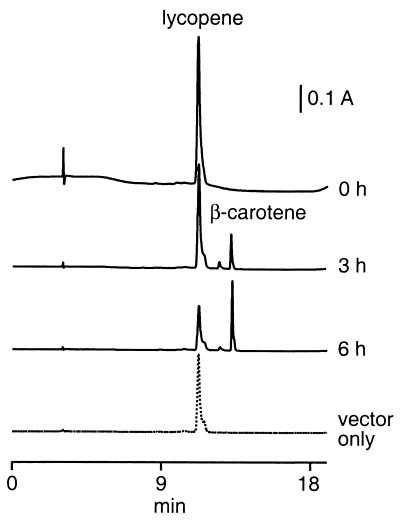
The experiments described above provide indirect evidence that the H. salinarum crtY encodes lycopene β-cyclase. To provide direct evidence that this is the case, the gene was introduced into an E. coli strain engineered to produce lycopene. The strain contains a pACYC derivative bearing carotenoid biosynthetic genes from E. uredovora. The H. salinarum crtY open reading frame was placed into a compatible pBAD expression vector to allow induction of crtY expression by arabinose. Upon induction, E. coli strains containing the crtY expression vector accumulated two carotenoid species which eluted at 12.5 and 13.2 min and were distinct from lycopene at 11.2 min (Fig. 6, 3 and 6 h traces).
FIG. 6.
Comparison of lycopene and β-carotene in lycopene-producing E. coli containing the pBAD expression vector only (dotted line) or harboring the H. salinarum crtY open reading frame (solid lines). Cells were grown to mid-log phase and expression was induced with the addition of arabinose to a concentration of 0.5%. Cells were harvested at 0, 3, or 6 h as indicated and total carotenoid was extracted and analyzed by HPLC as described in Materials and Methods. The vector-only sample was extracted 6 h after induction. Traces are shown normalized to total cell protein and offset along the vertical axis for clarity.
To confirm that the major carotenoid species was β-carotene, the E. coli samples were extracted and fractionated by HPLC. The HPLC fractions corresponding to the peak at ≈13 min contained a species with UV-visible spectral features identical to commercial β-carotene, including an absorbance maximum of 450 nm in isopropanol (data not shown). MALDI-TOF mass spectrometry revealed that the extracted species had a mass ion value of 536.42, identical to that expected for β-carotene (30). Therefore, the major species synthesized in lycopene-producing E. coli when H. salinarum crtY is expressed is β-carotene. The minor species may be γ-carotene, which results from cyclization at one end of lycopene. A minor species of similar mobility relative to lycopene and β-carotene has been confirmed to be γ-carotene in studies of other lycopene cyclases in E. coli (22, 40).
β-Carotene was not detected in E. coli prior to inducing expression of H. salinarum crtY (Fig. 6, 0 h trace), and levels of β-carotene were increased as the period of induction was prolonged (Fig. 6, 3 and 6 h traces). Moreover, a control strain containing the expression vector without the H. salinarum crtY insert did not produce β-carotene (Fig. 6, vector-only trace). These results demonstrate that conversion of lycopene to β-carotene in this strain is dependent on crtY expression and that the H. salinarum crtY encodes a functional lycopene β-cyclase.
DISCUSSION
We have identified an H. salinarum gene, crtY, that encodes a functional lycopene β-cyclase required for BR biogenesis. The function of the gene was tentatively assigned on the basis of similarities to recently identified lycopene β-cyclases found in fungi (2, 39, 40) and bacteria (22, 41). Our deletion analysis and heterologous expression studies of the gene confirmed this assignment.
Our results are consistent with a proposed carotenoid metabolic pathway in which lycopene is the immediate precursor of β-carotene, which in turn is a precursor of the retinal in BR. Several earlier experiments support this pathway. Inhibition of lycopene cyclization with nicotine leads to an increase in lycopene and a decrease in BR and retinal levels (37). β-Carotene production from lycopene is observed in cell-free lysates of Halobacterium cutirubrum, an extreme halophile closely related to H. salinarum (23). Finally, β-carotene accumulates when retinal and BR production is abolished by double deletion of brp and blh (30). Deletion of the crtY gene in the present study yielded results similar to those of the nicotine inhibition studies, except that the effects on BR and retinal levels were more pronounced. Thus, the proposed pathway for carotenoid metabolism during BR biogenesis is correct.
The step in this pathway catalyzed by CrtY may be the only means of synthesizing β-carotene in H. salinarum. First, a single crtY gene was identified in the Halobacterium sp. NRC-1 genome sequence. No orthologs of other lycopene cyclase genes were found. Second, deletion of the gene reduced the levels of β-carotene and BR to below detectable limits, indicating a complete block in β-carotene production. Third, the crtY deletion probably affects the synthesis of rhodopsins other than BR. In strains derived from H. salinarum S9, the levels of halorhodopsin and sensory rhodopsin I are ≈9% and ≈3% that of BR, respectively (8), and the level of sensory rhodopsin II is at least threefold lower than that of sensory rhodopsin I (34). Our strains lack the regulatory mutation in S9 (3) that leads to overproduction of BR, and possibly halorhodopsin and sensory rhodopsin I, but the ratio of these proteins is probably similar. If an alternative pathway existed to produce β-carotene for halorhodopsin , we would have detected a small retinal peak in the HPLC analysis of the ΔcrtY extracts, which was not observed (Fig. 5A). Thus, it seems likely that the same pathway is used to synthesize β-carotene for both BR and halorhodopsin. We cannot exclude the possibility that the sensory rhodopsins use an alternative carotenoid metabolic pathway, since the very low amount of retinal from these proteins may have been undetected by our HPLC analysis.
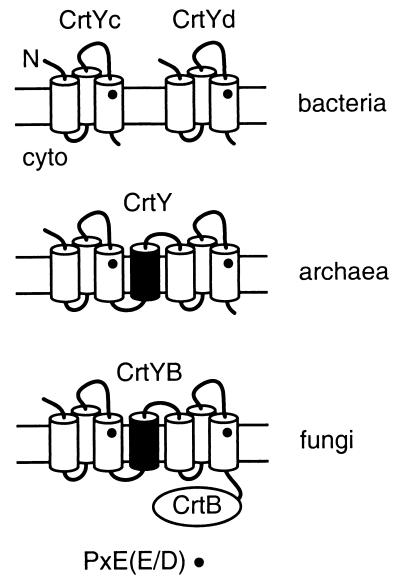
Comparative sequence analysis and topology predictions provide a model for lycopene cyclase evolution. Evolutionary relationships among the bacterial and fungal proteins have been noted previously (2, 22, 40). The bacterial crtYc and crtYd genes may have arisen from duplication of a gene that encoded a homodimeric lycopene cyclase. Our topological analysis indicates that each monomer of the bacterial enzymes contains three transmembrane segments and has the same N-out-C-in orientation (Fig. 7). The conserved, potentially catalytic motif PXE(E/D) is thereby situated toward the extracytoplasmic face of the membrane in each monomer (Fig. 7). Archaeal CrtY proteins found in H. salinarum and predicted in S. solfataricus contain two domains similar to the bacterial proteins linked by an additional transmembrane segment that allows the topology of each domain to be retained (Fig. 7). The fungal crtYB gene appears to have arisen from fusion of an archaeal-like gene with a phytoene synthase (crtB) gene. The predicted membrane topology of fungal CrtYB is identical to that of the archaeal proteins and results in a cytosolic location for the phytoene synthase domain (Fig. 7), which is expected since the CrtB substrate geranylgeranyl pyrophosphate is generated in the cytosol. Thus, carotenogenic archaea encode a lycopene cyclase that has a structural organization intermediate between that of bacteria and fungi. Further studies may reveal whether the relationship among members of this class of cyclases arose from lateral gene transfer or evolution from a common ancestor.
FIG. 7.
Model of CrtY topology and evolutionary relatedness. Transmembrane helices are shown as cylinders and the lipid bilayer is indicated by parallel lines. The conserved catalytic motif PXE(E/D) is denoted by the dot (•). CrtB indicates the phytoene synthase domain of the fungal protein.
H. salinarum CrtY and the related lycopene cyclases in bacteria and fungi may play a unique role in carotenoid metabolism. CrtY functions in the biogenesis of known integral membrane proteins, including BR and possibly other H. salinarum rhodopsins. Like CrtY, the other proteins implicated in H. salinarum BR biogenesis, including Brp and Blh, are predicted to be integral membrane proteins. Preliminary immunoblotting experiments of fractionated cells suggest that Brp is indeed localized to the H. salinarum membrane (M. P. Krebs, unpublished results). We propose that H. salinarum CrtY and related proteins are dedicated to producing β-carotene for use by other integral membrane proteins, such as Brp and Blh. Direct interaction of these proteins within the membrane may influence carotenoid and retinal metabolism during BR biogenesis in H. salinarum, which is known to be regulated (15, 36). Further experiments are in progress to test this model.
Acknowledgments
We thank T. Bergsbaken for technical assistance and C. Echavarri-Erasun for help with HPLC.
This research was supported by National Science Foundation grant MCB-9983120 to M.P.K.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrach, N., R. Fernández-Martin, E. Cerdá-Olmedo, and J. Avalos. 2001. A single gene for lycopene cyclase, phytoene synthase, and regulation of carotene biosynthesis in Phycomyces. Proc. Natl. Acad. Sci. USA 98:1687-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baliga, N. S., S. P. Kennedy, W. V. Ng, L. Hood, and S. DasSarma. 2001. Genomic and genetic dissection of an archaeal regulon. Proc. Natl. Acad. Sci. USA 98:2521-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Béjà, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 5.Béjà, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 6.Betlach, M., J. Friedman, H. W. Boyer, and F. Pfeifer. 1984. Characterization of a halobacterial gene affecting bacterio-opsin gene expression. Nucleic Acids Res. 12:7949-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieszke, J. A., E. N. Spudich, K. L. Scott, K. A. Borkovich, and J. L. Spudich. 1999. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry 38:14138-14145. [DOI] [PubMed] [Google Scholar]
- 8.Bogomolni, R. A., and J. L. Spudich. 1982. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc. Natl. Acad. Sci. USA 79:6250-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botella, J. A., F. J. Murillo, and R. Ruiz-Vazquez. 1995. A cluster of structural and regulatory genes for light-induced carotenogenesis in Myxococcus xanthus. Eur. J. Biochem. 233:238-248. [DOI] [PubMed] [Google Scholar]
- 10.Britton, G. 1985. General carotenoid methods. Methods Enzymol. 111:113-149. [DOI] [PubMed] [Google Scholar]
- 11.Cline, S. W., W. L. Lam, R. L. Charlebois, L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35:148-152. [DOI] [PubMed] [Google Scholar]
- 12.Dale, H., C. M. Angevine, and M. P. Krebs. 2000. Translocation order of the extracellular domains of bacterioopsin in vivo. Proc. Natl. Acad. Sci. USA 97:7847-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dale, H., and M. P. Krebs. 1999. Membrane insertion kinetics of a protein domain in vivo: the bacterioopsin N terminus inserts co-translationally. J. Biol. Chem. 274:22693-22698. [DOI] [PubMed] [Google Scholar]
- 14.DasSarma, S., U. L. RajBhandary, and H. G. Khorana. 1984. Bacterio-opsin mRNA in wild-type and bacterio-opsin-deficient Halobacterium halobium strains. Proc. Natl. Acad. Sci. USA 81:125-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshpande, A., and S. Sonar. 1999. Bacterioopsin-triggered retinal biosynthesis is inhibited by bacteriorhodopsin formation in Halobacterium salinarium. J. Biol. Chem. 274:23535-23540. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, M. L., and M. L. Dyall-Smith. 1990. A plasmid vector with a selectable marker for halophilic archaebacteria. J. Bacteriol. 172:756-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isenbarger, T. A., and M. P. Krebs. 1999. Role of helix-helix interactions in assembly of the bacteriorhodopsin lattice. Biochemistry 38:9023-9030. [DOI] [PubMed] [Google Scholar]
- 18.Isenbarger, T. A., and M. P. Krebs. 2001. Thermodynamic stability of the bacteriorhodopsin lattice as measured by lipid dilution. Biochemistry 40:11923-11931. [DOI] [PubMed] [Google Scholar]
- 19.Krebs, M. P., T. Hauss, M. P. Heyn, U. L. RajBhandary, and H. G. Khorana. 1991. Expression of the bacterioopsin gene in Halobacterium halobium using a multicopy plasmid. Proc. Natl. Acad. Sci. USA 88:859-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krebs, M. P., and T. A. Isenbarger. 2000. Structural determinants of purple membrane assembly. Biochim. Biophys. Acta 1460:15-26. [DOI] [PubMed] [Google Scholar]
- 21.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 22.Krubasik, P., and G. Sandmann. 2000. A carotenogenic gene cluster from Brevibacterium linens with novel lycopene cyclase genes involved in the synthesis of aromatic carotenoids. Mol. Gen. Genet. 263:423-432. [DOI] [PubMed] [Google Scholar]
- 23.Kushwaha, S. C., M. Kates, and J. W. Porter. 1976. Enzymatic synthesis of C40 carotenes by cell-free preparation from Halobacterium cutirubrum. Can. J. Biochem. 54:816-823. [DOI] [PubMed] [Google Scholar]
- 24.Lam, W. L., and W. F. Doolittle. 1989. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc. Natl. Acad. Sci. USA 86:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misawa, N., S. Kajiwara, K. Kondo, A. Yokoyama, Y. Satomi, T. Saito, W. Miki, and T. Ohtani. 1995. Canthaxanthin biosynthesis by the conversion of methylene to keto groups in a hydrocarbon β-carotene by a single gene. Biochem. Biophys. Res. Commun. 209:867-876. [DOI] [PubMed] [Google Scholar]
- 26.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oesterhelt, D. 1998. The structure and mechanism of the family of retinal proteins from halophilic archaea. Curr. Opin. Struct. Biol. 8:489-500. [DOI] [PubMed] [Google Scholar]
- 28.Oesterhelt, D., and W. Stoeckenius. 1974. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 31:667-678. [DOI] [PubMed] [Google Scholar]
- 29.Peck, R. F., S. DasSarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 30.Peck, R. F., C. Echavarri-Erasun, E. A. Johnson, W. V. Ng, S. P. Kennedy, L. Hood, S. DasSarma, and M. P. Krebs. 2001. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J. Biol. Chem. 276:5739-5744. [DOI] [PubMed] [Google Scholar]
- 31.Sapienza, C., and W. F. Doolittle. 1982. Unusual physical organization of the Halobacterium genome. Nature 295:384-389. [DOI] [PubMed] [Google Scholar]
- 32.Schmidhauser, T. J., F. R. Lauter, M. Schumacher, W. Zhou, V. E. Russo, and C. Yanofsky. 1994. Characterization of al-2, the phytoene synthase gene of Neurospora crassa. Cloning, sequence analysis, and photoregulation. J. Biol. Chem. 269:12060-12066. [PubMed] [Google Scholar]
- 33.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spudich, E. N., S. A. Sundberg, D. Manor, and J. L. Spudich. 1986. Properties of a second sensory receptor protein in Halobacterium halobium phototaxis. Proteins 1:239-246. [DOI] [PubMed] [Google Scholar]
- 35.Spudich, J. L., C.-S. Yang, K.-H. Jung, and E. N. Spudich. 2000. Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16:365-392. [DOI] [PubMed] [Google Scholar]
- 36.Sumper, M., and G. Herrmann. 1976. Biosynthesis of purple membrane: control of retinal synthesis by bacterio-opsin. FEBS Lett. 71:333-336. [DOI] [PubMed] [Google Scholar]
- 37.Sumper, M., H. Reitmeier, and D. Oesterhelt. 1976. Biosynthesis of the purple membrane of halobacteria. Angew Chem. Int. 15:187-194. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velayos, A., A. P. Eslava, and E. A. Iturriaga. 2000. A bifunctional enzyme with lycopene cyclase and phytoene synthase activities is encoded by the carRP gene of Mucor circinelloides. Eur. J. Biochem. 267:5509-5519. [DOI] [PubMed] [Google Scholar]
- 40.Verdoes, J. C., K. P. Krubasik, G. Sandmann, and A. J. van Ooyen. 1999. Isolation and functional characterisation of a novel type of carotenoid biosynthetic gene from Xanthophyllomyces dendrorhous. Mol. Gen. Genet. 262:453-461. [DOI] [PubMed] [Google Scholar]
- 41.Viveiros, M., P. Krubasik, G. Sandmann, and M. Houssaini-Iraqui. 2000. Structural and functional analysis of the gene cluster encoding carotenoid biosynthesis in Mycobacterium aurum A+. FEMS Microbiol. Lett. 187:95-101. [DOI] [PubMed] [Google Scholar]