Abstract
The MinC protein is an important determinant of septal ring positioning in Escherichia coli. The N-terminal domain (ZMinC) suppresses septal ring formation by interfering with FtsZ polymerization, whereas the C-terminal domain (DMinC) is required for dimerization as well as for interaction with the MinD protein. MinD oscillates between the membrane of both cell halves in a MinE-dependent fashion. MinC oscillates along with MinD such that the time-integrated concentration of ZMinC at the membrane is minimal, and hence the stability of FtsZ polymers is maximal, at the cell center. MinC is cytoplasmic and fails to block FtsZ assembly in the absence of MinD, indicating that recruitment of MinC by MinD to the membrane enhances ZMinC function. Here, we present evidence that the binding of DMinC to MinD endows the MinC/MinD complex with a more specific affinity for a septal ring-associated target in vivo. Thus, MinD does not merely attract MinC to the membrane but also aids MinC in specifically binding to, or in close proximity to, the substrate of its ZMinC domain. MinC-mediated division inhibition can also be activated in a MinD-independent fashion by the DicB protein of cryptic prophage Kim. DicB shows little homology to MinD, and how it stimulates MinC function has been unclear. Similar to the results obtained with MinD, we find that DicB interacts directly with DMinC, that the DMinC/DicB complex has a high affinity for some septal ring target(s), and that MinC/DicB interferes with the assembly and/or integrity of FtsZ rings in vivo. The results suggest a multistep mechanism for the activation of MinC-mediated division inhibition by either MinD or DicB and further expand the number of properties that can be ascribed to the Min proteins.
Assembly of the FtsZ ring (5) is the first visible event in formation of the septal ring organelle, which mediates cell constriction during cytokinesis in prokaryotes and eukaryotic organelles of prokaryotic origin (17, 33, 37, 38, 44, 49). The plane of cell division is determined by the plane of the initial Z ring (2). Therefore, a proper definition of the site of initial FtsZ assembly is crucial for the proper distribution of mother cell components to her progeny.
The MinC protein is an important negative regulator of FtsZ assembly in Escherichia coli (4, 10, 11, 29, 30). The peptide consists of 231 residues which fold into two domains of roughly equal sizes. The N-terminal domain (Z domain, designated here ZMinC) is responsible for inhibition of FtsZ assembly, whereas the C-terminal domain (D domain, DMinC) is responsible for both homodimerization of MinC and binding MinD (8, 26, 53). The effect of MinC on cell division is determined by its cellular location, which, in turn, is determined by the activities of the MinD and MinE proteins (28, 46).
MinD is an ATPase that associates with the membrane in a peripheral manner (9, 27, 48, 50). The protein directly binds MinC and MinE (30) and recruits both to the membrane (28, 46, 47). In the absence of MinD, overexpression of MinC (over 25-fold) is sufficient to block cell division (13). However, MinD is required for MinC function when the division inhibitor is present at physiological levels in the cell (10, 11, 13).
The MinE protein imparts topological specificity to MinC/MinD such that FtsZ assembly is not blocked at the normal division site at midcell. In the absence of MinE, MinC/MinD is found over the entire membrane, Z-ring assembly is blocked at all membrane sites, and cells form long nonseptate filaments (11, 28, 46, 48, 50). In wild-type cells, however, MinC/MinD action at midcell is prevented by MinE in an interesting way. MinE causes MinD to undergo a pole-to-pole oscillatory localization cycle during which the protein alternately accumulates on the membrane of one cell half every other 25 s or so (15, 22, 27, 48, 50). MinC is not required for oscillation (48) but oscillates along with MinD (28, 46). As a result, the concentration of membrane-associated MinC/MinD over time is maximal at the cell poles and minimal at the cell center. We proposed that it is this time-integrated concentration differential of MinC which limits stable assemblies of FtsZ to the middle of the cell (22, 46). The feasibility of such a mechanism is strongly supported by computer simulations of MinCDE and FtsZ dynamics, which closely resemble the behavior of these proteins observed in vivo (25, 41).
In addition to its normal role in division site placement, MinC is also involved in the division block which occurs upon induction of dicB. This gene is part of the dic operon, which resides on cryptic prophage Kim (Qin) (6). Under normal conditions, expression of dicB is actively repressed (3). When expression is induced, however, cell division rapidly ceases (34), and this division block is dependent on MinC (10, 35). In contrast to MinC/MinD-mediated division inhibition, MinC/DicB-mediated division inhibition does not require MinD and is resistant to suppression by MinE. Both MinC-dependent division blocks can be suppressed by overexpression of FtsZ, however (10, 35). These results indicate that, although MinD (30 kDa) and DicB (7 kDa) have little sequence homology, either protein can independently activate MinC-mediated division inhibition (10, 35). Because MinD recruits MinC to the membrane, we and others proposed that MinD might activate MinC simply by causing the division inhibitor to accumulate at the membrane to a local concentration that is sufficiently high for blocking FtsZ assembly (28, 46). How DicB activates MinC function has not yet been addressed.
In this paper we investigate the mechanism(s) whereby MinD and DicB stimulate MinC-dependent division inhibition. We find that, like MinD, DicB interacts directly with the C-terminal D domain of MinC and that MinC/DicB blocks cell division by preventing the formation of stable FtsZ rings. Using MinC derivatives which lack a functional Z domain, we further show that complexes between DMinC and either DicB or MinD have a high affinity for one or more septal ring components in vivo. The results suggest that both MinD and DicB activate MinC function by directing its D domain to, or close to, the substrate of its Z domain. For MinD, the bulk recruitment of MinC to the membrane is likely to contribute to its MinC-activating properties, and this step may normally precede the more specific targeting of MinC/MinD to membrane-associated septal ring components. For DicB, we find no evidence for binding of MinC/DicB to the membrane per se, suggesting that DicB stimulates MinC-mediated division inhibition primarily by directly targeting the inhibitor to its substrate.
MATERIALS AND METHODS
E. coli strains, phages, and plasmids.
Relevant E. coli strains, plasmids, and phages used in this study are listed in Table 1. Strain LL1 was obtained by P1-mediated transduction of lon::Tn10 from RC7 to PB114.
TABLE 1.
E. coli strains, phages, and plasmids used in this study
| Strain, phage, or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| E. coli strains | ||
| PB103 | dadR trpE trpA tna | 12 |
| PB147 | PB103, ΔminDE | 22 |
| PB114 | PB103, ΔminCDE::aph | 11 |
| RC7 | lon::Tn10 | 51 |
| LL1 | PB114, lon::Tn10 | This work |
| Phages | ||
| λDB175 | imm21bla+lacIq+ Plac::minDE | 11 |
| λDR155 | imm21bla+lacIq+ Plac::minD | 46 |
| λDR119 | imm21bla+lacIq+ Plac::gfp-t-minD | 48 |
| λDR120 | imm21bla+lacIq+ Plac::gfp-t-ftsZ | 21 |
| λDB182 | imm21bla+lacIq+ Plac::dicB | 10 |
| λJE39 | imm21bla+lacIq+ Plac::gfp-t-dicB | This work |
| Plasmids | ||
| pJE44 | cat+araC+ PBAD::dicB | This work |
| pJE75 | cat+araC+ PBAD::minE | This work |
| pJE80 | cat+araC+ PBAD::sfiA | This work |
| pLL18 | aadA+cl857Ts PλR::gfp-t-minC(5-231) | This work |
| pLL13 | aadA+cl857Ts PλR::gfp-t-minC(14-231) | This work |
| pJE46 | aadA+cl857Ts PλR::t-minC(14-231) | This work |
| pPC105 | aadA+cl857Ts PλR::gfp-t-minC(108-231) | This work |
| pLL14 | aadA+cl857Ts PλR::gfp-t-minC(141-231) | This work |
| pJE78 | aadA+cl857Ts PλR::gfp-t-minC(108-231)-h | This work |
| pJE79 | aadA+cl857Ts PλR::gfp-t-minC(108-208)-h | This work |
The positions of in-frame GFP (gfp), linker peptide (t), and His tag peptide (h) coding sequences, when these sequences are present in the genetic construct, are indicated.
Phage λJE39 was obtained after crossing λNT5 with plasmid pJE39 (see below), as described previously (11).
Plasmids pBAD33 (20), pDB182 (10), pDR120 (21), and pDR175 (46) were described previously. Vectors pUC18 (New England Biolabs), pBluescript KS (Stratagene), and pET21a to -d (Novagen) were obtained from commercial sources. Unless specified otherwise, PCRs were performed as described previously (23). PCR-derived portions of plasmid inserts were sequenced as described previously (23).
To create pJE39, we performed a PCR with primers 5′-GACGGATCCATGAAAACGTTATTACCAAACG-3′ and 5′-AATGGTCGACTCATTGTGAACATCCTTTTGG-3′. The fragment was treated with BamHI and SalI (sites underlined) and the 195-bp fragment was used to replace the 1,163-bp BamHI-SalI fragment of pDR120. This step yielded a plasmid (pDR148) encoding green fluorescent protein (GFP) fused to the product of a functional but mutant (G18C, A61S) allele of dicB. The product of a second PCR (using primers 5′-GACGGATCCATGAAAACGTTATTACCAAACG-3′ and 5′-AATGGTCGACTTATTCTGCACATCCTTTTGGCATC-3′ and pDB182 as the template) was next used to replace the mutant with the wild-type version of dicB, resulting in pJE39.
Plasmid pJE44 was constructed in two steps. First, plasmid pDB182 was digested with EcoRI and PstI, yielding a 252-bp fragment, which was ligated to similarly treated pBluescript KS. The 292-bp XbaI-HindIII dicB fragment of the resulting plasmid (pJE43) was next inserted into pBAD33, placing dicB downstream of PBAD.
For plasmid pJE75, a 400-bp HindIII minE fragment from plasmid pJE69 (to be described in detail elsewhere) was inserted into pBAD33, placing minE downstream of PBAD.
For pJE80, we performed a PCR with primers 5′-GCTGGGATCCATATGTACACTTCAGGCTATG-3′ and 5′-TAGTGTCGACTGAAAGCATTGGCTG-3′ to amplify sfiA. The product was treated with NdeI and SalI (sites underlined), and the 565-bp fragment was inserted into pET21b, yielding pDB276. The 611-bp XbaI-HindIII fragment of the latter was then inserted into pBAD33, placing sfiA downstream of PBAD.
The remaining plasmids listed in Table 1 are derivatives of pSC101 derivative pDR175 (46). Except for pJE46, which lacks gfp, all encode fusion proteins in which Gfpmut2 (GFP) is linked, via the T7.tag peptide (T; Novagen) to the N termini of various portions of MinC. As indicated in Table 1, expression of these fusions is under the control of PλR and cI857.
Creation of these plasmids involved the use of two other new constructs: pDB411, a pUC18/pET21b hybrid vector, and pJE13, which was used as template in PCR described below.
To obtain pDB411, pUC18 DNA was treated with EcoRI and XbaI, followed by incubation with deoxynucleoside triphosphates and Klenow enzyme to produce flush ends. Recircularization of the large fragment resulted in a pUC18 derivative (pDB410) in which EcoRI and XbaI sites were regenerated but in which a number of other restriction sites were deleted. Ligation of the 769-bp AlwNI-XbaI fragment of pDB410 to the 2,144-bp AlwNI -XbaI fragment of pET21b yielded pDB411.
Plasmid pJE13 encodes a fusion protein [STII-T-MinC(5-231)] in which the StreptagII (STII; Genosys) and T7.tag epitopes replace the first four residues of MinC. Oligonucleotides 5′-CATGGCTAGCTGGAGCCACCCGCAGTTCGAAAAAGGCG-3′ (sense) and 5′-AATTCGCCTTTTTCGAACTGCGGGTGGCTCCAGCTAGC-3′ (antisense) were annealed, resulting in a fragment encoding the STII epitope with NcoI- and EcoRI-compatible overhangs. This fragment was inserted into a pET21a-derived minD construct (pDR13), yielding pDB389, which encodes STII-MinD. pJE13 was obtained by replacing the 821-bp NdeI-HindIII minD fragment of pDB389 with an 885-bp NdeI-HindIII t-minC (5-231) fragment from another pET21a-derived construct (pJE3), encoding a His10-T-MinC(5-231) fusion.
For plasmid pLL13 we used primers 5′-CAGTGGATCCTGACTTTATCTGTGGTTCATCTGCATGAG-3′ and 5′-TAGGTCGACTTAATTTAACGGTTGAACGGTCAAAGCG-3′ in a PCR to amplify a fragment encoding amino acids (aa) 14 to 231 of MinC, flanked by BamHI and SalI sites (underlined). After digestion with BamHI and SalI, the 678-bp fragment was ligated to the large BamHI-SalI fragment of pJE13, producing plasmid pPC103 [PT7::stII-t-minC (14-231)], which encodes STII-T-MinC(14-231) under the control of the T7 promoter. The 696-bp NheI-SalI fragment from pPC103 was used to replace the 874-bp NheI-SalI fragment of pDR175 (46). The resulting plasmid, pLL13, encodes the fusion protein GFP-T-MinC(14-231).
Plasmid pLL18 was obtained by replacing the 921-bp NcoI-MluI fragment of pLL13 with the 948-bp NcoI-MluI fragment of pDR175.
For plasmid pPC105 we performed a PCR with primers 5′-TAGCGGATCCCAGCTCCCACACCGCAGGCTCCAGCG-3′ and 5′-TAGGTCGACTTAATTTAACGGTTGAACGGTCAAAGCG-3′. After digestion with BamHI and SalI (sites underlined) the resulting 379-bp fragment was ligated to the large BamHI-SalI fragment of pJE13, yielding plasmid pPC102 [PT7::stII-t-minC (108-231)], which encodes STII-T-MinC(108-231) under the control of the T7 promoter. The 411-bp NheI-SalI fragment of pPC102 was used to replace the 874-bp NheI-SalI fragment of pDR175 (46), producing plasmid pPC105 [PλR::gfp-t-minC (108-231)].
To construct pLL14, a PCR fragment was generated with primers 5′-ATCCGGATCCCACAATGTGATCTGATTGTTACAAGC-3′ and 5′-TAGGTCGACTTAATTTAACGGTTGAACGGTCAAAGCG-3′. Upon digestion of the product with BamHI and SalI (sites underlined), the resulting 287-bp fragment was used to replace the 665-bp BamHI-SalI fragment of pPC103, resulting in pLL3 [PT7::stII-t-minC (141-231)]. The 874-bp NheI-SalI fragment of pDR175 (46) was next replaced with the 312-bp NheI-SalI fragment of pLL3, yielding pLL14 [PλR::gfp-t-minC (141-231)].
Construction of plasmid pJE46 took several steps. The 969-bp XbaI-HindIII fragment of pJE13 and the 791-bp XbaI-HindIII fragment of pPC103 were inserted separately into pDB411, resulting in pLL1 and pLL2 encoding STII-T-MinC(5-231) and STII-T-MinC(14-231), respectively, under the control of the tac promoter. To obtain a version of pLL1 without any remaining minD sequences, the 449-bp MluI-NcoI fragment from pLL1 was used to replace the 422-bp MluI-NcoI fragment of pLL2. The STII tag was removed from the resulting plasmid (pLL11) by recircularization of the large fragment after treatment with NheI, yielding pLL12. The 767-bp XbaI-SalI fragment of pLL12 was then used to replace the 1,463-bp XbaI-SalI fragment of pLL18, thereby removing gfp. Finally, the 723-bp NheI-SalI fragment of the resulting plasmid (pJE45) was replaced with the 696-bp NheI-SalI fragment of pLL2, resulting in pJE46, which encodes T7.tag-MinC(14-231).
For pJE78, we performed a PCR with primers 5′-CATGGCTAGCTGGAGCCACCCGCAGTTCGAAAAAGGCG-3′ and 5′-TAGGTCCTCGAGATTTAACGGTTGAACGGTCAAAGCG-3′. The fragment was digested with BamHI (internal site) and XhoI (site underlined), yielding a 689-bp fragment, which was used to replace the 686-bp BamHI-XhoI fragment of pPC103 to produce pLL6. The 809-bp XbaI-XhoI fragment of pLL6 was inserted into pDB411, yielding pLL8, which encodes an STII-T-MinC(5-231)-H6 fusion under control of Ptac. pLL8 and pLL18 were digested with StyI and SalI, respectively, treated with Klenow enzyme and deoxynucleoside triphosphates to generate flush ends, and then digested with AgeI. The 584-bp fragment of pLL8 was then ligated to the large fragment of pLL18, resulting in pLL19. Finally, the 948-bp NcoI-MluI fragment of pLL19 was replaced with the 636-bp NcoI-MluI fragment from pPC105. Plasmid pJE78 encodes the fusion GFP-T-MinC(108-231)-H6.
To obtain pJE79, we performed a PCR with primers 5′-CATGGCTAGCTGGAGCCACCCGCAGTTCGAAAAAGGTCA-3′ and 5′-GGTATCCTCGAGTGGGATTTGATCACTCAGCCAGTATTCAC-3′. The product was treated with NheI and XhoI (sites underlined), and the resulting 651-bp fragment was used to replace the 720-bp NheI-XhoI fragment of pLL19, yielding pLL21. Last, the 948-bp NcoI-MluI fragment of pLL21 was replaced with the 636-bp NcoI-MluI fragment from pPC105. Plasmid pJE79 encodes the fusion GFP-T-MinC(108-208)-H6.
E. coli growth conditions.
To assess the effect of MinC/DicB on Z-ring assembly, strains PB147(λDR120)/pJE44 [ΔminDE (Plac::gfp-ftsZ)/PBAD::dicB] and PB147(λDR120)/pBAD33 [ΔminDE (Plac::gfp-ftsZ)/vector] were grown at 37°C in Luria-Bertani medium containing 25 μg of chloramphenicol/ml and 37 μM isopropyl β-d-thiogalactopyranoside (IPTG). At an optical density at 600 nm (OD600) of 0.1 to 0.2, arabinose was added to a final concentration of 0.05%, and growth was allowed to continue. Aliquots for microscopic analyses were removed from the culture at 60-min intervals.
All other strains were grown at 30 or 37°C in M9 minimal salts medium supplemented with 50 μg of tryptophan/ml, 0.2% Casamino Acids, 0.2% maltose, and IPTG and/or arabinose. Cultures to be analyzed by microscopy were typically grown to an OD600 of 0.1 to 0.4.
Yeast strains and two-hybrid plasmids.
Strain PJ69-4A (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) has been described previously (31).
Strain SL3004 (MATα trp1-901 leu2 ura3 his3 gal4 gal80 lys2-801 ade2-101) will be described elsewhere (K. Henry and S. Lemmon, unpublished data).
The two-hybrid plasmids listed in Table 3 are derivatives of the pGBDU-C(X) (URA3+) and pGAD-C(X) (LEU2+) vector series (31) and encode fusions of the Gal4 DNA binding domain (BD; pGBDU derivatives) or Gal4 activation domain (AD; pGAD derivatives) to the N terminus of MinC, MinD, or DicB. Transcription of the fusion in yeast is under the control of the ADH1 promoter in each case.
TABLE 3.
Yeast two-hybrid interactionsa
| Plasmids (BD/AD) | Protein fused to:
|
Activity (U) | |
|---|---|---|---|
| BD | AD | ||
| pJE30/pJE10 | MinC | MinC | 7 |
| pJE9/pJE10 | MinD | MinC | 19 |
| pJE55/pJE10 | DicB | MinC | 132 |
| pJE30/pJE54 | MinC | DicB | 113 |
| pJE9/pJE54 | MinD | DicB | 2 |
| pJE55/pJE54 | DicB | DicB | 4 |
| pJE9/pJE8 | MinD | MinD | 64 |
| pJE30/pGAD-C2 | MinC | —b | 4 |
| pJE9/pGAD-C2 | MinD | — | 3 |
| pJE55/pGAD-C2 | DicB | — | 2 |
LBD and AD indicate fusions to the yeast Ga14 binding domain and activating domain, respectively.
—, unfused AD.
Plasmids pJE8 and pJE9 were obtained by inserting the 827-bp BamHI-SalI minD fragment of pDR119 (Plac::gfp-minD) (48) into the multiple cloning sites (MCS) of pGAD-C2 and pGBDU-C2, respectively.
Plasmid pJE10 was made in two steps. First, primers 5′-AGGGGATCCAACACGCCAATCGAGCTTAAAGGG-3′ and 5′-TAGGTCGACTTAATTTAACGGTTGAACGGTCAAAGCG-3′ were used to generate a PCR fragment encoding amino acids 5 to 231 of MinC, flanked by BamHI and SalI sites (underlined). After digestion with BamHI and SalI, the 696-bp fragment was used to replace the 1,163-bp BamHI-SalI fragment of pDR120 (21). The same 696-bp fragment was next isolated from the resulting plasmid (pDB379) and inserted into the MCS of pGAD-C1.
For pJE30, the 708-bp EcoRI-SalI fragment of pJE10 was subcloned into the MCS of pGBDU-C1.
To generate pJE54 and pJE55, the 195-bp BamHI-SalI dicB fragment from pJE39 (see above) was inserted into the MCS of pGAD-C1 and pGBDU-C1, respectively.
Yeast methods and two-hybrid β-galactosidase assays.
Plasmids encoding BD fusions (pGBDU derivatives) were introduced into strain PJ69-4A, and those encoding AD fusions (pGAD derivatives) were introduced into SL3004 by the lithium acetate transformation method (16). Appropriate pairs of transformants were mated, and diploids were selected by growth on plates containing synthetic complete medium (19) lacking leucine and uracil (C-Leu-Ura).
Diploids were grown at 30°C to mid-log phase (OD600 = 0.7 to 1.0) in liquid C-Leu-Ura, and β-galactosidase activities were measured by using the yeast β-galactosidase assay kit essentially as recommended by the supplier (Pierce). Briefly, reactions were started by addition of 350 μl of cell culture to 350 μl of a solution containing equal parts of Y-PER reagent and 2× β-galactosidase assay buffer and mixtures were incubated at 37°C. Reactions were stopped by addition of 300 μl of stop solution (1 M Na2CO3), followed by 15 s of vortexing. Cell debris was removed by centrifugation at 14,000 × g for 1 min, and the absorbances (420 nm) of supernatants relative to that of the supernatant of a control reaction performed with cell-free culture medium were determined. The number of units were determined by the ratio (1,000 × A420)/(t × 0.35 × OD600), where t is the reaction time in minutes.
Microscopy and other methods.
Live cells were examined for most experiments. Sometimes, however, cells were chemically fixed as described previously (1), except that cells were incubated in the presence of formaldehyde, glutaraldehyde, and sodium phosphate buffer (pH 7.5) at final concentrations of, respectively, 2.40%, 0.04%, and 30 mM for cells grown in Luria-Bertani medium or 0.70%, 0.01%, and 9 mM for cells grown in M9-based medium. Immunofluorescence with anti-FtsZ antibodies was performed as previously described (21), except that PB114(λDB182)/pLL13 and PB114(λDR155)/pLL13 cells were grown to an OD600 of 0.4 to 0.7 at 37°C in M9 minimal salts medium supplemented with 50 μg of tryptophan/ml, 0.2% Casamino Acids, 0.2% maltose, and 100 μM IPTG and then fixed as described above. The procedure preserved a level of GFP-MinC(14-231) signal sufficient for imaging directly. For fluorescence and differential interference contrast (DIC) imaging, cells were applied to a microscope slide and viewed with a Zeiss Axioplan-2 microscope outfitted with a Hamamatsu C4742-95 progressive scan cooled charge-coupled device camera and a plan-NEOFLUAR (100×, numerical aperture = 1.3) oil immersion objective, by using Cy3-specific (580-nm dichroic mirror, 510- to 560-nm excitation filter, and 590-nm barrier filter) and/or GFP-specific (495-nm dichroic mirror, 450- to 490-nm excitation filter, and 500- to 550-nm barrier filter) filter sets for fluorescent images and Nomarski optics for DIC images. Images were captured with QED software and were further manipulated with Adobe Photoshop.
Cell lengths and positions of septa and fluorescent rings were measured by using Object Image, version 1.6, software (55). Length-to-ring (L/R) ratios were determined as described previously (21).
Western analyses were performed essentially as described previously (13) with the following modifications. Nitrocellulose blots were incubated overnight at 4°C with a commercially available rabbit anti-GFP antibody (Sigma) diluted 1:5,000. After three washes, blots were incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma) diluted 1:40,000. After four additional washes, blots were developed with the ECL Western blotting analysis system (Amersham Pharmacia Biotech). Digital images of blots were collected with a Bio-Rad Fluor-S-Max MultiImager using the accompanying Quantity One software.
RESULTS
MinC/DicB interferes with the assembly of FtsZ rings.
To study the effect of MinC/DicB expression on the integrity of FtsZ rings, we used strain PB147(λDR120) [ΔminDE(Plac::gfp-ftsZ)], harboring either plasmid pJE44 [PBAD::dicB] or vector pBAD33. Cells were grown in the presence of 37 μM IPTG (to induce expression of GFP-FtsZ) to an OD600 of 0.2. Arabinose was then added (to 0.05%) to induce expression of dicB, and aliquots were prepared for microscopic examination at 60-min intervals. The results are summarized in Table 2 and Fig. 1.
TABLE 2.
Quantitation of cell length and FtsZ rings after MinC/DicB-mediated filamentationa
| Plasmid | Time (min) | R− cells
|
R+ cells
|
R− + R+ cells
|
||||
|---|---|---|---|---|---|---|---|---|
| % (n) | Avg length (range) (μm) | % (n) | Avg length (range) (μm) | L/R (μm) | Avg length (range) (μm) | L/R (μm) | ||
| pJE44 [PBAD::dicB] | 10 | 20 (9) | 6.7 (3.2-20.0) | 80 (36) | 13.4 (4.4-33.5) | 5.5 | 12.0 (3.2-33.5) | 6.1 |
| 60 | 7 (2) | 21.9 (8.0-35.8) | 93 (25) | 15.1 (4.2-49.8) | 4.9 | 15.6 (4.2-49.8) | 5.4 | |
| 120 | 35 (9) | 39.0 (4.2-66.3) | 65 (17) | 30.3 (2.6-70.8) | 16.1 | 33.3 (2.6-70.8) | 27.1 | |
| 180 | 30 (17) | 51.4 (3.5-98.4) | 70 (39) | 11.0 (2.5-97.6) | 8.6 | 23.3 (2.5-98.4) | 26.0 | |
| pBAD33 (vector) | 0 | 16 (14) | 2.5 (1.9-3.5) | 84 (72) | 3.5 (1.9-15.9) | 2.7 | 3.4 (1.9-15.9) | 3.1 |
| 180 | 22 (13) | 2.5 (1.8-3.8) | 78 (47) | 2.8 (1.9-4.5) | 2.7 | 2.7 (1.8-4.5) | 3.4 | |
Cells of strain PB147(λDR120) [ΔminDE(Plac::gfp-ftsZ)] harboring the indicated plasmid or vector were treated and analyzed as described in the text. Parameters were calculated separately for cells without fluorescent ring structures (R− cells), cells with one or more rings (R+ cells), and all cells combined (R− + R+ cells). n, number of cells examined. Any visible accumulation of fluorescence resembling a ring was counted as one.
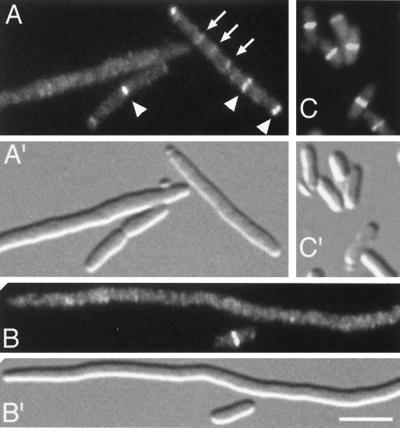
FIG. 1.
Inhibition of Z-ring assembly by MinC/DicB. Fluorescence (A to C) and corresponding DIC (A′ to C′) micrographs showing the distribution of GFP-FtsZ in cells which either were (A and B) or were not (C) subjected to MinC/DicB-mediated division inhibition. Cells of strain PB147(λDR120) [ΔminDE(Plac::gfp-ftsZ)] carrying either pJE44 [PBAD::dicB] (A and B) or vector pBAD33 (C) were grown at 37°C in the presence of 37 μM IPTG to an OD600 of 0.2. Arabinose was added to 0.05%, and growth was allowed to continue for 60 (A) or 180 (B and C) min. Arrowheads (A), positions of strongly fluorescent rings comparable to those observed in normally dividing cells (C); arrows, positions of some faintly fluorescent structures. Cells were chemically fixed before examination. Both bright and faint rings were included in the data shown in Table 2. Bar, 4 μm.
As expected, cells containing pBAD33 continued to divide normally upon addition of arabinose (Fig. 1C′), whereas those harboring pJE44 developed a pronounced division defect (Fig. 1A′ and B′). The average length of the latter cells was already 3.5-fold (12.0 μm) that of the former (3.4 μm) at the time of arabinose addition. This was likely due to a low level of dicB expression from the plasmid even before arabinose was added. After 3 h, the average dicB-expressing cell was about ninefold longer (23.3 μm) than the control cells (2.7 μm).
Throughout the experiment, about 80% of PB147(λDR120)/pBAD33 cells showed brightly fluorescent ring structures, and the L/R ratio of the population as a whole increased only slightly from 3.1 (0 min) to 3.4 μm (180 min). In comparison, the localization of GFP-FtsZ in PB147(λDR120)/pJE44 filaments differed significantly in that the level of fluorescence in the cytoplasm was noticeably elevated and far fewer ring structures were present. This is reflected in the L/R value for the population as a whole, which increased substantially from 6.1 μm at 0 min to 27.1 μm at 120 min (Table 2). By 180 min, this value had decreased slightly to 26.0 μm. At this time, the culture also contained a significant number of small, ring-containing cells (an example is shown in Fig. 1B), resulting in a decreased average cell length compared to that at 120 min. Since PB147 is Ara+, we presume this reemergence of small cells was due to consumption of the inducing sugar.
In addition to finding a high L/R value, we found that many of the ring-like accumulations still present in the PB147(λDR120)/pJE44 filaments were very dim (Fig. 1A), indicating that they contained relatively little FtsZ and suggesting that they represent poorly assembling and/or disintegrating Z rings. We conclude that MinC/DicB interferes with the assembly and/or integrity of FtsZ rings.
MinC and DicB interact in a yeast two-hybrid system.
To test whether MinC and DicB can interact directly, we used a yeast two-hybrid assay (31). As controls, we also tested the interaction between MinC and MinD and DicB and MinD, as well as the interaction of MinC, MinD, and DicB with themselves (Table 3). Consistent with previous reports, we observed a moderately strong interaction between MinC and MinD (24, 30, 39) and a weaker interaction of MinC with itself (26). Interestingly, MinC interacted very strongly with DicB in both genetic configurations, demonstrating a direct interaction between the two proteins. DicB did not appear to interact with itself to a significant degree. In contrast, MinD showed a strong interaction with itself (Table 3), as was recently also observed by Szeto et al. for both the Neisseria gonorrhoeae and the E. coli proteins (52). This self-interaction is consistent with the proposed ability of MinD to cooperatively assemble on the membrane during its dynamic localization cycle in vivo (22, 27, 41, 48).
Localization of MinC in DicB-induced filaments.
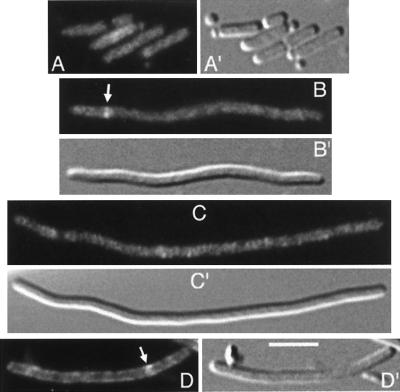
MinD recruits MinC to the cytoplasmic membrane (28, 46). To determine whether DicB might similarly affect the cellular distribution of MinC, we studied cells of strain PB114(λDB182)/pLL18 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-minC]. This strain lacks minD and is lysogenic for a phage which carries dicB downstream of the lac promoter. In addition, the strain harbors plasmid pLL18, which encodes a fully functional derivative of MinC in which the first 4 aa are replaced with GFP. Expression of the GFP-MinC fusion is under the control of the λR promoter and a temperature-sensitive allele of repressor cI. When grown at 37°C (GFP-MinC+) in the absence of IPTG (DicB−), cells displayed a Min− phenotype and GFP-MinC was present throughout the cytoplasm (Fig. 2A). Cells formed filaments in the presence of IPTG, but the bulk of GFP-MinC still appeared throughout the cytoplasm (Fig. 2B and C). This seemed to indicate that activation of MinC function by DicB was not accompanied by a major redistribution of MinC in the cell (but see below). In a minority of the filaments (<5%), however, one or more ring-like accumulations of varying intensity could be seen above the cytoplasmic signal (Fig. 2B). Although such rings were relatively rare, they were undetectable in cells that had grown without IPTG, implying that their formation was dependent on DicB.
FIG. 2.
Different effects of MinD and DicB on the distribution of GFP-MinC. Micrographs show the distribution of GFP-MinC in the presence of MinD (D) or DicB (B and C) or in the absence of either activator (A). Shown are cells of strain PB114/pLL18 [ΔminCDE/cI857, PλR::gfp-minC], lysogenic for either λDB182 [Plac::dicB] (A to C) or λDR155 [Plac::minD] (D). Cells were grown at 37°C in the presence of either 0.1% glucose (A) or 100 μM IPTG (B to D) and were chemically fixed prior to examination. Bar, 4 μm.
DicB-dependent association of DMinC with septal rings.
We reasoned that the presence of these ring-like accumulations of GFP-MinC in DicB-induced filaments, as well as their low number, could be explained if DicB was to act by concentrating MinC directly on septal ring structures. The high local concentration of MinC would then cause a rapid disassembly of the septal rings, resulting in a filamentous phenotype as well as a redistribution of MinC back into the cytoplasm.
To test this scenario, we used a fusion (GFP-DMinC) in which the first 13 aa of MinC are replaced by GFP. Whereas this fusion still interacts with DicB and MinD, it no longer inhibits cell division and, thus, lacks a functional Z domain (Table 4 and our unpublished data).
TABLE 4.
Functionality and localization of GFP-MinC fusionsa
| Protein (plasmid) | LL1 (λDB182) [ΔminCDE(Plac::dicB)]
|
LL1(λDR155) [ΔminCDE(Plac::minD)]
|
LL1 (λDB175) [ΔminCDE(Plac::minDE)]
|
|||
|---|---|---|---|---|---|---|
| Phen. | Loc. | Phen. | Loc. | Phen. | Loc. | |
| GFP-T-MinC(5-231) (pLL18) | Sep− | Cb | Sep− | Mb | WT | O |
| GFP-T-MinC(14-231) (pLL13) | Min− | R | Min− | R | Min− | O/R |
| GFP-T-MinC(108-231) (pPC105) | Min− | R | Min− | R | Min− | O/R |
| GFP-T-MinC(141-231) (pLL14) | Min− | C | Min− | C | Min− | C |
| GFP-T-MinC(108-231)-H (pJE78) | Min− | R | Min− | R | Min− | O/R |
| GFP-T-MinC(108-208)-H (pJE79) | Min− | C | Min− | C | Min− | C |
Lysogens containing the indicated plasmids were grown for ≈5 h at 37°C in the presence of 100 μM IPTG to an OD600 of 0.1. Live cells were observed through fluorescence optics to determine the localization pattern of the GFP-MinC fusion (Loc.) and through phase-contrast optics to determine the division phenotype (Phen.). C, cytoplasmic; M, along the periphery of the cell; O, oscillating; R, stably associated with rings; O/R, transiently associated with rings during oscillation cycle. When grown in the absence of IPTG, cells were Min− and fluorescence was cytoplasmic in all cases.
Ring-like accumulations were detected at a low frequency in these filaments (see text).
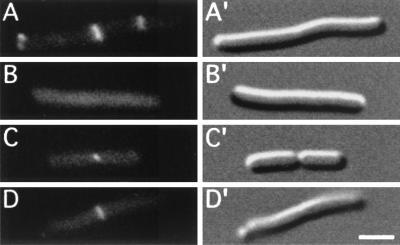
When strain PB114(λDB182)/pLL13 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-t-minC(14-231)] was grown at 37°C in the absence of inducer, cells displayed a Min− phenotype and the fluorescence signal was distributed throughout the cytoplasm (Fig. 3A and A′). This random distribution was not due to a possible release of the GFP moiety by proteolytic processing, as only the full-length fusion was detectable in Western analyses using anti-GFP antibodies (Fig. 4, lane 2). When grown in the presence of IPTG, the cells were still Min−. Strikingly, however, the expression of DicB caused the GFP-DMinC fusion to accumulate in bright rings (Fig. 3B and C; Table 4). Such rings were present in ∼80% of the population (189 of 230 cells), and their cellular locations corresponded to those expected for septal ring structures, including sites of active cell wall invagination. To confirm this location, cells were fixed and immunostained with anti-FtsZ antibodies. As illustrated in Fig. 5A, the GFP-DMinC rings indeed colocalized with FtsZ to septal rings in these cells. We next examined the localization of GFP-DMinC in cells expressing both DicB and division inhibitor SfiA (SulA), which interferes with the assembly and maintenance of septal rings by blocking FtsZ polymerization (42, 54) independently of MinC (10). To this end, strain PB114(λDB182)/pLL13 was transformed with either vector plasmid pBAD33 or with a derivative (pJE80) that carries sfiA downstream of the PBAD promoter. Cells carrying the control plasmid continued to divide and showed the DicB-dependent accumulation of GFP-DMinC in rings, regardless of the presence of arabinose in the growth medium (Fig. 6C). In contrast, cells carrying pJE80 ceased division upon addition of arabinose, and the resulting filaments were completely devoid of fluorescent rings (Fig. 6D). These results showed that, in the presence of DicB, MinC is targeted directly to one or more septal ring components.
FIG. 3.
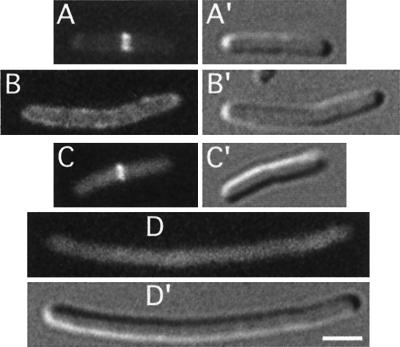
Mutually dependent accumulation of DMinC and DicB on rings. Micrographs show the distribution of GFP-DMinC (A to C) in the absence (A) and presence (B and C) of DicB and that of GFP-DicB (D to F) in the absence (D) and presence (E and F) of DMinC. (A to C) Fixed cells of strain PB114(λDB182)/pLL13 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-minC(14-231)] which were grown at 37°C in the absence (A) or presence (B and C) of 100 μM IPTG. (D to F) Fixed cells of strain PB114(λJE39)/pJE46 [ΔminCDE(Plac::gfp-dicB)/cI857, PλR::minC(14-231)] which were grown in the presence of 37 μM IPTG at 30°C (D) or 37°C (E and F). Bar, 2 μm.
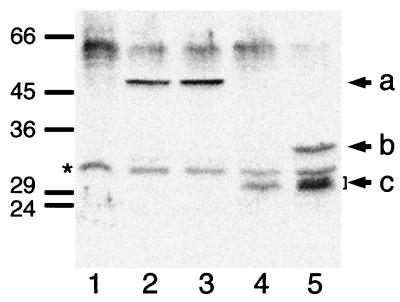
FIG. 4.
Detection of GFP-DMinC and GFP-DicB by immunoblotting. Immunoblots containing whole-cell extracts were probed with anti-GFP antibodies. a to c, positions of GFP-DMinC (a) and both full-length (b) and processed (c) forms of GFP-DicB. The positions of molecular mass standards (in kilodaltons) and of an unidentified cross-reacting antigen that is present in all extracts (∗) are indicated on the left. Lanes contained extracts of strains PB114 [ΔminCDE] (lane 1), PB114(λDB182)/pLL13 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-minC (14-231)] (lanes 2 and 3), and PB114(λJE39)/pJE46 [ΔminCDE(Plac::gfp-dicB)/cI857, PλR::minC(14-231)] (lanes 4 and 5). Cells were grown at 30°C (lane 4) or 37°C (lanes 1 to 3 and 5) in the presence of 0 (lane 2), 37 (lanes 4 and 5), or 100 (lanes 1 and 3) μM IPTG.
FIG. 5.
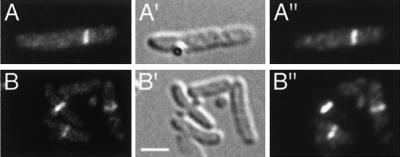
DicB- and MinD-mediated targeting of DMinC to septal rings. Micrographs showing colocalization of GFP-DMinC (A and B) and FtsZ (A" and B") in cells expressing either DicB (A) or MinD (B). Cells of strains PB114(λDB182)/pLL13 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-minC(14-231)] (A) and PB114(λDR155)/pLL13 [ΔminCDE(Plac::minD)/cI857, PλR::gfp-minC(14-231)] (B) were grown at 37°C in the presence of 100 μM IPTG, chemically fixed, and subjected to immunostaining with anti-FtsZ primary antibodies and Cy3-conjugated secondary antibodies. Cells were viewed through Nomarski (A′ and B′) and fluorescence optics with GFP-specific (A and B) or Cy3-specific (A" and B") filter sets. Bar, 2 μm.
FIG. 6.
Localization of DMinC in SfiA-induced filaments. Micrographs showing the location of GFP-DMinC in MinD+ (A and B) and DicB+ (C and D) cells in the absence (A and C) or presence (B and D) of SfiA. Strains used were PB114(λDR155)/pLL13/pBAD33 [ΔminCDE(Plac::minD)/cI857, PλR::gfp-minC(14-231)/vector] (A), PB114(λDR155)/pLL13/pJE80 [ΔminCDE(Plac::minD)/cI857, PλR::gfp-minC(14-231)/PBAD::sfiA] (B), PB114(λDB182)/pLL13/pBAD33 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-minC(14-231)/vector] (C), and PB114(λDB182)/pLL13/pJE80 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-minC(14-231)/PBAD::sfiA] (D). Cells were grown at 37°C in the presence of 100 μM IPTG to an OD600 of 0.1, arabinose was added to 0.1%, and growth was allowed to continue for 45 min before inspection by fluorescence (A to D) and DIC (A′ to D′) microscopy. Bar, 2 μm.
To evaluate what domain of the MinC peptide is required for DicB-dependent targeting, we used pLL18 derivatives encoding GFP fusions to various portions of MinC. As shown in Table 4, fusions containing residues 108 to 231 of MinC still accumulated in rings in a DicB-dependent fashion, whereas fusions containing aa 141 to 231 or aa 108 to 208 failed to do so. These results are consistent with genetic analyses of minC missense alleles (43) and indicate that the domain of MinC required for interaction with DicB overlaps that responsible for its interaction with MinD (see below) (26).
DMinC-dependent integrity and localization of DicB.
The DicB-mediated targeting of MinC to a septal ring ligand could occur in several ways. (i) Without itself being targeted to a specific cellular location, DicB might bind cytoplasmic MinC and modify it to a form with affinity for a septal ring component. (ii) DicB itself might have affinity for a septal ring factor and recruit MinC. (iii) The binding of DicB to MinC might create a site on the heteromeric complex with a high affinity for a septal ring-associated target.
To discriminate between these possibilities, we studied the location of a GFP-DicB fusion in the absence and presence of DMinC. The ΔminCDE strain PB114(λJE39)/pJE46 is lysogenic for phage λJE39 [Plac::gfp-dicB], which encodes GFP-DicB under the control of the lac promoter and which also harbors plasmid pJE46 [cI857, PλR::minC(14-231)], which expresses MinC (aa 14 to 231) in a temperature-dependent fashion. Upon growth in the presence of IPTG (DicB-GFP+) at 30°C (DMinC−), cells showed fluorescence throughout the cell body (Fig. 3D). Upon growth at 37°C (DMinC+), cells still showed a significant signal throughout the cytoplasm. However, the vast majority of cells also showed clear accumulations in rings, which were easily discernible above the cytoplasmic signal (Fig. 3E and F).
To evaluate the integrity of the GFP-DicB fusion in these experiments, we performed Western analyses using anti-GFP antibodies. Interestingly, whereas a significant amount of intact GFP-DicB (35 kDa) was present in cells coexpressing DMinC (Fig. 4, lane 5), the full-length species was almost undetectable in cells lacking DMinC (lane 4). In both cases, cell extracts contained multiple smaller products, which migrated as 30- to 31-kDa species during sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Such species might still contain an intact GFP moiety (27 kDa) and contribute to the significant fluorescence signal seen in the cytoplasm of these cells. We obtained very similar results with a DicB-GFP fusion in which the GFP tag is fused to the C terminus of DicB (data not shown). Given the size of the DicB fusion breakdown products and our experience that many other GFP fusions are much more stable (e.g., GFP-MinC; Fig. 4, lane 2), the likeliest explanation of these results is that the binding of DMinC to the DicB moiety of the fusion protects the latter from rapid proteolytic attack.
The Lon protease is a major protein-processing factor in E. coli, and the SfiA division inhibition protein is among its substrates (18). To assess whether Lon might be responsible for processing the GFP-DicB fusion, we also examined the integrity of the latter in LL1, a lon derivative of strain PB114. As in PB114, however, the concentration of intact GFP-DicB in LL1 was strongly dependent on the coexpression of DMinC (data not shown). Thus, although Lon may contribute to the degradation of GFP-DicB, it is clearly not the sole factor responsible for its instability.
Due to the instability of the DicB fusions in the absence of MinC, we are not yet able to conclusively dismiss the possibility that a stable form of DicB by itself may have some intrinsic affinity for the septal ring. Nevertheless, our results strongly favor the possibility that DMinC and DicB colocalize to the septal ring as part of a heteromeric complex.
Interdependent targeting of DMinC and MinD to the septal ring in MinE− cells.
In MinE− filaments, the bulk of both MinD and MinC is distributed along the membrane (28, 46, 48, 50). In many such filaments, however, this distribution is not completely even. Quite often, some small but distinct accumulations at the membrane are evident, especially in the smaller filaments. Figure 2D shows an example of such an accumulation in a filament of strain PB114(λDR155)/pLL18 [ΔminCDE(Plac::minD)/cI857, PλR::gfp-minC(5-231)]. As for the MinC/DicB filaments described above, we suspected that these accumulations could correspond to remnants of septal rings which had not yet been fully cleared by the action of ZMinC.
To test this idea, we introduced pLL13 into strain PB114(λDR155) and studied the localization of GFP-DMinC in the absence and presence of MinD by growing cells in the absence and presence of IPTG, respectively. As shown in Fig. 7A to D, the fusion accumulated in bright ring structures in a MinD-dependent manner. Again, these rings corresponded to septal rings, as shown by colocalization with FtsZ (Fig. 5B) and their sensitivity to SfiA (Fig. 6B). Furthermore, the minimal domain of MinC required for MinD-mediated targeting of DMinC to septal rings was the same as that for DicB-mediated targeting (aa 108 to 231; Table 4).
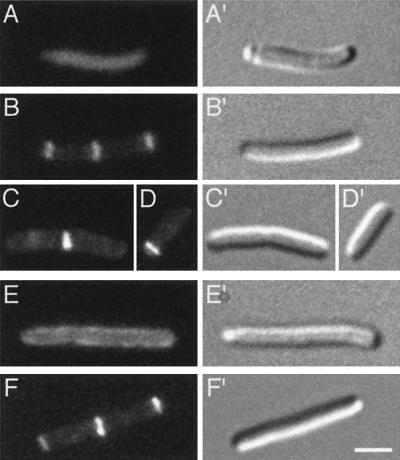
FIG. 7.
Mutually dependent accumulation of DMinC and MinD on rings. Micrographs show the distribution of GFP-DMinC (A to D) in the absence (A) and presence (B to D) of MinD and that of GFP-MinD in the absence (E) and presence (F) of DMinC. (A to D) Cells of strain PB114(λDR155)/pLL13 [ΔminCDE(Plac::minD)/cI857, PλR::gfp-minC(14-231)] which were grown at 37°C in the absence (A) or presence (B to D) of 100 μM IPTG. (E and F) Cells of strain PB114(λDR119)/pJE46 [ΔminCDE(Plac::gfp-minD)/cI857, PλR::minC (14-231)] which were grown in the presence of 37 μM IPTG at 30°C (E) or 37°C (F). Bar, 2 μm.
To evaluate whether, conversely, MinD would be attracted to septal rings in a DMinC-dependent manner, we used strain PB114(λDR119)/pJE46 [ΔminCDE(Plac::gfp-minD)/cI857, PλR::minC(14-231)]. Whereas at 30°C (DMinC−) cells showed GFP-MinD all along their peripheries (Fig. 7E), the fusion indeed accumulated in bright rings when cells were grown at 37°C (DMinC+). In the latter cells, furthermore, very little fluorescence remained on the membrane between rings (Fig. 7F).
We conclude that, similar to DMinC/DicB, the DMinC/MinD heteromer has a high affinity for a septal ring components(s) in vivo. Furthermore, these interactions of MinC with the division apparatus are separable from any interactions of the N-terminal Z domain of MinC responsible for FtsZ depolymerization.
Given that ZMinC activity can be counteracted by overproduction of FtsZ (10), we also examined the location of a fully functional GFP-MinC fusion in cells carrying ftsZ on a multicopy plasmid. The results were similar to those obtained with GFP-DMinC in cells with normal FtsZ levels in that GFP-MinC appeared cytoplasmic in the absence of MinD and DicB but decorated septal ring structures when either activator was supplied (not shown). Thus, activator-dependent targeting of MinC to septal rings can also be observed when the Z domain of MinC is intact and the rings are stabilized by an increased pool of FtsZ.
DicB competes with MinD for binding MinC in vivo.
Expression of dicB induces filamentation in both wild-type (WT) and minD cells, suggesting that MinC targeting by DicB is dominant over that by MinD (10, 35). One possibility is that DicB efficiently competes with MinD for binding MinC. Alternatively, DicB might be able to bind the MinC/MinD complex, after which the tripartite complex targets some septal ring component. To discriminate between these possibilities, we introduced plasmid pJE44 into strain PB114(λDR119)/pJE46. In the resulting transformants, expression of GFP-MinD can be regulated with IPTG, that of DMinC can be regulated with temperature, and that of DicB can be regulated with arabinose. Cells that were grown at 37°C in the presence of IPTG but without arabinose showed bright rings as described above (Fig. 8A). In contrast, rings were almost completely absent when cells were grown in the presence of 0.05% arabinose for 4 h. Instead, the GFP-MinD fusion was distributed more or less evenly over the entire membrane (Fig. 8C). In the presence of less arabinose (0.02%), cells with intermediate patterns predominated (Fig. 8B). These results indicate that DicB competes with MinD for complex formation with MinC.
FIG. 8.
DicB removes MinD from rings. Micrographs show the distribution of GFP-MinD in the presence of DMinC and in either the absence (A) or presence (B and C) of DicB. Cells of strain PB114(λDR119)/pJE46/pJE44 [ΔminCDE(Plac::gfp-minD)/cI857, PλR::minC (14-231)/PBAD::dicB] were grown at 37°C in the presence of 37 μM IPTG, and no (A), 0.02% (B), or 0.05% (C) arabinose. Bar, 2 μm.
Transient association of oscillating DMinC with septal rings.
The affinity of DMinC/MinD for septal rings described above was observed most clearly in cells lacking MinE (see above). In WT cells, the MinC/MinD complex oscillates from pole to pole in a MinE-dependent fashion and an accumulation of MinC/MinD in ring structures is not, or at best very rarely, observed (28, 46, 48, 50). A rationale for the latter is that MinE prevents an association of MinC/MinD with the septal ring in WT cells by sweeping the complex away from the cell center and that noncentral septal rings simply are not present, or are highly unstable, due to the action of the intact Z domain of MinC (15, 22, 41). It can be predicted, therefore, that, even in the presence of MinE, MinC/MinD should be capable of decorating noncentral septal rings provided they are sufficiently stable.
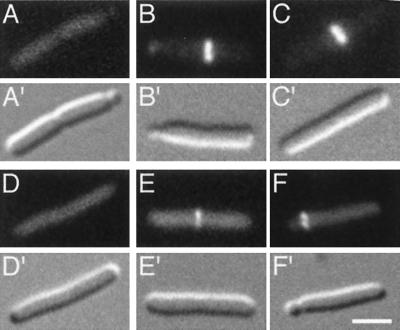
To test this prediction, we compared the localization of fully functional MinC with that of DMinC, in cells expressing both MinD and MinE. For this purpose, pLL18 [cI857, PλR::gfp-minC (5-231)] or pPC105 [cI857, PλR::gfp-minC (108-231)] was introduced into strain LL1(λDB175) [ΔminCDE(Plac::minDE)] and cells were grown in the presence of IPTG at 37°C. Cells expressing the GFP-MinC fusion showed a WT division phenotype, and the fusion oscillated as described before (46) with a cycle time of about 45 s and without any obvious propensity for accumulating on ring structures (Table 4; Fig. 9A). As expected, cells expressing the GFP-DMinC fusion were Min−, and the fusion showed oscillation similar to that of the fully functional fusion. However, the GFP-DMinC fusion also showed an obvious affinity for rings with which it transiently associated while moving from one cell pole to the other (Fig. 9B and C). To determine the positions of transiently decorated rings, we collected time-lapse images of individual cells and measured cell length and the distance between (transiently) fluorescent rings and the proximal cell pole. For comparison, we also measured the positions of division septa in a parallel culture. As shown in Fig. 10, transient association of GFP-DMinC/MinD occurred almost exclusively at non-centrally positioned rings, providing further support for the proposed role of MinE in keeping MinC/MinD away from septal ring structures located at the cell center (15, 22, 41).
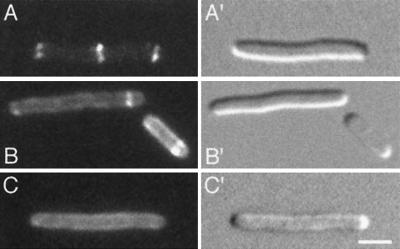
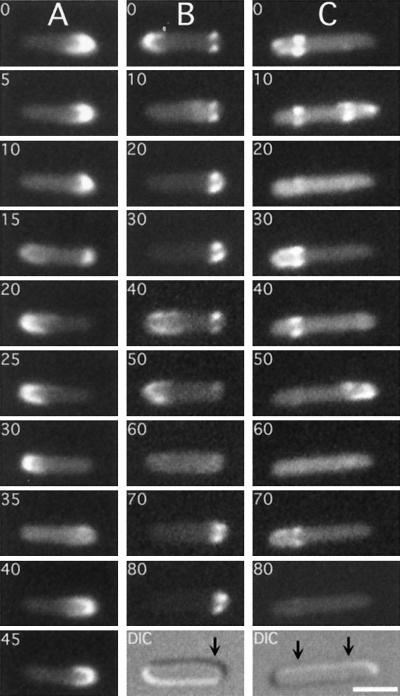
FIG. 9.
Transient association of oscillating DMinC with rings. Time-lapse images show the oscillation of GFP-MinC (A) and GFP-DMinC (B and C) in the presence of MinD and MinE. Shown are cells of strain LL1(λDB175) [ΔminCDE(Plac::minDE)] harboring either pLL18 [cI857, PλR::gfp-minC(5-231)] (A) or pPC105 [cI857, PλR::gfp-minC(108-231)] (B and C). Cells were grown at 37°C in the presence of 100 μM IPTG. Times in seconds are indicated. Arrows in the DIC panels (B and C) mark the positions of rings to which GFP-DMinC transiently associated as it moved from one end of the cell to the other. Such transient associations are not observed when the Z domain of MinC is functional (A). Bar, 2 μm.
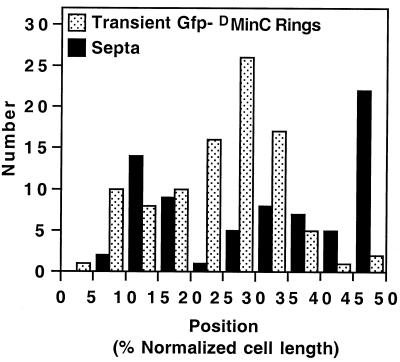
FIG. 10.
Positions of rings transiently decorated by oscillating DMinC. Time-lapse and DIC images of randomly chosen cells of strain LL1(λDB175)/pPC105 (see legend to Fig. 9) were collected, and the positions of transiently fluorescent ring structures and division septa were determined. x axis, position of structures relative to the proximal cell pole and midcell after normalization of cell length; y axis, number of structures at each position. The distributions of transiently fluorescent rings and septa represent data from time-lapse images of 77 live cells and DIC images of 260 fixed cells, respectively. Data from cells >7.0 μm were not included because the majority of these showed a multizonal oscillation pattern, similar to that described before (46, 48), rather than the pole-to-pole pattern seen in smaller cells.
Decoration of septal rings by DMinC/DicB is unaffected by MinE.
In contrast to a MinC/MinD-induced division block, MinC/DicB-induced filamentation is not suppressed by MinE (10), suggesting that MinE should have little effect on the targeting of MinC/DicB to septal ring structures. To test this prediction, plasmid pJE75 [PBAD::minE] or vector pBAD33 was introduced into strains PB114(λDR155)/pLL13 [ΔminCDE(Plac::minD)/cI857, PλR::gfp-minC(14-231)] and PB114(λDB182)/pLL13 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-minC (14-231)]. Transformants were grown at 37°C in the presence of both IPTG (100 μM) and arabinose (0.1%), and cells were examined for the localization of GFP-DMinC.
Interestingly, PB114(λDB182)/pLL13 cells harboring pJE75 showed fluorescent rings (Fig. 11D) at about the same frequency (∼80% of cells) as cells of either strain carrying the control plasmid (Fig. 11A and C). In contrast, the GFP-DMinC fusion appeared almost completely cytoplasmic in virtually all PB114(λDR155)/pLL13/pJE75 cells (Fig. 11B). We note that although MinC oscillates from pole to pole when MinE is present at physiological levels (see above), MinC becomes cytoplasmic at high levels of MinE (J. E. Johnson and P. A. J. de Boer, unpublished data), which may be related to the ability of MinE to interfere with the interaction between MinC and MinD (30).
FIG. 11.
GFP-DMinC/DicB rings are resistant to MinE. Fluorescence (A to D) and DIC (A′ to D′) micrographs show the localization of GFP-DMinC in MinD+ DicB− (A and B) and MinD− DicB+ (C and D) cells in the absence (A and C) or upon (over)expression (B and D) of MinE. Shown are cells of strains PB114(λDR155)/pLL13/pBAD33 [ΔminCDE(Plac::minD)/cI857, PλR::gfp-minC (14-231)/vector] (A), PB114(λDR155)/pLL13/pJE75 [ΔminCDE(Plac::minD)/cI857, PλR::gfp-minC (14-231)/PBAD::minE] (B), PB114(λDB182)/pLL13/pBAD33 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-minC(14-231)/vector] (C), and PB114(λDB182)/pLL13/JE75 [ΔminCDE(Plac::dicB)/cI857, PλR::gfp-minC (14-231)/PBAD::minE] (D). Cells were grown for 3.5 h to an OD600 of 0.3 at 37°C in the presence of 100 μM IPTG and 0.1% arabinose before examination. Bar, 2 μm.
We conclude that, in contrast to the localization of DMinC/MinD, the targeting of DMinC/DicB to septal rings is indeed unaffected by MinE.
DISCUSSION
This study further elucidates the mechanism by which MinD and DicB stimulate the division-inhibitory activity of MinC. Both MinD and DicB were found to interact directly with the C-terminal D domain of the MinC peptide, and both the DMinC/MinD complex and the DMinC/DicB complex were shown to have a high affinity for septal ring structures in vivo. These observations indicate that the Z and D domains of MinC participate in separate interactions with components of the division apparatus. The Z domain is thought to interact directly with FtsZ polymers, resulting in depolymerization (26, 29). In contrast, the interaction of the D domain with division components requires the binding of this domain to MinD or DicB, and it appears to be the complex of DMinC with either activator which interacts with some septal ring factor(s).
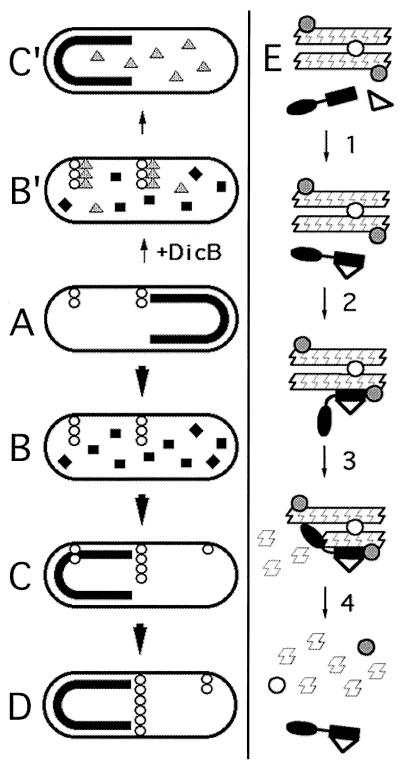
These results suggest a three-step mechanism for MinC-mediated division inhibition as outlined in Fig. 12E. In the first step, the D domain of MinC binds to either MinD or DicB. Under normal conditions, only the interaction with MinD is relevant because dicB transcription is actively repressed (3). When transcription is induced, however, the DicB protein effectively competes with MinD for binding MinC. In the next step, the complex of MinC with either MinD or DicB binds a target which is closely associated with FtsZ polymers, bringing the Z domain of MinC in close proximity to its substrate. Finally, the Z domain stimulates depolymerization of FtsZ, which simultaneously leads to dispersal or destruction of the target recognized by the D domain complex.
FIG. 12.
Models for MinD- and DicB-dependent activation of MinC-mediated division inhibition. Under conditions of normal growth (A to D), MinC and MinD co-oscillate from the membrane on one cell half to the other in a MinE-dependent fashion (MinC/MinD is indicated by a thick black line or squares; MinE is omitted for simplicity). FtsZ polymerization (chains of open circles) is allowed to initiate on any site of the membrane not occupied by MinC/MinD and not subject to nucleoid occlusion (38). As soon as MinC/MinD returns to occupy that half of the membrane, nascent ring structures that formed in the previous half cycle and that are positioned off-center are the substrate for MinC/MinD-mediated destruction (C and D), while structures that may have formed at the cell center are protected because the oscillating behavior of the division inhibitor causes its time-averaged concentration to be minimal at the cell's middle (22, 25, 41). Upon induction of dicB expression in WT cells (B′ and C′), DicB efficiently competes with MinD for binding MinC to form MinC/DicB complexes (shaded triangles). The latter do not oscillate and destroy nascent septal ring structures at all sites in the cell, resulting in filamentous growth of the bacterium. At a molecular level (E), MinC-mediated division inhibition in vivo is likely to occur in several steps. In step 1, MinD or DicB (open triangle) binds the C-terminal D domain of MinC (DMinC; black rectangle portion of molecule). This binding event produces a complex with a high affinity for a target that is part of both nascent and mature septal ring structures. In the simplest scenario, the inhibitor complex directly recognizes polymers of FtsZ (chain of Z-shaped monomers). Alternatively, additional septal ring components (open and shaded circles) may first need to associate with FtsZ polymers to create target sites. In step 2, DMinC/MinD or DMinC/DicB binds the target. This event is independent of any interaction involving the N-terminal domain of MinC (ZMinC; black oval portion of molecule). Rather, step 2 serves to bring ZMinC in close proximity to its substrate, allowing efficient disassembly of FtsZ polymers in step 3. Disassembly of FtsZ polymers by ZMinC simultaneously destroys the septal ring ligand(s) recognized by the DMinC complex (step 4). As a result, MinC/DicB is instantly released into the cytoplasm, whereas MinC/MinD remains associated with the surrounding membrane until it is forced off by the action of the next MinE wave (15, 22, 27).
Although the purified Z domain of MinC was shown to stimulate FtsZ depolymerization in vitro (26, 29), one recent report suggested that MinC might block cell division in vivo primarily by interfering with the interaction between FtsZ and FtsA rather than by acting on FtsZ polymers directly (32). Subsequent localization studies, however, using either GFP-tagged FtsZ in live E. coli (45) (C. Hale and P. A. J. de Boer, unpublished data) or anti-FtsZ antibodies in Bacillus subtilis (36) quite clearly show that MinC/MinD-induced filamentation coincides with a failure to assemble Z rings. Here we showed that an intact Z domain of MinC is also required for MinC/DicB-induced filamentation and that Z rings fail to assemble in such filaments as well. Therefore, it is highly likely that both MinC-dependent division blocks, whether stimulated by MinD or DicB, result from a direct interference with FtsZ polymerization by the Z domain of MinC.
It remains to be determined what septal ring component(s) is recognized by DMinC/MinD and DMinC/DicB and whether both complexes bind the same or different targets. In WT cells, the MinC/MinD complex is rarely, if ever, seen to decorate a ring. Rather the proteins rapidly move from the membrane at one cell end to the other in an oscillatory membrane association-dissociation cycle, which requires the activities of both MinD and MinE. Due to the action of MinE, MinC/MinD complexes are kept away from the assembling or assembled septal ring at the cell center, while they prevent assembly of complete Z rings at noncentral sites due to the activity of ZMinC in the complex (15, 22, 26-29, 46, 48, 50). Blocking the assembly of functional Z rings near cell poles requires a minimum oscillation frequency of at least 0.25 full cycles per min (22, 48). From this observation it can be inferred that FtsZ must be capable of rapidly initiating polymerization on the noncentral portion of the membrane soon after MinC/MinD has departed. Therefore, we expect that the physiological targets of MinC/MinD during normal growth are small septal ring intermediates that assembled within the previous 20 to 25 s (one-half oscillation cycle) on that half of the cell membrane from which MinC was temporarily absent (Fig. 12A to D). We imagine that such incipient structures consist of FtsZ polymers and factors that rapidly associate with the polymers. Associated factors could include FtsA and ZipA, which bind FtsZ directly, as well as one or more other division factors which can join the assembly subsequent to ZipA and FtsA (7). In the simplest scenario, the DMinC/MinD complex is directly attracted to (polymers of) FtsZ. However, any other septal ring component which associates with FtsZ before the assembly of a stable Z ring is completed might provide, or contribute to the formation of, a specific binding surface for DMinC/MinD (Fig. 12E).
It will also be interesting to determine what determinants within the DMinC/MinD and DMinC/DicB complexes provide specificity for the septal ring target(s). Binding specificity may be provided by a (sub)domain on DMinC which is exposed only upon binding to either MinD or DicB. Alternatively, specificity may be provided by domains on MinD and DicB that become exposed upon binding to DMinC or by a combination of determinants present on both partners in the complexes. In the crystal structure of MinC from Thermotoga maritima, the D domain is folded in a triangular, right-handed β-helix, one side of which provides the dimer interface (8). How MinD and DicB bind such a structure and how their binding might affect this structure are intriguing questions.
Especially with regard to the possibility that MinD might provide binding specificity to a septal ring component(s), it is interesting to compare our results with those obtained with B. subtilis, where the binding of the MinC/MinD complex to the septal ring at midcell is an essential step in the normal localization cycles and functions of these proteins. In this organism, MinE is absent and MinC/MinD does not oscillate from pole to pole. Rather, the complex associates with the mature central septal ring just prior to cell constriction. The DivIVA protein also joins the septal ring at about the same time, though independently of MinC/MinD, and is required for retaining MinC/MinD at the newly formed cell poles long after constriction has completed and the septal ring which initially attracted MinC/MinD has dissipated (14, 39, 40). As in E. coli, the location of MinC is dictated by that of MinD (39). However, the B. subtilis MinD protein shows a significant intrinsic affinity for septal rings, even in the absence of MinC and DivIVA (39). In contrast, we have not observed any obvious intrinsic affinity of E. coli MinD or DMinC for the septal ring when either is expressed without its partner. Unfortunately, whether or not DicB by itself possesses an intrinsic affinity for any specific target in the cell could not be determined due to the instability of the DicB-GFP fusion (and, we suspect, native DicB as well) when DMinC is absent.
This work further expands the number of known molecular interactions involving the MinC and MinD proteins. MinC is now known to interact with FtsZ (polymers) through its Z domain and with itself, as well as with MinD and DicB, through its D domain (see above) (8, 26, 28-30, 39, 46, 53). MinD interacts with itself, MinC, and MinE, as well as with ATP and the cell membrane (see above) (9, 27, 30, 47, 48, 50, 52). In addition, the MinC/MinD complex binds to some as yet undefined septal ring-associated target. Understanding how all these interactions take place, how they are regulated, and how they contribute to the functions and remarkable properties of the Min proteins in the cell remains a major challenge for the future.
Acknowledgments
We thank Cynthia Hale for technical support and comments on the manuscript, David Raskin and Piyali Chatterjee for help in plasmid construction, Sandra Lemmon for the gift of materials, and Kenneth Henry, Dan Gelperin, and other members of the Lemmon lab for advice on two-hybrid analyses.
This work was supported by NIH grant GM-57059. J.E.J. and L.L.L. were supported by NIH NRSA Institutional Training Grant T32GM08056.
REFERENCES
- 1.Addinall, S. G., E. Bi, and J. Lutkenhaus. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall, S. G., and J. Lutkenhaus. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22:231-237. [DOI] [PubMed] [Google Scholar]
- 3.Béjar, S., F. Bouché, and J.-P. Bouché. 1988. Cell division inhibition gene dicB is regulated by a locus similar to lambdoid bacteriophage immunity loci. Mol. Gen. Genet. 212:11-19. [DOI] [PubMed] [Google Scholar]
- 4.Bi, E., and J. Lutkenhaus. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 6.Cam, K., S. Béjar, D. Gil, and J.-P. Bouché. 1988. Identification and sequence of division inhibitor gene dicB suggests expression from an internal in-frame translation start. Nucleic Acids Res. 16:6327-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 8.Cordell, S. C., R. E. Anderson, and J. Lowe. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer, P. A. J., R. E. Crossley, A. R. Hand, and L. I. Rothfield. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10:4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1990. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc. Natl. Acad. Sci. USA 87:1129-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 12.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1988. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J. Bacteriol. 170:2106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, D. H., and J. Errington. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905-915. [DOI] [PubMed] [Google Scholar]
- 15.Fu, X., Y. L. Shih, Y. Zhang, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 17.Gilson, P. R., and P. L. Beech. 2001. Cell division protein FtsZ: running rings around bacteria, chloroplasts and mitochondria. Res. Microbiol. 152:3-10. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 19.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hale, C. A., and P. A. J. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ, and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hale, C. A., H. Meinhardt, and P. A. J. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in E. coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hale, C. A., A. C. Rhee, and P. A. J. de Boer. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi, I., T. Oyama, and K. Morikawa. 2001. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 20:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howard, M., A. D. Rutenberg, and S. de Vet. 2001. Dynamic compartmentalization of bacteria: accurate division in E. coli. Phys. Rev. Lett. 87:278102. [DOI] [PubMed] [Google Scholar]
- 26.Hu, Z., and J. Lutkenhaus. 2000. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu, Z., and J. Lutkenhaus. 2001. Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7:1337-1343. [DOI] [PubMed] [Google Scholar]
- 28.Hu, Z., and J. Lutkenhaus. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34:82-90. [DOI] [PubMed] [Google Scholar]
- 29.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, J., C. Cao, and J. Lutkenhaus. 1996. Interaction between FtsZ and inhibitors of cell division. J. Bacteriol. 178:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Justice, S. S., J. Garcia-Lara, and L. I. Rothfield. 2000. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol. Microbiol. 37:410-423. [DOI] [PubMed] [Google Scholar]
- 33.Kuroiwa, T. 2000. The discovery of the division apparatus of plastids and mitochondria. J. Electron Microsc. 49:123-134. [DOI] [PubMed] [Google Scholar]
- 34.Labie, C., F. Bouché, and J.-P. Bouché. 1989. Isolation and mapping of Escherichia coli mutations conferring resistance to division inhibition protein DicB. J. Bacteriol. 171:4315-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labie, C., F. Bouché, and J.-P. Bouché. 1990. Minicell-forming mutants of Escherichia coli: suppression of both DicB- and MinD-dependent division inhibition by inactivation of the minC gene product. J. Bacteriol. 172:5852-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin, P. A., R. L. Schwartz, and A. D. Grossman. 2001. Polymer stability plays an important role in the positional regulation of FtsZ. J. Bacteriol. 183:5449-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 38.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 39.Marston, A. L., and J. Errington. 1999. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33:84-96. [DOI] [PubMed] [Google Scholar]
- 40.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meinhardt, H., and P. A. J. de Boer. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. USA 98:14202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee, A., C. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulder, E., C. L. Woldringh, F. Tétart, and J.-P. Bouché. 1992. New minC mutations suggest different interactions of the same region of division inhibitor MinC with proteins specific for minD and dicB coinhibition pathways. J. Bacteriol. 174:35-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osteryoung, K. W., and R. S. McAndrew. 2001. The plastid division machine. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:315-333. [DOI] [PubMed] [Google Scholar]
- 45.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raskin, D. M., and P. A. J. de Boer. 1999. MinDE dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raskin, D. M., and P. A. J. de Boer. 1997. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91:685-694. [DOI] [PubMed] [Google Scholar]
- 48.Raskin, D. M., and P. A. J. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothfield, L., S. Justice, and J. García-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 50.Rowland, S. L., X. Fu, M. A. Sayed, Y. Zhang, W. R. Cook, and L. I. Rothfield. 2000. Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J. Bacteriol. 182:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sen, M., and L. I. Rothfield. 1998. Stability of the Escherichia coli division inhibitor protein MinC requires determinants in the carboxy-terminal region of the protein. J. Bacteriol. 180:175-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szeto, J., S. Ramirez-Arcos, C. Raymond, L. D. Hicks, C. M. Kay, and, J. A. Dillon. 2001. Gonococcal MinD affects cell division in Neisseria gonorrhoeae and Escherichia coli and exhibits a novel self-interaction. J. Bacteriol. 183:6253-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szeto, T. H., S. L. Rowland, and, G. F. King. 2001. The dimerization function of MinC resides in a structurally autonomous C-terminal domain. J. Bacteriol. 183:6684-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trusca, D., S. Scott, C. Thompson, and D. Bramhill. 1998. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J. Bacteriol. 180:3946-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vischer, N. O. E., P. G. Huls, and C. L. Woldringh. 1994. Object-Image: an interactive image analysis program using structured point collection. Binary 6:160-166. [Google Scholar]