Abstract
The nematode-bacterium complex of Heterorhabditis-Photorhabdus is pathogenic to insect larvae. The bacteria undergo a form of phenotypic switching whereby the primary form, at the stationary phase of the growth cycle, makes a range of products and has the capacity to support nematode growth, whereas the secondary form does not express these phenotypes. The work described here investigated the mechanism regulating phenotypic variation by transforming the primary cells with secondary-form DNA on a low-copy-number vector and screening for colonies which did not produce the yellow pigment characteristic of primaries. Four transformants all carrying the same gene were found to loose primary-form-specific characteristics, and the gene was sequenced and identified as ner, a regulatory gene in gram-negative bacteria and their phages. Unexpectedly, inactivation of the endogenous gene in the secondaries did not cause them to revert to the primary phenotype, and the gene was expressed in the primary form as well as the secondary form during exponential but not stationary phase and deregulated in the plasmid-bearing primary form. These and other pieces of evidence indicate that the endogenous ner gene is not responsible for the secondary phenotype, but that ner, when overexpressed, can repress expression of primary phenotypes at stationary phase. Inactivation of the endogenous ner gene in the primary form affected the outer membrane protein profile. A number of outer membrane proteins displayed differential accumulation in the primary and secondary forms at stationary phase, and two of the primary-form-specific proteins were absent from the ner primary strain.
Photorhabdus and Xenorhabdus spp. are bacteria (family Enterobacteriaceae) carried symbiotically in the gut of nematodes of the genera Heterorhabditis and Steinernema, respectively. The nematode and bacteria act together to kill a range of insect prey (pathogenicity mechanisms reviewed in reference 10) and are used in biological pest control. In addition to producing an insecticidal toxin, the bacteria are needed for growth and reproduction of the nematodes (27). The strain used in this work fell originally into the species Xenorhabdus luminescens, which was then reclassified as a new genus with a single species, Photorhabdus luminescens. Recent taxonomic work (13) has divided Photorhabdus into three species, and the K122 strain used in this work probably falls into the P. temperata species (36).
Photorhabdus and Xenorhabdus can undergo a form of phenotypic switching, which has been called phase variation between primary (phase 1) and secondary (phase 2) forms. The bacteria isolated from the nematode or insect carcass are the primary variant, whereas secondary cells can be generated by prolonged growth at stationary phase or in low osmolarity medium, at a frequency which is strain dependent. There are a large number of primary-form-specific characteristics such as extracellular enzyme and crystal protein production, pilus and glycocalyx synthesis, bioluminescence, dye uptake, and motility (9, 16), which are lacking or present in decreased amounts in secondary cells. Moreover, the secondary form is unable to support nematode growth and reproduction (14). The primary-form-specific traits are induced at stationary phase, and many of them could derive from changes in membrane properties or secretion pathways. For example, the Photorhabdus lipase gene is expressed and lipase protein is produced in both variants, but only the primary form secretes active lipase enzyme (41).
This phenomenon has been labeled phase variation, but it probably does not result from reversible DNA instability, as is the case for the classical forms of phase variation (23). For example, several genes whose expression is primary-form specific have identical restriction patterns in the two forms (18, 20, 40, 41), and Southern cross (2) and amplified fragment length polymorphism (D. M. Roche and B. C. A. Dowds, unpublished data) analyses have failed to identify any difference in band pattern between primary and secondary total chromosomal DNA. Furthermore, reversion from the secondary to the primary form is not observed. This suggests that the secondary form may be a mutant rather than a phase variant. Some evidence for this is found in the experiments of Krasomil-Osterfeld (24), who found that low-osmolarity growth generated reversible secondaries during the first subculture, followed by stable secondaries after prolonged subculturing. The unstable isolates may be true phase variants, whereas the stable phase 2 cells may be mutants selected during prolonged culturing under stressful conditions.
Based on differences in levels of respiratory enzymes and lag times, it has been suggested that secondary cells of Xenorhabdus may be better adapted to survival as free-living organisms in the soil (34), and indeed protein profiles of primary and secondary cultures of Photorhabdus are identical during exponential phase but strikingly different at stationary phase (29), suggesting a role for the secondary form under starvation conditions. It has been proposed that the stable secondaries are an artifact of in vitro conditions, and that it is the reversible forms that have a function in vivo (24). The manner in which the secondary variant is usually isolated—prolonged incubation at stationary phase—provides the conditions for selection of multiple mutations, suggesting the possibility that the secondary is more analogous to a growth-advantage-in-stationary-phase (GASP) mutant (42) than a phase variant.
Whether or not the secondary form is a spontaneous mutant, it is clear that the primary-phase-specific phenotypes are coordinately regulated, and the induction of phase shift by multiple independent transposon insertions in X. nematophilus (39) implies that a complex pathway regulates this kind of phenotypic variation. In the work described here, we hypothesize that phenotypic variation in Photorhabdus is controlled by a cascade of regulatory genes and ask whether a repressor inhibits expression of the phase-specific phenotypes in the secondary form. We describe a gene which transforms primary cells of Photorhabdus sp. strain K122 into a secondary-like form, and which was identified as one of the ner family which codes for a DNA binding protein in a number of phages and bacteria (3). In phage Mu, Ner inhibits expression of the lysogenic repressor gene from the Pc promoter and, in conjunction with integration host factor, negatively autoregulates early transcription from the Pe promoter to ensure low-level expression of genes needed for replicative transposition (reviewed in reference 21).
MATERIALS AND METHODS
Strains and plasmids
Strains and plasmids are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype and/or description | Source or reference |
|---|---|---|
| Strains | ||
| Photorhabdus K122 primary | Wild isolate from Ireland | 22 |
| Photorhabdus K122 secondary | Isolated from prolonged stationary-phase culture of primary cells | 41 |
| Photorhabdus KN1 | Primary K122 with ner disrupted | This study |
| Photorhabdus KN2 | Secondary K122 with ner disrupted | This study |
| E. coli Tg-1 | supE44 hsdΔ5 thi-1 Δ(lac-proAB) F′(traD36 proAB+lacIqlacZΔM15) | Stratagene |
| E. coli XL1-Blue | F′ proAB lacIqZΔM15 Tn10(Tetr) | Stratagene |
| Plasmids | ||
| pBR322 | Apr Tetr with BamHI site, 4.4 kb | Promega Corporation |
| pGEM-7Zf(−) | Apr, multiple cloning site, 3 kb | Promega Corporation |
| pHP45Ω-Km | Source of Kmr interposon | 12 |
| pBS115 | Source of sacB gene; same as pBS101 (4) except sacB/sacR not under control of the lac promoter | Todd Ciche |
| pK2B | 1.5-kb insert containing ner in BamHI site of pBR322 | This study |
| pK2N101 | 1.2-kb ner-containing insert from pK2B in EcoRI site of pGEM7Zf(−) | This study |
| pK2N102 | 0.85-kb insert (without ner) from pK2B between EcoRI and SphI sites in pGEM7Zf(−) | This study |
| pDR100 | pK2B with ner disrupted by Kmr interposon from pHP45Ω-Km | This study |
| pDR101 | pDR100 with sacB gene from pBS115 in EcoRV site | This study |
Media and phenotypic analysis.
Luria-Bertani (LB) and other media were prepared as described by Sambrook et al. (32). Dye indicator media were McConkey agar (MCA, Oxoid); McConkey broth purple agar (MCBPA, Oxoid); 2% proteose peptone no. 2 agar with eosin Y (400 mg/liter) and methylene blue (65 mg/liter) (EBA); nutrient agar with bromothymol blue (25 mg/liter) and 2,3,5-triphenyltetrazoleum chloride (30 mg/liter (NBTA); and Congo Red medium (Congo Red [0.01%] in LB).
Antibiotic activity against Micrococcus luteus ATCC 4698 was determined by the method of Akhurst (1); motility on swim agar was determined by the method of Givaudan et al. (19); and lipase activity was assessed on Tween 80 agar plates (5). Protease was assayed in culture supernatant from stationary-phase cultures (33) or on gelatin nutrient agar (6). Hemolysin activity was determined on plates containing blood agar base (bioMerieux) supplemented with 7% horse blood. The accumulation of crystal proteins was assessed by growing Photorhabdus cultures in LB at 30°C with shaking at 200 rpm for 30 h followed by separation of the total intracellular proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (20%) (32).
Preparation and screening of a secondary-phase library.
Chromosomal DNA from K122 secondary cells (Table 1) was partially digested with Sau3A and enriched in fragments of 2 to 10 kb by sucrose gradient centrifugation. These were ligated into BamHI-cut pBR322 and transformed into Escherichia coli Tg-1. The library of recombinant plasmids was transformed into phase 1 cells of K122 by electroporation as follows. An early-exponential-phase culture of K122 (optical density at 600 nm [OD600] = 0.2) was chilled on ice for 90 min and harvested by centrifugation. The cells were washed three times with ice-cold 1 mM HEPES (pH 7.0). The cells were resuspended in an equal volume, 0.5× volume and 0.01× volume of HEPES after each successive wash. All apparatus and solutions in immediate contact with the cells during electroporation were cooled on ice.
Up to 100 ng of DNA in a 1- to 5-μl volume was gently mixed with 40 μl of cells, and the mixture was transferred to an electroporation cuvette (0.1-cm gap width) which was placed in a GenePulser (Bio-Rad) The cell-DNA mix was subjected to an electric pulse of 21 kV/cm with a time constant of 4.5 ms (settings of 2.1 kV, 100 Ω, and 25 μF). Luria broth (1 ml) was then gently mixed with the cells, and the suspension, transferred to a test tube, was shaken at 200 rpm for 3 h at 28°C. The cells were then plated on LB plus ampicillin (150 μg/ml), and 3 × 104 transformants were screened for white colonies in a background of yellow-pigmented primary colonies.
DNA sequencing and sequence analysis.
DNA sequencing of the ner gene and its flanking region was carried out on the insert of the pK2B clone and on chromosomal DNA from K122 primary and secondary cells. Chromosomal DNA was sequenced using primers DR100 (5′-CTGGACCATGGTCGAAAGACACGATTGGC-3′) and DR101 (5′-GGCGTGGATCCTCTGCTCGTACCTCAGAC-3′) to obtain the sequence of the coding region of the gene and KON1 (5′-CCGAACAGTGAGATAAAC-3′) and KON2 (5′-CAGCGCCGCAATAATATCAG-3′) to sequence 500 bp upstream of the gene. Restriction sites and open reading frames were identified using DNAsis analysis software for Macintosh (version 1.0). Homology searching was carried out using the National Center for Biotechnology Information (NCBI) databases from the website of Baylor College of Medicine, Texas (http://dot.imgen.bcm.tmc.edu:9331). Protein sequences were aligned using ClustalW alignment software, and the helix-turn-helix motif was identified using the MotifFinder program of the GCG sequence analysis suite (University of Wisconsin, Madison).
ner gene disruptions.
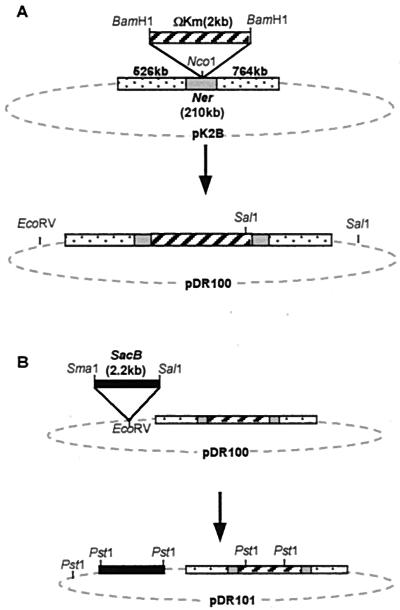
The ner gene was disrupted on pK2B as follows. pK2B (Fig. 1A) was cleaved with NcoI, which cuts the plasmid once, within the ner gene (Fig. 2). pHP45Ω-Km was cut with BamHI, which removes the interposon. The linearized plasmid and the interposon had their 5′ protruding ends filled in using the Klenow fragment of DNA polymerase (Promega), and the blunt-ended fragments were ligated to generate pDR100 (Fig. 1A). The plasmid was transformed into E. coli XL1-Blue, and transformants were selected on LB plus kanamycin (50 μg/ml) plus ampicillin (100 μg/ml).
FIG. 1.
Structure of pK2B and construction of pDR100 (A) and pDR101 (B).
FIG. 2.
Nucleotide sequence of the insert in pK2B. The ner ORF is underlined, and the predicted sequence of the Ner protein is shown underneath. The boxes indicate the locations of two perfect 13-bp direct repeats.
The pDR100 construction was confirmed in two ways. First, primers DR100 and DR101 were used to amplify a fragment including the ner gene. A 300-bp PCR product was obtained as expected from pK2B, whereas this band was not generated from pDR100, and the anticipated fragment of 2.3 kb was too large to be amplified under these conditions. Second, pDR100 and pK2B were cut with EcoRV and SalI, revealing that an extra SalI site had been introduced with the interposon, as expected; in addition, the band sizes from the single and double digests were as expected.
The endogenous ner gene was disrupted by marker exchange-eviction mutagenesis (31). First, the sacB gene was introduced into pDR100 as follows. pDR100 was cleaved with EcoRV, which linearized the plasmid outside the ner region. pBS115 was cut with SmaI and SalI, which removes the sacB gene. This fragment had its protruding end filled in with Klenow as above and was blunt-end ligated into linearized pDR100. The resultant pDR101 (Fig. 1B) was transformed into E. coli XL1-Blue, and Kmr sucrose-sensitive (Sucs) colonies were obtained after replica plating on LB plus kanamycin (50 μg/ml) plus ampicillin (100 μg/ml) with and without sucrose (5%). Plasmid DNA was prepared and found to have the appropriate restriction sites.
K122 primary and secondary cells were transformed with pDR101 by electroporation. The electroporated cells were grown in liquid LB overnight. Transformants were then selected on LB plus kanamycin (50 μg/ml) plus sucrose (5%). Colonies which grew are expected to have lost the sacB gene and taken up the Kmr gene by recombination onto the chromosome via a double crossover mediated by the ner sequences flanking the interposon. The loss of the plasmid was confirmed by finding that the Sucr Kmr colonies were also Aps by replica plating on LB with and without ampicillin (100 μg/ml).
To confirm that the endogenous ner gene was indeed disrupted by the interposon, the chromosomal DNA was subjected to PCR (using the same procedure as for pDR100, above) and Southern blotting, and in both cases the expected band sizes were obtained. For the Southern blots, chromosomal DNA was prepared from K122 phases 1 and 2, and the two putative disrupted strains KN1 (ner disrupted in primary cells) and KN2 (ner disrupted in secondary cells). The DNA was cleaved with EcoRV, an enzyme that does not cut within the ner-interposon region. Two blots were made and probed with (i) the ner gene (PCR product from primers DR100 and DR101) and (ii) the Kmr interposon cleaved from pHP45Ω-Km with BamHI. The ner probe revealed a band in KN1 and KN2 that was 2 kb larger than the band in K122 phase 1 or 2 cells, as expected from the size of the interposon fragment being 2 kb. The interposon probe hybridized to a band, the same size as the ner-hybridizing band, that was present in KN1 and KN2, but not in K122 phase 1 or 2 DNA.
Southern and Northern blots.
Southern blotting was carried out as described by Sambrook et al. (32). A 453-bp restriction fragment carrying the ner gene was labeled with digoxigenin (Boehringer Mannheim), and hybridization followed the manufacturer's instructions. For the Northern blot, K122 primary, secondary, and primary/pK2B cultures were grown to OD600s of 0.3, 0.6, 1.0 (exponential phase), and 4 to 5 (stationary phase), and RNA was extracted using Trizol reagent (Gibco-BRL). Then 10 μg of RNA of each sample was loaded onto a 1% formaldehyde gel (32) alongside RNA molecular weight markers (Sigma). The gel was blotted onto Hybond N+ membranes after electrophoresis. The probe was a PCR product of the ner gene made by using the DR100 and DR101 primers. Labeling and detection were carried out with the ECL system from Amersham, and the film used was BioMax Light-1 (Kodak). Relative intensities of the chemiluminescent signals were determined by scanning the 2-min and 2-h exposure films using an Eagle Eye 11 still video system (Stratagene).
Outer membrane protein preparation and electrophoresis.
Outer membrane proteins were made by an adaptation of the procedure of Forst et al. (15). Cultures were grown in 50 ml of LB to OD600 values of 0.8 for exponential-phase samples and 4.0 to 4.5 for stationary-phase samples. Cells were pelleted, resuspended in 20 ml of ice-cold 20 mM sodium phosphate buffer (pH 7.5), and lysed in a French press (SLM-Aminco) at a pressure of 2,000 lb/in2. After removing the cell debris, the supernatant was ultracentrifuged to precipitate the outer membrane proteins as described by Forst et al. (15). The pellets were resuspended in 15 ml of 0.5% sodium N-lauryl sarcosinate and incubated for 30 min at room temperature. The samples were again spun in the ultracentrifuge as above, and the pellets were resuspended in 150 to 200 μl (depending on the pellet size) of gel loading buffer containing 1% SDS. The samples were boiled for 5 min before running 4 to 6 μl per lane on SDS-10% PAGE with or without 8 M urea (28).
Nucleotide sequence accession number.
The sequence of the 1,497-bp insert of pK2B has been deposited with GenBank and given accession number AF157489.
RESULTS
Screening for a phase variation regulatory gene.
A library of secondary-phase Photorhabdus sp. strain K122 DNA was constructed. The bank consisted of 2- to 10-kb enriched Sau3A fragments of phase 2 DNA inserted into the tetr gene of pBR322 at the BamHI site. The library was transformed into primary-phase K122, and 3 × 104 transformant colonies were screened for loss of the yellow pigment specific to primary colonies. The average insert size of the library was 2.5 kb, and 80% of vector molecules carried inserts. Thus, the entire genome (5.35 × 106 bp) was represented 11 times.
Four nonpigmented colonies were identified, and in each case the recombinant plasmid was isolated and retransformed into primary cells. In all four cases, transformation resulted in loss of pigment, while control cells transformed with pBR322 retained the yellow pigmentation characteristic of primary cells. In addition, the colonies had a nonmucoid consistency, did not bioluminesce, and were lipase negative on Tween agar, all characteristics of the secondary form. Two of the clones had inserts of 1.5 kb and two of 1.55 kb, and restriction maps revealed that all had the same sites for the restriction endonucleases AatII, EcoRI, and HpaI, so it was apparent that the four clones contained separate isolates of the same gene.
DNA sequence.
The insert of one of the clones, pK2B (Fig. 1A), was sequenced on both strands (Fig. 2). It was 1,497 bp in length, and contained an open reading frame (ORF) of 210 bp and a number of smaller ORFs, most too small to be protein-coding sequences. The 210-bp ORF displayed homology to genes specifying a DNA-binding protein, ner of phage Mu and nlp (ner-like protein) of E. coli. The same result was obtained when the DNA sequence was translated in all six possible reading frames and compared with the nonredundant protein sequence database. The most closely related organisms were Mu for the DNA sequence and E. coli for the protein sequence.
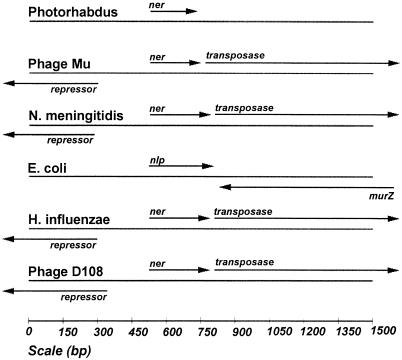
The sequences flanking ner do not show any significant homology to any sequences in the database and do not contain protein-coding regions. In contrast, the regions flanking ner/nlp in other species do contain other genes, and the gene organization falls into two classes, neither of which matches the arrangement in Photorhabdus (Fig. 3). One group includes phages Mu and D108 as well as Haemophilus influenzae and Neisseria meningitidis, suggesting that nlp is carried on a prophage in these bacterial genomes. A second group contains E. coli and Salmonella enterica serovar Typhimurium, which have different genes upstream but retain the murZ gene (coding for the Mu transposase) downstream, though in reverse orientation.
FIG. 3.
Genes flanking ner in phages and bacteria.
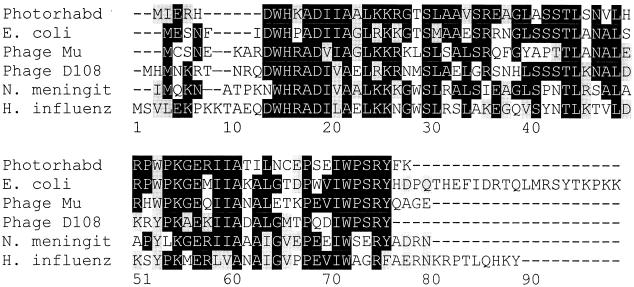
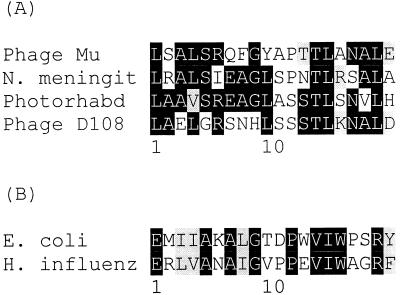
Figure 4 shows the best alignment of the predicted Ner protein sequence with other sequenced Ner homologues. The closest sequence is Nlp of E. coli, which displays 68% identity (77% conserved substitutions). A helix-turn-helix motif was identified in Ner, spanning amino acids 23 to 42. This region has been aligned with the helix-turn-helix in the other proteins (Fig. 5), and based on similarity scores, the six proteins can be divided into two groups: one includes Photorhabdus, Mu, D108, and N. meningitidis, and the other contains E. coli and H. influenzae. The two groups also differ somewhat in the size of the proteins, the first having proteins of 70 to 76 amino acids and the second having proteins of 89 to 92 amino acids.
FIG. 4.
Best-score alignment of predicted Ner protein sequences. Photorhabd, Photorhabdus sp. strain K122; N. meningit, N. meningitidis; H. influenz, H. influenzae.
FIG. 5.
Alignment of helix-turn-helix motifs of predicted Ner protein sequences. N. meningit, N. meningitidis; Photorhabd, Photorhabdus sp. strain K122; H. influenz, H. influenzae.
Instability of ner-containing subclone.
The insert of pK2B with a small amount of flanking vector DNA was excised as two fragments by cleavage of pK2B with EcoRI [cuts once each in vector and insert (see Fig. 2 for insert site)] and SphI (cuts once in vector). The two fragments were subcloned into pGEM7Zf(−) as inserts of 1,212 bp, including the 210-bp ner ORF and an 847-bp insert, to generate pK2N101 and pK2N102, respectively. Phase 1 cells transformed with pK2N102 were primary-like in every test, whereas transformants with pK2N101 divided into two populations. One population of nonpigmented colonies were secondary-like in every test (Table 2), while the second population of yellow colonies remained primary-like. The ratio of nonpigmented to pigmented transformants was 7:1.
TABLE 2.
Phenotypes of phase 1 cells transformed with ner-containing plasmid pK2N101 (white colonies)
| Phenotype | Primary | Secondary | Primary/pK2N101 (white transformants) |
|---|---|---|---|
| Pigment (on LB) | Yellow | White | White |
| Colony consistency | Mucoid | Nonmucoid | Nonmucoid |
| Bioluminescence | +++ | − | − |
| Lipase | +++ | − | − |
| Protease | +++ (6.2)a | − | + (2.4)a |
| Antibiotics | +++ | − | − |
| Crystal proteins | + | − | − |
| Dye uptake | |||
| NBTA | Green | Red | Red |
| MCA | Red | Pink | Pink |
| EBA | Black | Purple | Purple |
| MCBPA | Green | Purple | Purple |
| Congo Red | ++ | − | − |
| Hemolysin | ++ | − | − |
Values represent units of protease per milliliter of supernatant from cultures grown to an OD600 of 3.
The white transformants remained stable when subcultured, and the plasmid could be isolated (albeit at low yields) from these cultures. On the other hand, attempts to isolate the plasmid from pK2N101 yellow transformants failed to yield DNA. However, it was clear that the plasmid had not been lost, because subculturing of cells from the yellow pK2N101 transformants yielded white colonies, yellow colonies, and colonies that had sections of yellow pigment as well as nonpigmented sections. It appears that the instability is associated with the loss of the 660 bp downstream of ner that is present in the stable pK2B construct but absent from pK2N101.
One possible cause of the instability is unequal division of plasmid copies between daughter cells following cell division. Whatever the reason, it is clear that the region of secondary DNA in pK2N101 carrying the ner ORF could induce a primary to secondary transition (Table 2).
Plasmid curing.
To further demonstrate that the ner gene was responsible for phenotypic switching when transformed into primary cells, primary/pK2B and primary/pK2N101 (white) cells were cured of their respective plasmids, and the phenotype was assessed with respect to pigmentation. The two cultures were grown in LB medium without ampicillin for three subcultures of 4 days each, until pigment was detected in the flasks. Samples were then plated on LB agar without ampicillin, and a mixture of white and yellow colonies grew. Twenty white and 20 yellow colonies from each culture were spotted onto LB agar with and without ampicillin, and all of the yellow colonies were found to be luminescent and ampicillin sensitive, while all the white colonies were nonluminescent and ampicillin resistant. Thus, there was 100% correlation between loss of the plasmid and primary pigmentation and luminescence.
Insertional inactivation of ner on pK2B.
The ner gene was disrupted with an interposon on pK2B (Fig. 1A), and the resultant plasmid, pDR100, was transformed into primary and secondary cells. Primaries, secondaries, and transformants were subjected to plate assays for pigmentation, bioluminescence, lipase protease, color on NBTA and Congo Red media, motility on swim and swarm agars, and the production of antibiotics. Neither the original ner+ plasmid nor the ner-disrupted plasmid had any effect on the phenotypes of secondary cells. On the other hand, the ner+ plasmid converted primary cells into secondary variants, as already described, but the disrupted plasmid failed to generate a phenotypic shift. This is further confirmation that the phenotypic switching is caused by the ner gene on the plasmid.
ner gene structure and expression in primary and secondary cells.
DNA from primary and secondary cells was cleaved with HindIII, EcoRV, AvaI plus ClaI, and BamHI plus EcoRI and hybridized to a probe containing the ner gene. In all digests, just one band (smallest, 1.4 kb in size) hybridized to the probe, implying that there is a single copy of the ner gene in strain K122. The same size bands hybridized in DNA from the primary and secondary. Thus, the crude arrangement of the DNA around the ner gene appears to be the same in the two forms. This was confirmed by sequencing primary and secondary chromosomal DNA including the ner gene plus 500 bp upstream and 60 bp downstream of the coding region. The sequences on the two chromosomes were identical, and identical to that of the secondary gene insert in pK2B shown in Fig. 2.
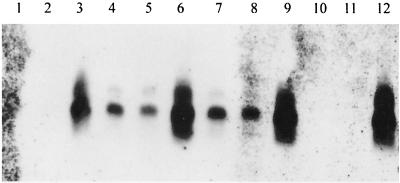
It might be expected that the ner gene would be expressed in secondary but not in primary cells. However, a Northern blot revealed that a 350- to 400-nucleotide RNA complementary to the gene was present in both forms in approximately equal amounts (Fig. 6). A larger minor band was also complementary to the probe, suggesting that there is a second transcription initiation site upstream of the main start point. The ner transcript became detectable between culture OD600 values of 0.3 and 0.6 and then doubled in amount between OD600 = 0.6 and OD600 = 1.0 (late exponential phase), before declining in stationary phase (OD600 = 4). This pattern of gene expression was confirmed with quantitative reverse transcriptase-PCR (Q-RT-PCR) (method in reference 8), where the levels of ner RNA were measured relative to 16S rRNA (data not shown).
FIG. 6.
Expression analysis (Northern blot) of the ner gene. The gel contained RNA from primary (lanes 1, 4, 7, and 10), secondary (lanes 2, 5, 8, and 11), and primary/pK2B (lanes 3, 6, 9, and 12) cells grown to culture OD600 values of 0.3 (lanes 1 to 3), 0.6 (lanes 4 to 6), 1.0 (lanes 7 to 9), and 4 to 5 (lanes 10 to 12).
Primary-form phenotypes are expressed at stationary phase and not during the exponential phase of the growth cycle (14, 16, 29). One possible explanation of the data so far is that Ner is responsible for this exponential-phase repression in the primary. The effects of ner-carrying plasmids on the primary form might be to maintain a high level of Ner at stationary phase so that the primary phenotypes remain repressed at this stage of the growth cycle. Thus, the levels of ner mRNA were measured in the primary strain carrying the pK2B plasmid. It was indeed found that the levels of ner RNA as a proportion of total RNA rose steadily from early to late exponential phase and continued to rise during stationary phase (Fig. 6). In fact, expression levels were much higher (80-fold higher in primary/pK2B than in the primary at late exponential phase) than expected based on the copy number (copy number, 2 to 5) of the plasmid (data not shown), implying that Ner may positively regulate its own expression or that multiple copies of the gene titrate out a negative regulator.
The data suggest that high-level expression of ner at stationary phase in primary/pK2B overrides stationary-phase-specific derepression of primary phenotypes. It is clear from Fig. 6 that the wild-type regulation of ner gene expression (i.e., switched off at stationary phase) does not pertain to the primary carrying pK2B. Rather than expression falling to an undetectable level, it actually increases twofold between the late exponential (OD600 = 1.0) and stationary (OD600 = 4.0) phases. The multiple copies of the gene may titrate out a stationary-phase-specific repressor.
The possibility remains that despite having the same RNA levels, the primary and secondary forms have different levels of Ner protein. Attempts were made to prepare antiserum for use in comparing levels of Ner protein. Ner protein was purified (results not shown) and injected into rabbits, but unfortunately, the protein was insufficiently antigenic, and the antiserum produced failed to detect Ner on Western blots. Thus, a tentative conclusion about Ner levels in the two forms and the recombinant strain must be based on ner RNA levels. Since the level of ner RNA is the same in the primary and secondary forms, we conclude that expression of the endogenous ner gene is probably not sufficient to generate the secondary phenotype. This was investigated further by inactivating the ner gene in the two forms (see next section).
ner mutants.
The endogenous ner gene on the chromosome was disrupted in the primary and secondary strains by allelic exchange with ner::ΩKm on pDR101 (Fig. 1B) to generate strains KN1 and KN2, respectively, and Q-RT-PCR showed that the ner gene is not expressed at a detectable level in either strain (data not shown). The cultures were assayed for a number of phase variant characteristics (Table 3), and in both cases, the disruption did not affect the viability of the cells. KN2 was identical to the secondary form for all phenotypes tested, showing that the lack of intact ner in phase 2 cells does not cause them to revert to a primary-like phenotype. These results are consistent with the expression results, where the lack of ner expression in the secondary at stationary phase did not cause the cells to express primary-form-specific characteristics.
TABLE 3.
Phenotypes of K122 with ner gene disrupted
| Phenotype | Primary (ner+) | Secondary (ner+) | KN1 (ner::ΩKm) | KN2 (ner::ΩKm) |
|---|---|---|---|---|
| Pigment (on LB) | Yellow | White | Yellow | White |
| Colony consistency | Mucoid | Nonmucoid | Mucoid | Nonmucoid |
| Bioluminescence | +++ | − | +++ | − |
| Lipase | +++ | − | +++ | − |
| Protease | +++ | − | +++ | − |
| Antibiotics | +++ | − | +++ | − |
| Crystal proteins | + | − | + | − |
| Dye uptake | ||||
| NBTA | Green | Red | Green | Red |
| Congo Red | ++ | − | ++ | − |
| Hemolysin | ++ | − | ++ | − |
In the previous section, it was hypothesized that ner regulates growth phase-specific expression of the primary phenotypes. If this hypothesis is correct, then the primary form with a nonfunctional ner gene might be expected to display exponential-phase expression of primary-form-specific phenotypes. This was tested by sampling strain KN1 and the primary form at various stages of the growth cycle (culture OD600 of 0.2, 0.4, 0.6, 1.2, and 1.8) and assaying for protease activity, bioluminescence, and pigment concentration. The two strains showed identical patterns of undetectable levels of these activities until stationary phase (culture OD600 = 1.8). Thus, the ner gene in single copy does not appear to play a role in growth phase regulation of phenotypic variation. Its repression of primary characteristics at stationary phase requires a higher than normal level of expression of the gene, as observed in the primary carrying pK2B.
In order to find a possible role for the endogenous ner gene, the primary and secondary forms were compared to their respective ner knockout strains, KN1 and KN2, for a range of phenotypes. No difference was found between the secondary and KN2, while the primary was identical to KN1 for all characteristics tested (Table 3) except for some differences in outer membrane proteins.
Outer membrane protein conformation.
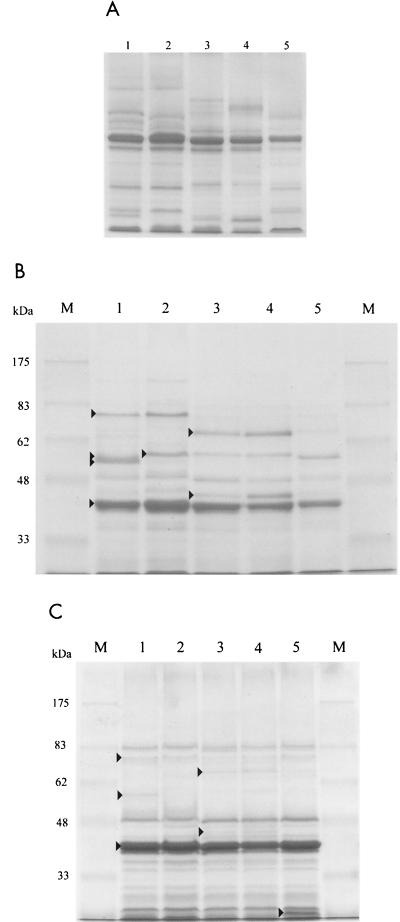
Outer membrane proteins were prepared from cultures grown to stationary phase. It is clear that both the protein conformation, as determined on urea gels (Fig. 7A), and the protein composition, as shown on SDS-PAGE (Fig. 7B), are quite different in the primary (lanes 1) and secondary (lanes 3) forms. In particular, the primary has major bands of 77, 58, and 56 kDa as well as some less intense bands which are missing from the secondary, while the secondary has proteins of 69, 59, and 46 kDa which are absent from the primary. The outer membrane protein difference is the only positive secondary-form-specific phenotype found that is not simply an absence of a primary gene product. While pK2B apparently transformed the primary into a secondary with respect to all other phenotypes tested, it did not have this effect on the outer membrane protein pattern; this suggests that the effect of pK2B is to eliminate primary-form-specific gene expression rather than to convert the primaries into secondaries. The primary/pK2B (Fig. 7A and B, lanes 5) transformant had a depleted outer membrane protein profile. It lacked the 77- and 56-kDa primary-form-specific bands as well as the 69-, 59-, and 46-kDa bands specific to the secondary.
FIG. 7.
Gels of outer membrane proteins from primary (lanes 1), KN1 (lanes 2), secondary (lanes 3), KN2 (lanes 4), and primary/pK2B (lanes 5) strains. (A) Urea gel of stationary-phase outer membrane proteins. (B) SDS-PAGE of stationary-phase outer membrane proteins. Bands marked with arrows have molecular masses of 77, 58, 56, and 42 kDa (lane 1), 59 kDa (lane 2), and 69 and 46 kDa (lane 3). (C) SDS-PAGE of exponential-phase outer membrane proteins. Bands marked with arrows have molecular masses of 77, 58, and 42 kDa (lane 1) and 69 and 44 kDa (lane 3). The arrow in lane 5 indicates a band that is unique to primary/pK2B. Lanes marked M contained molecular size markers.
With regard to the effect of ner deletion on outer membrane proteins, there were no reproducible differences between the wild-type secondary (Fig. 7B, lane 3) and the secondary with the ner gene disrupted (KN2) (Fig. 7B, lane 4). However, there were significant and reproducible differences between the wild-type primary (Fig. 7B, lane 1) and the primary with the ner gene disrupted (KN1) (Fig. 7B, lane 2). The 58- and 56-kDa primary-form-specific bands were absent, and the secondary-form-specific 59-kDa protein was present in KN1 (Fig. 7B). Other than these differences, KN1 looked identical to the primary. One possible explanation is that Ner is required for production of a protease that removes fragments from the 59-kDa protein to generate the 58- and/or 56-kDa proteins. On the urea gel (Fig. 7A), one primary band was missing from KN1, while a cluster of bands above the major porin protein migrated to different positions in the two strains. Thus, the small difference in protein composition (Fig. 7B) generated a somewhat larger effect on protein conformation (Fig. 7A).
The samples analyzed in Fig. 7A and 7B were extracted from cells grown to stationary phase. To determine which of the differentially expressed proteins were growth phase specific, outer membrane proteins were prepared from cells grown to exponential phase (OD600 = 0.8) and separated by SDS-PAGE (Fig. 7C). It can be seen that the outer membrane protein profile is similar in the primary, secondary, KN1, and KN2 strains at exponential phase, though with some minor bands displaying differential accumulation (see arrows in lane 3 and middle arrow in lane 1). Interestingly, the 77-kDa protein, which is primary-form specific at stationary phase, was present in all strains at a low level during exponential phase. There was one major difference between the primary carrying pK2B and the other strains at exponential phase in that the former contained an additional low-molecular-weight outer membrane protein (Fig. 7C).
With the exception of the major 42-kDa protein, the outer membrane protein profiles were quite different during exponential and stationary phases, and none of the proteins which were differentially expressed between strains at stationary phase (Fig. 7B) were major bands during exponential phase (Fig. 7C). The fact that all of these primary- and secondary-form-specific outer membrane proteins are induced at stationary phase is another indication of the link between growth phase and phenotypic variation.
DISCUSSION
Primary-form cells of Photorhabdus produce a large number of activities that are absent or present in greatly reduced amounts in the secondary form. Thus, a working hypothesis was made that secondary cells might contain an active repressor gene that is inactivated or not expressed in the primary form. The lack of a transposon mutagenesis system for this species at the time led to attempts to identify the proposed repressor by screening primary cells transformed with secondary DNA for colonies that had undergone a shift in pigmentation. Screening of primary colonies transformed with secondary DNA on a low-copy-number plasmid yielded four colonies out of 30,000 that exhibited phenotypic switching. All four contained the ner gene, which is therefore likely to be the only gene with the capacity to repress primary-form-specific phenotypes.
The ner gene codes for a DNA-binding protein which was first identified in phage Mu, where it regulates Mu transposition and repressor synthesis (21). Ner functions as a repressor in phages Mu and D108; it inhibits expression from the Pc promoter, and in conjunction with the E. coli integration host factor, it regulates expression from the Pe promoter (25). The Ner protein has a helix-turn-helix DNA recognition motif (35), and the structure of the operator to which it binds has been elucidated (26). A ner homologue has also been found in E. coli (7) and in the genomes of other gram-negative bacteria such as Neisseria spp. (37). The ner homologue is nonessential for E. coli viability (3), and its function in bacterial cells is unknown. When overexpressed, it stimulates expression of maltose- and lactose-utilizing enzymes in a mutant strain that lacks adenylyl cyclase but has partial cyclic AMP receptor protein activity in the absence of cyclic AMP (7). It appears likely that Ner functions as a DNA-binding regulatory protein, probably a repressor, in Photorhabdus spp., but the sequences to which it binds have not yet been identified. Thus, the effects reported in this paper may be exerted directly or indirectly.
It is unlikely that phenotypic shifting is mediated by ner in the normal situation because (i) the gene has the same sequence in the two variants; (ii) insertional inactivation of the gene in secondary cells did not cause them to switch into the primary form; and (iii) the gene is expressed in primary as well as secondary cells. This implies that ner only represses primary-form-specific phenotypes when present in additional copies. The copy number of pK2N101 in K122 was estimated at 2 to 5 (data not shown), setting an upper limit on the number of additional copies. It is probable that more than one copy of plasmid-encoded ner in addition to the endogenous copy is needed to effect the phase shift, since the yellow pK2N101 transformants were unstable, revealing that they contained at least one copy of the plasmid, which nevertheless failed to induce phase shifting. Expression of the ner gene was found to be deregulated in the strain containing the plasmid, so that the concentration of ner mRNA was much higher than expected based on copy number (e.g., 80-fold higher in primary/pK2B than in the primary at late exponential phase and an even greater difference at stationary phase).
The very high levels of ner mRNA in primary/pK2B at stationary phase suggests that overexpression of ner may repress stationary-phase-specific expression of the primary phenotypes. Nevertheless, ner in single copy does not regulate the growth phase effect, since the ner-disrupted strain KN1 was found not to display exponential phase expression of primary-form-specific phenotypes. Nor does single-copy ner play a role in differential expression between primary and secondary forms. This suggests that the effect of ner on primary cells depends on its overexpression at stationary phase leading to repression of stationary-phase-, primary-form-specific gene expression.
The finding that ner causes stationary-phase repression of primary-form-specific phenotypes only when overexpressed suggests that the gene or gene product is titrating a protein which exerts global regulation of primary characteristics or that Ner protein in excess is affecting expression of a global regulator gene or the genes coding for the primary-form-specific products, e.g., by competing for an operator. One protein known to destabilize repression by Ner is the histone-like protein integration host factor (25), so it could conceivably play a role in phenotypic variation. However, no genes other than ner were found to repress primary-form-specific phenotypes, so it must be asked why this proposed global regulator was not picked up in the screen. It is possible that the hypothesized regulator is a repressor that is lethal in multiple copies. Alternatively, it may be an activator that would only be identified by a reverse screen, i.e., transformation of a primary bank into secondary cells and screening for activation of pigment production. UV mutagenesis of a secondary culture yielded one primary form, but only after screening 150,000 colonies (29), suggesting the possibility that the mutant strain might contain multiple mutations. Thus, the secondary repressor model for phase switching may not be valid, or inactivation of the putative repressor gene may render the cells nonviable. Work is currently under way in the laboratory to identify such regulators of phase variation by transposon mutagenesis of primary and secondary cells.
Outer membrane proteins have not been analyzed before now in Photorhabdus spp. but have been studied extensively by Forst and colleagues in the related nematode symbiont Xenorhabdus nematophilus. In this species, the predominant outer membrane protein is the 30-kDa OmpF-like porin OpnP (17). By analogy, the major 42-kDa protein observed in K122 may also be a porin protein related to OmpF; this was the major protein present in all strains examined during both the exponential and stationary phases of the growth cycle.
The outer membrane protein profiles of the primary, secondary, KN1, and KN2 strains showed only minor differences during exponential phase, and primary/pK2B had the same pattern except for major accumulation of an additional low-molecular-weight protein. At stationary phase, no differences were detected between wild-type and ner-disrupted secondary cells (KN2), but there were differences between the wild-type and ner mutant primary cells (KN1). This is the first indication of a function for Ner in Photorhabdus in single copy. The outer membrane protein profile of the primary with ner insertionally inactivated differed from the wild-type primary in the replacement of the 58- and 56-kDa bands in the primary with a 59-kDa band in KN1 (the same band substitution is also found in the secondary). One possible explanation for this replacement is that Ner directly or indirectly controls expression of a specific protease which processes the 59-kDa outer membrane protein. Work is currently under way in the laboratory to screen a bank of promoter-lacZ transcriptional fusions for expression in the presence of an inducible ner gene. It is expected that this approach will reveal the genes directly controlled by ner.
The stationary-phase outer membrane proteins that differ between the primary and KN1 are a subset of the differences between primary and secondary cells. Therefore, outer membrane proteins are controlled independently by ner and another regulator of phenotypic variation. This was confirmed by finding that the primary containing multiple copies of ner (on pK2B) lacked some of the primary-form-specific and some of the secondary-form-specific proteins. The absence of the 77- and 56-kDa primary-form-specific bands from the primary/pK2B as well as the secondary strain and the induction of both of these proteins at stationary phase suggest a possible role for them in regulation of phenotypic variation. Alternatively, the secondary-form-specific outer membrane proteins of 69, 59, and 46 kDa may block induction of primary-form-specific phenotypes at stationary phase. Such regulation by outer membrane proteins is hypothesized to constitute control, not of gene expression, but of uptake into or release from the cell.
The outer membrane is a selective diffusion barrier whose constituent proteins alter in response to environmental change, such as temperature, osmolarity, oxygen availability, and growth phase in the case of X. nematophilus (16, 17). A large proportion of primary-form-specific characteristics are surface associated (e.g., pili, glycocalyx, flagella, the dye uptake) or secreted (e.g., lipase, protease, lecithinase, DNase, hemolysin, and antibiotics) and might conceivably be affected by the porin properties of the outer membrane. A more likely possibility is that the effect of multiple copies of ner on outer membrane protein production affects phenotypic variation indirectly by affecting environmental sensing of a signal which induces expression of these phenotypes at stationary phase in the primary. The signal pathway might be a quorum-sensing pathway or a two-component regulatory system.
Few genes involved in regulating phase variation have been identified from Photorhabdus, but two interesting candidates have been found. These are the cipA and cipB genes, which code for the crystal proteins produced by P. luminescens in the primary form only. The crystal proteins have no significant similarity to other known proteins, and their function is not known, though some possibilities, such as insecticidal toxin activity, have been ruled out. Insertional inactivation of both of these genes rendered the cells nonviable. However, inactivation of either gene alone in primary cells generated mutants that exhibited a partial phenotypic switch (4), including loss of the primary-form-specific ability to support nematode growth and reproduction. The mechanism—direct or indirect—whereby cipA and cipB regulate phase variant phenotypes is not known.
Xenorhabdus spp. undergo a similar range of phenotypic switching whose mechanism is equally poorly understood. Recently, a gene was inactivated in primary cells of X. nematophilus which generated a strain that was secondary-like for most phenotypes tested (39). In addition, the mutant was defective in its packaging or survival in or release from the nematode. The disrupted gene was sequenced and found to code for a novel protein 121 amino acids long. Other independent transposon insertions which led to phase shifting have not yet been characterized. However, the existence of a large number of transposon mutants all exhibiting a phase shift (39; unpublished observations) implies that a complex regulatory cascade controls expression of these genes. Genes which have been shown not to play a role in controlling phase variation in Photorhabdus or Xenorhabdus include ompR (B. Boylan and S. Forst, unpublished observationsl cited in reference 14), recA (30), hin (R. J. Akhurst and A. Smigielski, Abstracts of the V1th International Colloquium on Invertebrate Pathology, Montpellier, France, 1994, p. 1), and rpoS (38).
The coordinate control of the phase variant genes by a single or multiple regulatory networks in the primary form does not preclude the secondary variant's deriving from an accumulation of mutations, similar to the GASP mutants. The first GASP mutant identified had attenuated RpoS function, but a mutation in this gene in X. nematophilus has revealed that it is not involved in regulation of the primary-form-specific traits (38). Another GASP mutation identified recently was a null allele of the lrp gene, which codes for a DNA-binding protein that can act as an activator or a repressor (42). If the secondary variant of K122 is a spontaneous mutant arising in a primary culture, the mutation is not located in the ner gene, since the sequence for this gene was identical in the primary and secondary variants.
Acknowledgments
This work was supported by grants from Forbairt, Enterprise Ireland, and the Higher Education Authority (PRTLI scheme) of Ireland.
The first two authors contributed equally to this work. Names appear in alphabetical order.
We thank Antonia Volgyi and Philip Daborn for advice with the outer membrane protein and Q-RT-PCR experiments, respectively.
REFERENCES
- 1.Akhurst, R. J. 1982. Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J. Gen. Microbiol. 128:3061-3065. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst, R. J., A. J. Smigielski, J. Mari, N. Boemare, and R. G. Mourant. 1992. Restriction analysis of phase variation in Xenorhabdus spp. (Enterobacteriaceae), entomopathogenic bacteria associated with nematodes. Syst. Appl. Microbiol. 15:469-473. [Google Scholar]
- 3.Autexier, C., and M. S. DuBow. 1992. The Escherichia coli Mu/D108 phage ner homologue gene (nlp) is transcribed and evolutionarily conserved among the Enterobacteriaceae. Gene 114:13-18. [DOI] [PubMed] [Google Scholar]
- 4.Bintrim, S. B., and J. C. Ensign. 1998. Insertional inactivation of genes encoding the crystalline inclusion proteins of Photorhabdus luminescens results in mutants with pleiotropic phenotypes. J. Bacteriol. 180:1261-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleakley, B., and K. H. Nealson. 1988. Characterization of primary and secondary forms of Xenorhabdus luminescens strain Hm. FEMS Microbiol. Ecol. 53:241-250. [Google Scholar]
- 6.Boemare, N. E., and R. J. Akhurst. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae). J. Gen. Microbiol. 134:751-761. [Google Scholar]
- 7.Choi, Y.-L., T. Nisida, M. Kawamukai, R. Utsumi, H. Sakai, and T. Komano. 1989. Cloning and sequencing of an Escherichia coli gene, nlp, highly homologous to the ner genes of bacteriophages Mu and D108. J. Bacteriol. 171:5222-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daborn, P. J., N. Waterfield, M. A. Blight, and R. H. ffrench-Constant. 2001. Measuring virulence factor expression by the pathogenic bacterium Photorhabdus luminescens in culture and during infection. J. Bacteriol. 183:5834-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowds, B. C. A. 1997. Photorhabdus and Xenorhabdus—gene structure and expression, and genetic manipulation. Symbiosis 22:67-83. [Google Scholar]
- 10.Dowds, B. C. A., and A. Peters. 2002. Virulence mechanisms, p. 79-98. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, United Kingdom.
- 11.Ehlers, R.-U., and I. Niemann. 1998. Molecular identification of Photorhabdus luminescens strains by amplification of specific fragments of the 16S ribosomal DNA. Syst. Appl. Microbiol. 21:509-519. [DOI] [PubMed] [Google Scholar]
- 12.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 13.Fischer-Le-Saux, M., V. Viallard, B. Brunel, P. Normand, and N. E. Boemare. 1999. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subs P. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subs P. temperata subsp. nov. and P. asymbiotica sp. nov. Int. J. Syst. Bacteriol. 49:1645-1656. [DOI] [PubMed] [Google Scholar]
- 14.Forst, S., and D. Clarke. 2002. Nematode-bacterium symbiosis, p. 57-77. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, United Kingdom.
- 15.Forst, S., J. Delgado, G. Ramakrishnan, and M. Inouye. 1988. Regulation of ompC and ompF expression in Escherichia coli in the absence of envZ. J. Bacteriol. 170:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 17.Forst, S., J. Waukau, G. Leisman, M. Exner, and R. W. Hancock. 1995. Functional and regulatory analysis of the OmpF-like porin, OpnP, of the symbiotic bacterium Xenorhabdus nematophilus. Mol. Microbiol. 18:779-789. [DOI] [PubMed] [Google Scholar]
- 18.Frackman, S., M. Anhalt, and K. H. Nealson. 1990. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J. Bacteriol. 172:5767-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Givaudan, A., S. Baghdiguian, A. Lanois, and N. Boemare. 1995. Swarming and swimming changes concomitant with phase variation in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 61:1408-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Givaudan, A., A. Lanois, and N. Boemare. 1996. Cloning and nucleotide sequence of a flagellin encoding genetic locus from Xenorhabdus nematophilus: phase variation leads to differential transcription of two flagellar genes (fliCD). Gene 183:243-253. [DOI] [PubMed] [Google Scholar]
- 21.Goosen, N., and P. van de Putte. 1987. Regulation of transcription, p. 41-52. In N. Symonds, A. Toussaint, P. van de Putte, and M. M. Howe (ed.), Phage Mu. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Griffin, C. T., J. F. Moore, and M. J. Downes. 1991. Occurrence of insect-parasitic nematodes (Steinernematidae, Heterorhabditidae) in the Republic of Ireland. Nematologica 37:92-100. [Google Scholar]
- 23.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the on and off of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 24.Krasomil-Osterfeld, K. 1995. Influence of osmolarity on phase shift in Photorhabdus luminescens. Appl. Environ. Microbiol. 61:3748-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kukolj, G., and M. S. DuBow. 1992. Integration host factor activates the Ner-repressed early promoter of transposable Mu-like phage D108. J. Biol. Chem. 267:17827-17835. [PubMed] [Google Scholar]
- 26.Kukolj, G., and M. S. DuBow. 2000. The bacteriophage D108 Ner repressor binds a conformationally distinct operator. Mol. Gen. Genet. 263:592-600. [DOI] [PubMed] [Google Scholar]
- 27.Lunau, S., S. Stoessel, A. J. Schmidt-Peisker and R.-U. Ehlers. 1993. Establishment of monoxenic inocula for scaling up in vitro cultures of the entomopathogenic nematodes Steinernema spp. and Heterorhabditis spp. Nematologica 39:385-399. [Google Scholar]
- 28.Mizuno, T., and M. Kageyama. 1978. Separation and characterization of the outer membrane of Pseudomonas aeruginosa. J. Biochem. 84:179-191. [DOI] [PubMed] [Google Scholar]
- 29.O'Neill, K. 1999. Phase variation in Photorhabdus luminescens. Ph.D. thesis. National University of Ireland, Maynooth, Ireland.
- 30.Pinyon, R. A., F. H. Hew, and C. J. Thomas. 2000. Xenorhabdus bovienii T228 phase variation and virulence are independent of RecA function. Microbiology 146:2815-2824. [DOI] [PubMed] [Google Scholar]
- 31.Ried, J., and A. Collmer. 1987. An npt1-sacB-sacR cartridge for constructing unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schmidt, T. M., B. Bleakley, and K. H. Nealson. 1988. Characterization of an extracellular protease from the insect pathogen Xenorhabdus luminescens. Appl. Environ. Microbiol. 54:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smigielski, A. J., R. J. Akhurst, and N. E. Boemare. 1994. Phase variation in Xenorhabdus nematophilus and Photorhabdus luminescens: differences in respiratory activity and membrane energization. Appl. Environ. Microbiol. 60:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strzelecka, T. E., G. M. Clore, and A. M. Gronenborn. 1995. The solution structure of the Mu ner protein reveals a helix-turn-helix DNA recognition motif. Structure 3:1087-1095. [DOI] [PubMed] [Google Scholar]
- 36.Szallas, E., R. Pukall, H. Pamjav, G. Kovacs, Z. Buzas, A. Fodor, and E. Stackebrandt. 2000. Passengers who missed the train: comparative sequence analysis, PhastSystem PAGE RFLP and automated RiboPrint Phenotypes of Photorhabdus strains. In C. Griffin (ed.) Entomopathogenic nematode bacterial complexes—current achievements and prospects for the future (proceedings of the COST 819 Symposium) National University of Ireland, Maynooth, Ireland.
- 37.Tinsley, C. R., and X. Nassif. 1996. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc. Natl. Acad. Sci. USA 93:11109-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vivas, E. I., and H. Goodrich-Blair. 2001. Xenorhabdus nematophilus as a model for host-bacterium interactions: rpoS is necessary for mutualism with nematodes. J. Bacteriol. 183:4687-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volgyi, A., A. Fodor, and S. Forst. 2000. Inactivation of a novel gene produces a phenotypic variant cell and affects the symbiotic behavior of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 66:1622-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, H., and Dowds, B. C. A. 1991. Molecular cloning and characterization of the lux genes from the secondary form of Xenorhabdus luminescens, K122. Biochem. Soc. Trans. 20:68.S. [DOI] [PubMed] [Google Scholar]
- 41.Wang, H., and B. C. A. Dowds. 1993. Phase variation in Xenorhabdus luminescens: cloning and sequencing of the lipase gene and analysis of its expression in primary and secondary phases of the bacterium. J. Bacteriol. 175:1665-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zinser, E. R., and R. Kolter. 2000. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J. Bacteriol. 182:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]