Abstract
Pseudomonas azelaica HBP1 can use 2-hydroxybiphenyl (2-HBP) and 2,2′-dihydroxybiphenyl as sole carbon and energy sources by means of the hbp regulon. This regulon is composed of three genes, hbpCA and hbpD, coding for enzymes of a meta-cleavage pathway and the hbpR gene, which codes for a XylR/DmpR-type transcription regulator. It was previously shown that HbpR activates transcription from two σ54-dependent promoters, PhbpC and PhbpD, in the presence of 2-HBP. In this study, by using gel mobility shift assays with a purified fusion protein containing calmodulin binding protein (CBP) and HbpR, we detected two binding regions for HbpR in PhbpC and one binding region in PhbpD. DNase I footprints of the proximal binding region of PhbpC and of the binding region in PhbpD showed that CBP-HbpR protected a region composed of two inverted repeat sequences which were homologous to the binding sites identified for XylR. Unlike the situation in the XylR/Pu system, we observed simultaneous binding of CBP-HbpR on the two upstream activating sequences (UASs). Fragments with only one UAS did not show an interaction with HbpR, indicating that both pairs of UASs are needed for HbpR binding. The addition of both ATP and 2-HBP increased the DNA binding affinity of HbpR. These results showed for the first time that, for regulators of the XylR/DmpR type, the effector positively affects the recruitment of the regulatory protein on the enhancer DNA.
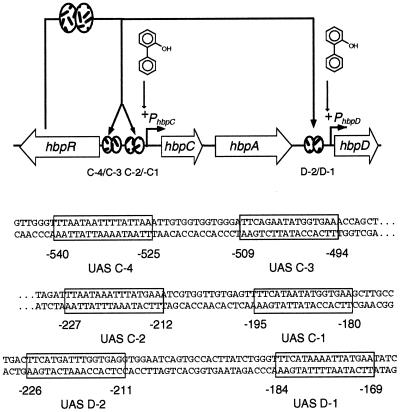
Pseudomonas azelaica HBP1 metabolizes 2-hydroxybiphenyl (2-HBP) and 2,2′-dihydroxybiphenyl through a meta-cleavage pathway (14, 15, 29). The enzymes involved in the first degradation steps are encoded by the hbpCA and hbpD genes (Fig. 1). Two promoters, designated PhbpC and PhbpD (13), control the expression of the hbpCA and hbpD genes, respectively. In the presence of 2-HBP, transcriptional activation from both promoters is mediated by the HbpR regulatory protein (11). On the basis of sequence similarities, HbpR has been identified as a member of the XylR/DmpR subclass of the NtrC family of prokaryotic enhancer binding proteins. Proteins of this family activate transcription at a distance from their cognate promoter through an intrinsic ATPase activity in concert with RNA polymerase containing the alternative sigma factor σ54 (6, 10). The process of activation by proteins of the XylR/DmpR subclass is initiated by a direct interaction with aromatic compounds which (mostly) are the substrates for the pathways to be controlled (32). It has been shown that for proper activation, the regulatory protein needs to bind at specific nucleotide sequences in its cognate promoter. These sequences are formed by two (imperfect) palindromic sequences of approximately 16 bp and with a spacing of 29 to 42 bp between the centers of the palindromes. They are usually called bacterial enhancer-like elements or upstream activating sequences (UASs) and are located 100 to 200 bp upstream of the −12/−24 target promoter (17, 20). This distance does not allow direct contact between the regulator and the σ54 RNA polymerase; therefore, looping of the DNA is required, bringing together both partners. Looping is facilitated by induced bending of the DNA by integration host factor or HU or by intrinsically curved DNA sequences (21). Changing the relative positions of the −12/−24 motif, the integration host factor binding site, and the UASs was shown to disturb the optimal promoter geometry and to lead to a decrease in transcriptional activation (1, 2, 8, 9, 20).
FIG. 1.
Genetic organization of the hbp genes in P. azelaica HBP1. Arrows depict the orientations and sizes of the genes; the solid line indicates noncoding DNA. The hbpR gene codes for the regulatory protein, which activates transcription from the hbpC and hbpD promoters upon exposure to 2-HBP. Regions containing the binding sites for HbpR (UAS) are shown within boxes in the sequence. Sequence numbers refer to the locations of the transcriptional start sites of hbpC and hbpD.
Proteins of the NtrC family have an intrinsic binding affinity for their UASs. In the absence of an effector (for regulators of the XylR type) or phosphorylation (for NtrC-type regulators), two dimers of the regulatory protein bind to the UASs (22, 26, 30). In contrast, the activated regulatory protein (for NtrC, its phosphorylated form) forms larger protein complexes at the UAS DNA. The latter finding was determined by atomic force microscopy and analytical ultracentrifugation, which showed that phosphorylated NtrC in the presence of the UAS DNA oligomerizes to the size of hexamers or octamers (26, 37). This oligomerization is a requirement for transcriptional activation. Although some conflicting data exist on this topic, binding of the dimers in the inactive state to each of the UASs in the native glnA promoter occurs with different affinities (22). Upon activation of the regulatory protein, the differences in the binding affinities for each of the UASs increase strongly (25, 31). In the current activation model, it is assumed that there is continuous cycling among the inactive dimer in solution, the inactive two-dimer pair on the UAS DNA, and the octameric (active) complex (7). XylR itself is supposed to follow more or less the same model as NtrC, except that activation of the protein takes place through effector binding and not through phosphorylation. However, most of the studies on XylR binding and activation have been made with a protein with a deletion of its N-terminal effector binding domain (ΔAXylR) (18-20). Since this protein is constitutively active, the exact changes in DNA binding affinity upon effector binding could not be shown directly.
The HbpR regulatory system has already demonstrated several differences with respect to the DmpR/Po and XylR/Pu systems. First, the HbpR protein has only 40% amino acid similarity with XylR and DmpR, whereas XylR and DmpR share 67% similar amino acids (13). Second, HbpR is the only member of the XylR/DmpR subclass described so far which is activated by biaromatic compounds, such as 2-HBP and 2,2′-dihydroxybiphenyl. Third, HbpR activates transcription from two promoters within a rather small cluster of only three genes (hbpCA and hbpD), an unusual scenario (11). Finally, the proposed binding sites for HbpR have slightly larger spacing between the UASs (13). Because of these differences, we were interested in investigating whether the interaction of HbpR with its binding regions also distinguishes it from the other members of the XylR/DmpR subclass. Furthermore, rather than working with an HbpR protein containing an N-terminal A-domain deletion (like most of the DNA binding studies with DmpR and XylR), our goal was to determine the DNA binding characteristics for the entire protein. For this purpose, we set out to produce and purify a fusion protein consisting of calmodulin binding protein (CBP) fused to the N terminus of HbpR (CBP-HbpR). This purified protein was used to study the initial interactions of HbpR on its cognate binding sites in the presence and absence of either ATP or 2-HBP. The characteristics of binding of HbpR to the three pairs of UASs were assessed by using DNase I footprinting and gel mobility shift assays (GMSAs). Our data indicate that HbpR binds simultaneously to both palindromic sequences and that both palindromes are required for HbpR binding. An increase in DNA binding affinity was observed in the presence of 2-HBP and ATP.
MATERIALS AND METHODS
Strains and medium.
Escherichia coli DH5α (27) was used as a host strain in routine cloning experiments. E. coli BL21(DE3)(pLysS) (Stratagene, La Jolla, Calif.) was used for protein overexpression. E. coli strains were grown at 25, 30, or 37°C on Luria-Bertani (LB) medium (27). When required, the medium was supplemented with the following antibiotics at the indicated concentrations: ampicillin, 100 μg·ml−1, and chloramphenicol, 25 μg·ml−1. All strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | endA1 hsdR17 (rK− mK−) supE44 thi-1 recA1 gyrA96 relA1 F− Δ(argF-lacZYA)U169 (φ80dlacZΔM15)λ− | Gibco BRL |
| BL21(DE3)(pLysS) | ompT lon hsdSB (rB− mB−) gal dcm (DE3) pLysS Cmr | Stratagene |
| Plasmids | ||
| pGEM-T-Easy | Apr | Promega |
| pCAL-n-FLAG | Apr; expression vector | Stratagene |
| pHBP130 | Apr ColE1; contains a 7.8-kb MluI-SalI fragment from P. azelaica HBP1 with hbpR and hpbC | (29) |
| pHYBP109 | Apr ColE1; pJAMA8 carrying hbpR under the control of its native promoter (PhbpR) | (13) |
| pHYBP132 | Apr ColE1; pET3d containing hbpR under the control of the φ10 promoter | (13) |
| pJAMA8 | Apr ColE1; luxAB-based promoter-probe vector | (13) |
| pHB150 | Apr; pCAL-n-EK vector Stratagene carrying the 0.5-kb PCR fragment containing the start of hbpR | This study |
| pHB151 | Apr; pHB150 carrying the 1.5-kb NsiI-SalI fragment from pHYBP132 which generated a complete hbpR | This study |
| pHB152 | pGEM-T-Easy containing a 117-bp fragment with UAS C-2 obtained by PCR with primers hbpC11 and hbpC12 | This study |
| pHB153 | pGEM-T-Easy containing an 85-bp fragment obtained by PCR with primers hbpC11 and hbpC15 | This study |
| pHB154 | pGEM-T-Easy containing an 86-bp fragment obtained by PCR with primers hbpC10 and hbpC13 | This study |
| pHB155 | pGEM-T-Easy containing a 118-bp fragment with UAS C-1 obtained by PCR with primers hbpC10 and hbpC14 | This study |
| pHB156 | pHB152 containing a 450-bp NaeI-BamHI fragment from pHB154; contains only UAS C-2 | This study |
| pHB157 | pHB153 containing a 480-bp NaeI-BamHI fragment from pHB155; contains only UAS C-1 | This study |
| pHB158 | pGEM-T-Easy containing a 118-bp fragment with UAS C-2 obtained by PCR with primers hbpC11 and hbpC16 | This study |
| pHB160 | pGEM-T-Easy containing a 110-bp fragment with UAS C-1 obtained by PCR with primers hbpC10 and hbpC17 | This study |
| pHB161 | pGEM-T-Easy containing a 115-bp fragment with UAS C-1 obtained by PCR with primers hbpC10 and hbpC18 | This study |
| pHB162 | pHB158 containing the 470-bp NaeI-BamHI fragment from pHB160 with UAS C-1; contains UASs C-1 and C-2 + 5 bp | This study |
| pHB163 | pHB158 containing the 475-bp NaeI-BamHI fragment from pHB161 with UAS C-1; contains UASs C-1 and C-2 + 10 bp | This study |
| pHB164 | pHB151 containing a 2.75-kb BamHI fragment from pHB109 with an hbpR-hbpC intergenic region-luxAB fusion; complete cbp-hbpR | This study |
| pHB165 | pHB150 containing a 2.75-kb BamHI fragment from pHB109 with an hbpR-hbpC intergenic region-luxAB fusion; incomplete cbp-hbpR | This study |
| pHB207 | Apr; pHB150 carrying the 2-kb BglII-SalI fragment from pHYBP132 which generated a complete hbpR without the CBP tag fusion | This study |
| pHB208 | pHB207 containing a 2.75-kb BamHI fragment from pHB109 with an hbpR-hbpC intergenic region-luxAB fusion; complete hbpR | This study |
Recombinant DNA techniques.
DNA sequencing, plasmid DNA isolation, ligation, transformation, and other DNA manipulations were carried out according to well-established procedures (27). Restriction endonucleases and other DNA-modifying enzymes were obtained from Amersham International plc (Little Chalfont, United Kingdom), Roche Biochemicals (Mannheim, Germany), and New England Biolabs Inc. (Beverly, Mass.) and used according to the specifications of the manufacturers. DNA fragments were isolated from agarose gels by using a PEQLAB kit (Biotechnologie GmbH, Erlangen, Germany). Double-stranded template sequencing on plasmids was performed by using a modified dideoxy-chain termination method (28) with primers that were labeled with the fluorescent dye IRD-800 at the 5′ end as described elsewhere (23).
Cloning of the hbpR overexpression vector.
Plasmid pCAL-n-FLAG (Stratagene) was used for the production and purification of the HbpR protein in E. coli. To clone hbpR into pCAL-n-FLAG, its first 494 nucleotides were amplified by using PCR with plasmid pHBP130 and primers LIChbpR1 (5′-GACGACGACAAGATGAAATCAAATAAAAATAATAG) and LIChbpR2 (5′-GGAACAAGACCCGTTACGCAACGGAAAACCAA). The PCR product was separated on an agarose gel, purified, and treated with Pfu polymerase (Stratagene) in the presence of dATP. The 3′-5′ exonuclease activity of Pfu polymerase removes nucleotide residues from both 3′ ends of the PCR product and stops on the first adenine residue because of the dATP present in the reaction. Thereby, 5′ single-stranded overhangs were generated (5′-GACGACGACAAGAT and 5′-GGAACAAGACCCGT) that were complementary to those in the prepared vector. Next, the hbpR fragment was ligated with pCAL-n-FLAG according to the manufacturer's recommendations. Transformation resulted in plasmid pHB150. The complete hbpR gene was assembled as follows: a 1.475-kb NsiI-SalI fragment from pHYBP132 containing the remaining sequence of hbpR was used to replace a 65-bp fragment of pHB150 cut with NsiI and SalI (yielding pHB151). Plasmid pHB151 produced an HbpR protein with an N-terminal CBP tag.
Overexpression and purification of CBP-HbpR.
E. coli BL21(DE3)(pLysS) containing pHB151 was grown at 30°C in LB medium to an optical density at 600 nm of 0.6. To induce T7 RNA polymerase-directed expression, isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added at a concentration of 0.1 mM, and cultures were further incubated for 3 h at 25°C. Bacteria were collected from 1 liter of culture, washed in 50 mM Tris-HCl buffer (pH 7.5), and centrifuged. After the supernatant was discarded, the cell pellet was stored frozen at −80°C. To disrupt cells, the bacterial pellet was thawed in 15 ml of loading buffer (loading buffer is 50 mM Tris-HCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, and 10% [vol/vol] glycerol [pH 8.0]) containing 400 mM NaCl and then subjected to ultrasonication four times for 1 min each time at 50% and 40 W (Branson 450 Sonifier). All subsequent steps were carried out at 4°C. The cell extract obtained was centrifuged for 30 min at 35,000 × g to remove cell debris. The supernatant was loaded on a 3-ml column of calmodulin resin equilibrated with loading buffer. After loading, the column was washed with 20 bed volumes of loading buffer containing 400 mM NaCl and then with 10 bed volumes of loading buffer containing 150 mM NaCl. Proteins binding to calmodulin were removed with elution buffer (elution buffer is 50 mM Tris-HCl, 0.2 mM EGTA, 150 mM NaCl, and 10% [vol/vol] glycerol [pH 8.0]). Fractions eluted from the column were collected in 1-ml portions and frozen in 30-μl aliquots at −80°C. The method of Bradford was used to determine the total protein concentration in our samples. Protein samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and the intensities of the different bands were quantified by laser densitometry scanning (300S computing densitometer; Molecular Dynamics, Sunnyvale, Calif.) by using the program Image-Quant (Molecular Dynamics). The proportion of CBP-HbpR was determined as the band intensity relative to the intensity of all bands. From the proportion and the total protein concentration, the concentration of CBP-HbpR was calculated.
Construction of plasmids for DNA binding studies.
Various DNA fragments were generated by using PCR in order to determine the locations of the HbpR binding regions. The sequences and locations of the primers used for this step are listed in Table 2. Furthermore, the UAS C-1 and C-2 regions were modified by deleting either one of the UASs or adding additional nucleotides between the two UASs. This step was also performed by using PCR (Table 2). For example, to remove the proximal site (UAS C-1), primers hbpC11 and hbpC12 were used. This step amplified a fragment with only the distal site (UAS C-2). Primers hbpC10 and hbpC13 were used to amplify the region downstream of UAS C-1. Both PCR fragments were separately cloned in pGEM-T-Easy to give pHB152 and pHB154, respectively. The amplified sequence was recovered from pHB154 as an NaeI-BamHI fragment and inserted into pHB152 (yielding pHB156). Using the same strategy, we constructed plasmids pHB157, pHB162, and pHB163, which contained a binding region with a deletion of UAS C-2, a binding region with 5 bp inserted between the UASs, and a binding region with 10 bp inserted between the UASs, respectively.
TABLE 2.
Primers used in this study
| Primer | Nucleotide sequencea | Position from the 5′ endb |
|---|---|---|
| LIChbpR1 | 5′-GACGACGACAAGATGAAATCAAATAAAAATAATAG | hbpR start codon |
| LIChbpR2 | 5′-GGAACAAGACCCGTTACGCAACGGAAAACCAA | 494 bp downstream of hbpR start codon |
| hbpC2 | 5′-CTGGCTAGGCGACAGCC | 459 bp upstream of hbpC start codon |
| hbpC4 | 5′-CCTGGCATGAGCTATCA | 322 bp upstream of hbpC start codon |
| hbpC6 | 5′-AATGAGCGCCAGAAAGCCT | 156 bp upstream of hbpC start codon |
| hbpC8 | 5′-TACCCGAGATTTGAAATCATTG | 20 bp downstream of hbpC start codon |
| hbpCA | 5′-ATTTTTATTTGATTTCATGGCGA | 672 bp upstream of hbpC start codon |
| hbpCB | 5′-GGCATCTGCCGACGGATC | 534 bp upstream of hbpC start codon |
| hbpCC | 5′-AACGATGGTGCGGTTTTCAT | 385 bp upstream of hbpC start codon |
| hbpCD | 5′-GACCGCGGAAGGGGTTTAC | 206 bp upstream of hbpC start codon |
| hbpC10 | 5′-CGGGCATATGGCGCCAGAAAGCCTAGCTCC | 162 bp upstream of hbpC start codon |
| hbpC11 | 5′-CGGGAAGCTTTGGTGCGGTTTTCATGGTCCTTA | 390 bp upstream of hbpC start codon |
| hbpC12 | 5′-CGCGGGATCCAACTCACAACCACGATTTCATAAA | 264 bp upstream of hbpC start codon |
| hbpC13 | 5′-CGCGGGATCCGCTTGCCCGCCATGGCAAG | 248 bp upstream of hbpC start codon |
| hbpC14 | 5′-CGCGGGATCCATCGTGGTTGTGAGTTTTCATAATA | 280 bp upstream of hbpC start codon |
| hbpC15 | 5′-CGCGGGATCCATCTACGAGAACCCTATCTATCTACTC | 296 bp upstream of hbpC start codon |
| hbpC16 | 5′-CGCGGGATCCCCACGATTTCATAAATTTATTAAATC | 273 bp upstream of hbpC start codon |
| hbpC17 | 5′-CGCGGGATCCTGTGAGTTTTCATAATATGGTGAAG | 271 bp upstream of hbpC start codon |
| hbpC18 | 5′-CGCGGGATCCGTGGTTGTGAGTTTTCATAATATG | 276 bp upstream of hbpC start codon |
| hbpD1 | 5′-CATCCTTGGGAGGGCGTAAC | 850 bp upstream of hbpD start codon |
| hbpD2 | 5′-CTTCAGAGCACTCGCCAC | 561 bp upstream of hbpD start codon |
| hbpD3 | 5′-TGGATCTGCAGTTGCCCTAAG | 622 bp upstream of hbpD start codon |
| hbpD4 | 5′-ATGAAGAGCGCGCGCGCTCTC | 366 bp upstream of hbpD start codon |
| hbpD5 | 5′-GCATTCCTCCTCCAGATGAG | 423 bp upstream of hbpD start codon |
| hbpD8 | 5′-CAGTGTACTTTGGCATTGGTC | 15 bp downstream of hbpD start codon |
Restriction sites for BamHI (5′-GGATCC-3′), NdeI (5′-CATATG-3′), and HindIII (5′-AAGCTT-3′) are shown in italic type. Mismatched residues at the 5′ end which resulted from the introduction of restriction or ligation-independent cloning overhangs are underlined.
When there were mismatches at the 5′ ends, the position of the residue downstream of the first mismatch is given.
DNase I footprinting.
The DNA fragments used for DNase I footprinting were amplified by PCR. For each PCR, one oligonucleotide was end labeled by phosphorylation with [γ-32P]ATP, allowing the specific labeling of one strand. The end-labeled fragments were mixed with various amounts of HbpR (0 to 450 nM) in binding buffer (binding buffer is 10 mM Tris-HCl, 20 mM KCl, 1 mM EDTA, and 10% [vol/vol] glycerol), to which 1 μg of poly(dI-dC) was added. DNA-protein complexes were allowed to form at 33°C for 15 min in a total volume of 50 μl for each footprinting reaction. DNase I (0.05 U; Roche Biochemicals) was then added to the reaction mixture, as were 1 mM MgCl2 and 0.5 mM CaCl2. The reaction mixture was incubated for 1 min at 30°C, and the reaction was stopped by the addition of 140 μl of stop mix solution (stop mix solution is 770 mM sodium acetate, 130 mM EDTA, and 256 μg of yeast tRNA ml−1). The footprinting mixture was subsequently extracted once with phenol and chloroform (1:1 [vol/vol]) and once with chloroform, and finally the DNA was precipitated with ethanol. The DNA was washed once with 70% ethanol, dried, resuspended in 5 μl of sequence loading buffer (sequence loading buffer is deionized formamide containing 10 mM EDTA, 0.3% [wt/vol] bromophenol blue, and 0.3% xylene cyanol), and loaded on a 6% polyacrylamide gel which contained 8 M urea and which had been prerun for 1 h. The gel was run at 1,800 V for 3 h in Tris-borate-EDTA (TBE) buffer (27). As size markers, DNA fragments which had been generated in a dideoxy sequencing reaction (28) with a Sequenase (version 2.0) DNA sequencing kit (Amersham) were loaded. After the run, the gel was soaked in a solution of 10% acetic acid, dried, and exposed overnight to Biomax film (Kodak) at −80°C.
GMSAs.
For GMSAs, the 32P-end-labeled fragments and the binding buffer were the same as those used in the footprinting assays. The reaction volume was reduced to 30 μl. When tested, 2-HBP and ATP were added to the reaction mixture at 10 and 5 mM, respectively. After binding, 5 μl of GMSA loading buffer (GMSA loading buffer is 40% glycerol [vol/vol], 50 mM EDTA, 0.1% [wt/vol] bromophenol blue, and four-times-concentrated TBE buffer) was added, and the mixture was loaded on a 5% polyacrylamide gel in TBE buffer. Separation was done for 2 h at 50 V in a Mini-Protean vertical electrophoresis chamber (Bio-Rad). The gel was dried and exposed overnight to Kodak X-Omat film at −80°C. Autoradiograms of GMSAs were quantitatively analyzed by densitometric scanning. Relative densities were calculated by comparing the measured densities of shifted bands to that in the lane in which CBP-HbpR had completely bound all DNA.
In vivo HbpR activation.
To determine the in vivo activity of the CBP-HbpR fusion protein, we used HbpR-mediated activation of the luxAB genes transcriptionally fused to the hbpC promoter as described before (11). The CBP-HbpR fusion of pHB151 was completed with the 2.75-kb BamHI fragment of plasmid pHYBP109 (containing the native hbpRC intergenic region fused to the luxAB genes). This BamHI fragment was inserted at the single BglII site of plasmid pHB151. After transformation, plasmids in which cbp-hbpR was expressed from the native hbpR promoter were selected (yielding pHB164). Similarly, pHB165 was constructed starting with pHB150. In plasmid pHB165, the hbpR gene has a deletion in the region coding for the C-terminal portion of the protein. This plasmid served as a negative control for 2-HBP-dependent luciferase activation. As a positive control, the hbpR gene was cloned under the control of the T7 gene φ10 promoter but without the CBP tag fusion. hbpR was cloned as a 2-kb BglII -SalI fragment of plasmid pHB132 and inserted into pHB150 cut with the same enzymes (yielding plasmid pHB207). Plasmid pHB207 was completed with the 2.75-kb BamHI fragment of plasmid pHYBP109 (yielding pHB208).
Luciferase assays.
E. coli DH5α with pHB164, pHB165, or pHB208 was induced in 7-ml glass vials that were closed with screw caps with a polytetrafluoroethylene (PTFE) liner (Supelco, Bellefonte, Pa.). The assay mixture contained 1.95 ml of LB medium, 30 μl of E. coli culture (at an optical density at 600 nm of 0.45), and 20 μl of dimethyl sulfoxide solution with 2-HBP. The final concentration of 2-HBP in the assay mixture was 25 μM. The negative control contained 20 μl of dimethyl sulfoxide. The glass vials were incubated at 30°C on a rotary shaker at 200 rpm for 2 h. After induction, samples of 0.2 ml were removed and transferred to a microtiter plate. Bioluminescence was measured at 30°C with a final n-decanal concentration of 2 mM in a MicroLumat LB 96 P luminometer (Berthold AG, Regensdorf, Switzerland) as described previously (33).
Synthetic oligonucleotides and chemicals.
Primers labeled with the fluorescent dye IRD-800 at the 5′ end were purchased from MWG-Biotech GmbH (Ebersberg, Germany). All other primers were obtained from Microsynth GmbH (Balgach, Switzerland). Ultrapure agarose, ammonium persulfate, N,N,N′,N′-tetramethylethylenediamine, Tris, and urea were purchased from Life Technologies. Rapid Gel-XL-40% acrylamide solution was obtained from Amersham. IPTG and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were obtained from Biosynth AG (Staad, Switzerland), and n-decanal was obtained from Sigma Chemical Co. (St. Louis, Mo.). Nutrient broth, yeast extract, and tryptic casein were purchased from Biolife S.r.l. (Milan, Italy), and ultrapure agar was obtained from Merck (Darmstadt, Germany). Antibiotics, inorganic salts, and all other organic chemicals were obtained from Fluka AG (Buchs, Switzerland).
RESULTS
Expression and purification of a CBP-HbpR fusion protein.
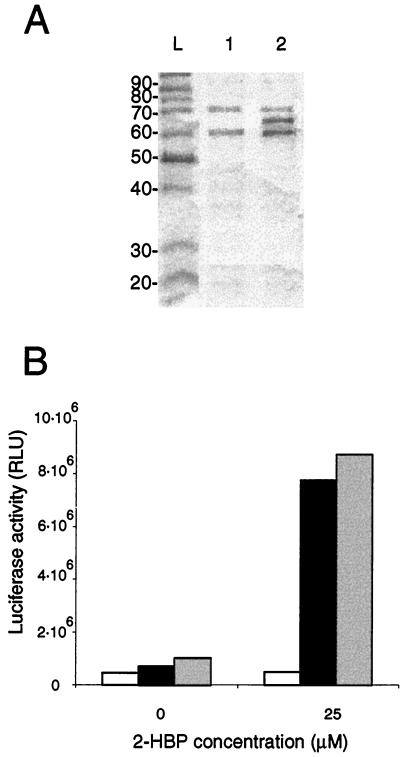
In order to study the characteristics of in vitro binding of HbpR to its DNA binding sites, we decided to express HbpR as a fusion protein in E. coli and then purify it. Expression as a fusion protein would most likely not result in its precipitation, a common phenomenon encountered with XylR (5), a protein related to HbpR. A fusion with CBP was chosen, since this choice in principle allowed complete cleavage of CBP from HbpR through an enterokinase cleavage site (36). CBP-HbpR was expressed in E. coli at 25°C for 3 h to reduce the formation of insoluble fusion protein observed at 37 or 30°C (data not shown). SDS-PAGE of cell extracts of E. coli BL21(pHB151) indeed revealed a protein of 67 kDa, which corresponds to the molecular mass of HbpR (63 kDa) plus that of CBP (4 kDa). This protein band was not detected in extracts of E. coli BL21(pHB150), which produced a truncated protein, CBP-HbpRΔ (Fig. 2A). The CBP-HbpR concentration in the preparation eluted from the calmodulin resin was estimated to be 200 μg·ml−1. Unfortunately, after binding and elution with calmodulin resin, the CBP-HbpR fusion protein always eluted in the presence of two contaminants, of 60 and 75 kDa (Fig. 2A). These contaminants, however, were also eluted from cell extracts of E. coli BL21(pHB150) and E. coli BL21, indicating that they were not of HbpR origin. Despite repeated attempts, it was not possible to purify the CBP-HbpR fusion protein from the contaminating proteins by use of the calmodulin resin. Unfortunately, CBP-HbpR was also not stable upon cleavage with enterokinase. We therefore had to use the CBP-HbpR preparation with both contaminants and with a CBP tag in our subsequent studies.
FIG. 2.
(A) Analysis by SDS-PAGE of the CBP-HbpR protein fraction after the single calmodulin resin purification step. Lanes: L, protein markers (sizes given in kilodaltons); 1, protein fraction purified from E. coli BL21(pHB150), expressing CBP-HbpRΔ; 2, protein fraction purified from E. coli BL21(pHB151), expressing CBP-HbpR (67 kDa). The contaminating proteins have sizes of 60 and 75 kDa. (B) Expression of luciferase activity from the hbpC promoter of P. azelaica reproduced in E. coli harboring plasmid pHB164 (black bars) and incubated with 25 μM 2-HBP. Plasmid pHB164 contains the cbp-hbpR fusion gene, the hbpC promoter, and the luxAB genes. As a negative control, E. coli harboring plasmid pHB165 (white bars) was used. pHB165 is identical to pHB164, except for a frameshift in hbpR. As a positive control, E. coli harboring plasmid pHB208 was used (grey bars). pHB208 is identical to pHB164, except that it contains the native hbpR gene. Luciferase activity was measured after 2 h of induction at 37°C. RLU, relative light units.
To check for true activity of the CBP-HbpR protein, we always conducted negative control experiments with purified cell extracts from E. coli BL21(pHB150), producing CBP-HbpRΔ. Second, we determined whether CBP-HbpR was capable of in vivo activation of the hbpC promoter. For this purpose, E. coli DH5α containing pHB164 was induced with 25 μM 2-HBP. The luciferase activity detected in E. coli expressing the CBP-HbpR fusion protein was similar to that found in E. coli(pHB208) expressing the native protein (Fig. 2B). From this result, we concluded that CBP-HbpR protein activates the hbpC promoter like the native HbpR protein. In contrast, no inducible expression of luciferase activity was observed in E. coli DH5α(pHB165), which expresses a CBP-HbpRΔ fusion protein with only the first 165 amino acids of HbpR (Fig. 2B). This result showed that inducible luciferase expression obtained with E. coli DH5α(pHB164) in the presence of 2-HBP was not due to proteins from E. coli DH5α itself.
DNA binding of the CBP-HbpR protein.
In a previous study, it was shown that HbpR activates transcription from two promoters, PhbpC and PhbpD, localized in the hbpRC and hbpAD intergenic regions, respectively (11). When tested in GMSAs, the CBP-HbpR purified fraction indeed bound both the hbpRC and the hbpAD intergenic regions but not the hbpCA region, which has no demonstrated HbpR-dependent promoter (data not shown). This result showed that binding to the hbpRC and hbpAD intergenic regions was specific and confirmed our previous findings that HbpR-dependent promoters were located in these regions. Furthemore, no binding was observed with purified cell extracts from E. coli producing CBP-HbpRΔ. This result showed that binding of the hbpRC and hbpAD intergenic regions was mediated by the CBP-HbpR fusion protein.
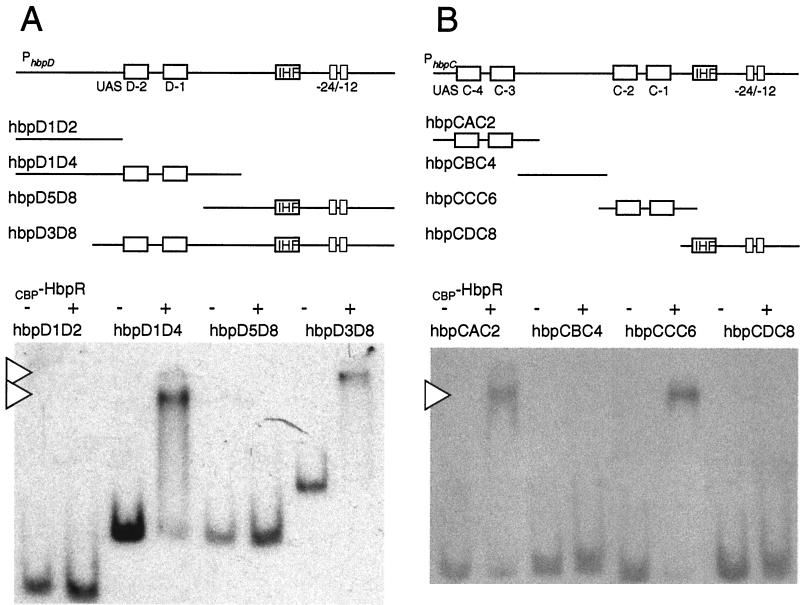
To determine the locations of the HbpR binding sites more precisely, a series of overlapping DNA fragments were generated and tested for the ability to be bound by CBP-HbpR (Fig. 3). GMSAs showed that fragments hbpD1D4 and hbpD3D8 (Fig. 3A) retained the ability to be bound by HbpR. These fragments shared a 256-bp region located at positions −359 to −103 with respect to the hbpD transcriptional start site. No binding was observed with fragments hbpD1D2 (−587 to −298) and hbpD5D8 (−160 to +278). This result showed that the sequences necessary for binding by HbpR were located between positions −160 and −298 relative to the hbpD transcriptional start site. Similarly, the locations of the HbpR binding sites in the hbpRC intergenic region were determined with overlapping DNA fragments covering the region between −604 and +76 bp with respect to the hbpC transcriptional start site. In this situation, two fragments, namely, hbpCAC2 (−604 to −391) and hbpCCC6 (−317 to −88), were bound by CBP-HbpR. No binding was observed with fragments hbpCBC4 (−468 to −256) and hbpCDC8 (−140 to +86) (Fig. 3B). In contrast to the hbpD promoter, therefore, the hbpC promoter contained two HbpR binding sites, located between −604 to −468 and −256 to −140. These results confirmed the results of previous studies which had indicated that these regions act as sites for HbpR-mediated transcriptional activation (12).
FIG. 3.
CBP-HbpR binding to different fragments from PhbpD and PhbpC. (A) GMSA of PhbpD fragments incubated with or without 1 μM CBP-HbpR (plus or minus at the top of the gel). The locations and relative sizes of the fragments are depicted in the diagram. Arrowheads indicate DNA-HbpR complexes. The name of the fragment corresponds to the primer name used in the PCR. (B) GMSA of PhbpC fragments incubated with or without CBP-HbpR. Conditions and symbols are as described for panel A. IHF, integration host factor.
CBP-HbpR simultaneously binds two palindromic sequences in its binding region.
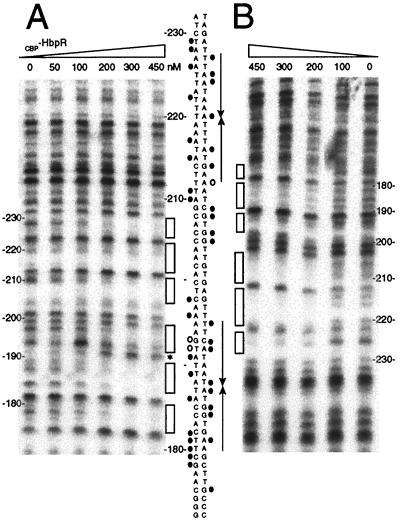
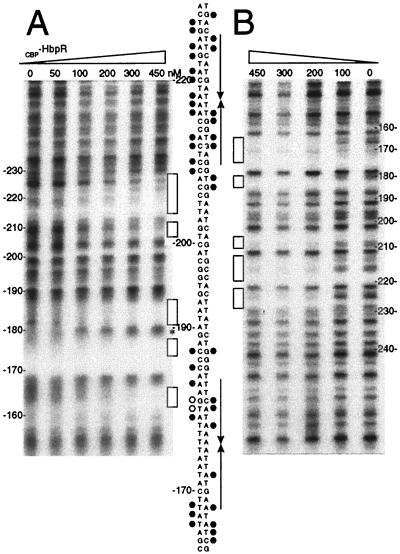
Previous promoter fusion studies and the GMSAs identified coarsely those regions on the DNA with which HbpR interacted. Sequence comparisons had suggested the presence within those regions of pairs of palindromic sequences which were similar to the so-called UASs of transcription activators of the XylR/DmpR type (11). To identify whether HbpR was indeed binding to these putative UASs, we performed DNase I footprinting analyses with CBP-HbpR and with labeled fragments containing UASs C-1 and C-2 (fragment hbpCCC6, within the hbpC promoter) or UASs D-1 and D-2 (fragment hbpD3D4, within the hbpD promoter). Both top and bottom strands of these fragments were subjected to DNase I nicking in the presence of increasing amounts of CBP-HbpR (Fig. 4 and 5). When the top-strand-labeled hbpCCC6 fragment was incubated with CBP-HbpR, protection appeared more or less at the predicted UASs, C-1 and C-2, or in directly neighboring nucleotides. The protection of these sites was confirmed by DNase I footprinting analysis of the bottom strand. The interaction of CBP-HbpR with the UASs became visible at concentrations of between 100 and 200 nM. Furthermore, no preferential or sequential protection of one or the other UAS was observed, suggesting that CBP-HbpR simultaneously binds to both UASs. As far as the resolution of the DNase I digestion pattern allowed, the base pairs contacted by CBP-HbpR within both UASs were very similar; basically, three regions within each UAS were protected, alternating with nonaffected base pairs and two neighboring hypersensitive base pairs (at positions −192 and −193 on the bottom strand) (Fig. 4). The protection pattern for the hbpD promoter fragment (hbpD3D4) (Fig. 5) was very similar to that for UASs C-1 and C-2. Both hbpD and hbpC promoters revealed a hypersensitive site located at the same base pair (TG) of both proximal UASs. The formation of hypersensitive sites probably reflected torsion of the DNA helix upon CBP-HbpR binding.
FIG. 4.
DNase I footprinting analysis of CBP-HbpR binding to UASs C-1 and C-2. The 229-bp 32P-end-labeled fragment hbpCCC6 containing UASs C-1 and C-2 was incubated with increasing amounts of CBP-HbpR (0 to 450 nM). (A) CBP-HbpR-mediated DNase I protection pattern for the bottom strand. (B) CBP-HbpR-mediated DNase I protection pattern for the top strand. Boxes indicate the regions protected from DNase I digestion upon the addition of CBP-HbpR. Between the panels, the sequences of UASs C-1 and C-2 and the positions that were contacted by CBP-HbpR are shown. Black circles indicate protection from DNase I digestion, while open circles and the asterisk indicate increased sensitivity to DNase I. The positions of the palindromes are indicated by arrows. Nucleotide numbering was relative to the transcriptional start site of hbpC.
FIG. 5.
DNase I footprinting analysis of CBP-HbpR binding to UASs D-1 and D-2. The 256-bp 32P-end-labeled fragment hbpD3D4 containing UASs D-1 and D-2 was incubated with increasing amounts of CBP-HbpR (0 to 450 nM). (A) CBP-HbpR-mediated DNase I protection pattern for the bottom strand of hbpD3D4. (B) CBP-HbpR-mediated DNase I protection pattern for the top strand of hbpD3D4. Nucleotide numbering was relative to the hbpD transcriptional start site. For symbols and further explanations, see the legend to Fig. 4.
Affinity of CBP-HbpR for its binding sites.
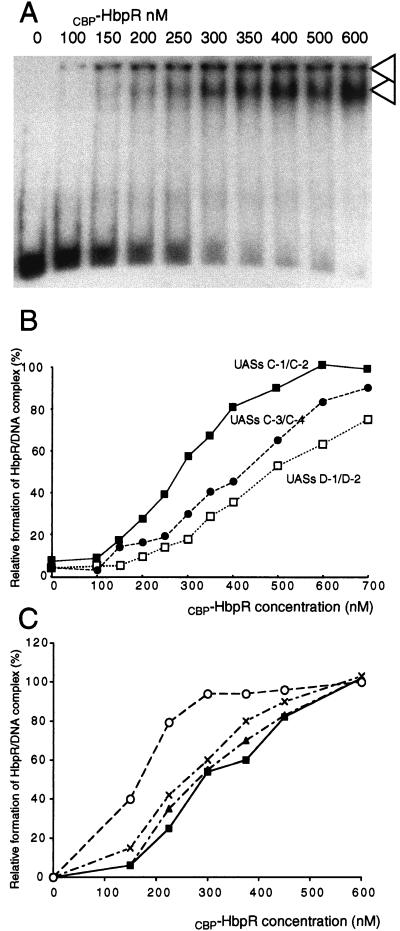
The affinity of CBP-HbpR for its three pairs of UASs in the hbp gene region was studied by GMSA titration. Increasing amounts of CBP-HbpR (between 0 and 600 nM) were allowed to contact DNA fragments with either UASs C-1 and C-2 (hbpCCC6) or UASs C-3 and C-4 (hbpCAC2) in the hbpC promoter or UASs D-1 and D-2 (hbpD3D4) in the hbpD promoter. In the presence of CBP-HbpR, two protein-DNA complexes were observed; one of these had not migrated into the gel at all (called complex 1), whereas the other had migrated to just below the wells (called complex 2) (Fig. 6A). Neither complex was present when the CBP-HbpRΔ purified fraction was incubated with the fragments containing each of the pairs of UASs (data not shown). This result indicated that the complexes were caused by HbpR-mediated binding. With increasing amounts of CBP-HbpR, the band corresponding to protein-DNA complex 2 increased in intensity (Fig. 6A), whereas the amount of complex 1 varied little. With laser densitometry, the relative densities of complex 2 obtained with increasing amounts of HbpR were calculated (Fig. 6B). Such affinity curves were made for all three pairs of UASs (Fig. 7B) and showed that CBP-HbpR had the highest affinity for UASs C-1 and C-2 (50% binding at 270 nM CBP-HbpR). Both UASs C-3 and C-4 and UASs D-1 and D-2 were bound with less affinity (50% binding at 430 and 490 nM CBP-HbpR, respectively). The reason for this affinity difference may lay in differences among the palindromic sequences which constitute the UASs or in the different amounts of spacing between the pairs of palindromic sequences.
FIG. 6.
Affinity characteristics of CBP-HbpR. (A) GMSA of fragment hbpCCC6 containing UASs C-1 and C-2 with increasing concentrations of CBP-HbpR (indicated above lanes). The amount of radiolabeled DNA fragment in the assay corresponded to 80 fmol. The arrowheads point to complex 1 (top) and complex 2 (bottom) (see text). (B) Graphic representation of the relative densities of the CBP-HbpR-DNA complexes formed with increasing CBP-HbpR concentrations with fragments containing UASs C-1 and C-2, UASs C-3 and C-4, and UASs D-1 and D-2. Relative densities were calculated by laser scanning densitometric analysis of band darkness on autoradiograms (as shown in panel A) and represent the darkness of the protein-DNA complex in each lane compared to that in the incubation with 600 nM HbpR. (C) Graphic representation of the relative densities of the CBP-HbpR-DNA complexes formed with increasing CBP-HbpR concentrations with fragments containing UASs C-1 and C-2 in the presence of ATP (multiplication signs), 2-HBP (triangles), and ATP and 2-HBP (open circles) or in the absence of an inducer (closed squares). ATP was used at a concentration of 5 mM, and 2-HBP was used at 10 μM. Relative densities were calculated as explained for panel B.
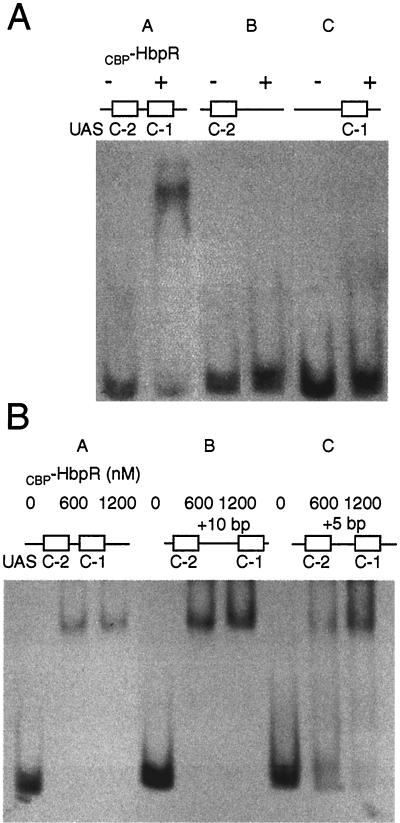
FIG. 7.
GMSAs with fragments containing modified UASs C-1 and C-2 and CBP-HbpR. (A) Effect of deleting one UAS on the binding of CBP-HbpR. Lanes: A, incubation of DNA fragment hbpCCC6, containing the native configuration of UASs C-1 and C-2, with (+) or without (−) 600 nM CBP-HbpR; B, incubation with the insert of pHB156 containing only the proximal UAS, C-1; C, incubation with the insert of pHB157 containing only the distal UAS, C-2 (12). (B) Effect of adding 5 and 10 bp between the two palindromes comprising UASs C-1 and C-2. Lanes: A, incubation with fragment hbpCCC6; B, incubation with the insert of pHB163, containing the 10-bp insertion; C, incubation with the insert of pHB162, containing the 5-bp insertion. The amount of CBP-HbpR added to each binding assay mixture is indicated above each lane.
Affinity of CBP-HbpR for UASs C-1 and C-2 changes in the presence of ATP and 2-HBP.
When increasing concentrations of CBP-HbpR were incubated with UASs C-1 and C-2 in the presence of ATP and/or 2-HBP, the following changes in binding affinity were observed in GMSAs. When tested separately, ATP and 2-HBP did not significantly change the binding affinity of CBP-HbpR for UASs C-1 and C-2 (50% binding at 270 nM CBP-HbpR) (Fig. 6C). However, the addition of both ATP and 2-HBP resulted in the formation of a CBP-HbpR-DNA complex at lower CBP-HbpR concentrations (50% binding at 170 nM CBP-HbpR). The distances to which the CBP-HbpR-DNA complex migrated in the gel in the presence or absence of ATP and 2-HBP did not differ, suggesting that the compositions of the protein-DNA complexes were similar under all four tested conditions. Increased binding affinity of CBP-HbpR was optimal at 2-HBP concentrations of between 10 and 100 μM. At higher 2-HBP concentrations, the binding affinity of CBP-HbpR decreased and was fully abolished at 1 mM 2-HBP (data not shown). At high 2-HBP concentrations, the proteins may become denatured.
The presence and the conformation of the UASs C-1 and C-2 are critical for the binding of HbpR.
We next addressed whether cooperative interactions were necessary for HbpR binding to UASs C-1 and C-2. We used GMSAs with fragments that contained either one or both pairs of palindromic sequences making up the UASs. Only the DNA fragment containing both UASs C-1 and C-2 was bound by CBP-HbpR (at 600 nM HbpR) (Fig. 7A). In contrast, DNA fragments containing only UAS C-1 or UAS C-2 were not bound by CBP-HbpR (Fig. 7A, lanes B and C). The addition of 2-HBP and ATP to assays with CBP-HbpR and DNA fragments with only UAS C-1 or UAS C-2 did not result in a stable protein-DNA complex visible on GMSAs either, not even at 600 nM CBP-HbpR (data not shown). These results demonstrated that both pairs of palindromes were needed for HbpR binding and suggested that some sort of cooperativity is needed between different HbpR protein complexes (most likely dimers bound to each UAS).
We next addressed whether the 32-bp spacing between the centers of UASs C-1 and C-2 is critical for cooperative interactions. We constructed fragments in which the intervening sequences between the two palindromes were increased by 5 and 10 bp (half helical and full helical insertions, respectively). Both the DNA with the original configuration and a DNA fragment with an additional 10 bp between the palindromes were completely bound in GMSAs at 600 nM CBP-HbpR (Fig. 7B, lanes A and B). However, the DNA fragment with an additional 5 bp between the palindromes did not form a stable protein-DNA complex at 600 nM CBP-HbpR. Rather, a smear was visible (Fig. 7B, lane C), suggesting that some binding by CBP-HbpR occurred but that the complex was not stable under the conditions of the gel analysis. Even at 1,200 nM, the DNA fragment was still not completely bound by CBP-HbpR. These results indicated that increasing the spacing between the palindromes by 5 bp (half helical insertion) destabilized the formation of the HbpR-DNA complex, whereas the insertion of 10 bp did not. Furthermore, these results showed that the affinity differences between UASs C-1 and C-2 and UASs D-1 and D-2 (with spacings of 32 and 42 bp between the centers of the palindromes, respectively) were not necessarily caused by spacing differences between the pairs of palindromes.
DISCUSSION
It was previously shown that HbpR, the transcription activator for the 2-HBP pathway in P. azelaica, mediates transcription from two σ54-dependent promoters, one of which is in front of the hbpC gene and the other of which is in front of the hbpD gene (11). HbpR has moderate sequence similarity to XylR but still belongs to the same subclass of the NtrC family of transcription activators (11). Here, we confirmed part of the working hypothesis for HbpR-mediated activation by showing the in vitro DNA binding of a partially purified CBP-HbpR fusion protein to promoter fragments. In addition, we discovered the binding of HbpR to a region (UASs C-3 and C-4) which does not function as a promoter for hbp transcription but which earlier had been noted as a potentially intact HbpR binding site (12). DNase I footprinting located the HbpR binding sites to two pairs of palindromic sequences which previously had been predicted as potential binding sites from DNA sequence comparisons with the XylR and DmpR binding sites (11). Unfortunately, we were not able to completely purify CBP-HbpR to homogeneity, nor were we able to cleave the CBP tag from HbpR without causing the loss of its activity. In principle, therefore, the presence of two other proteins in the fractions used for the DNA binding studies could have been responsible for the binding, and the CBP tag could have influenced the behavior of HbpR. From experiments in which we used purified protein extracts from E. coli producing CBP-HbpR with a large C-terminal deletion, we concluded that the contaminating proteins were not causing the observed DNA binding. Since the expression of the bacterial luciferase from PhbpC in the presence of CBP-HbpR was similar to that in the native HbpR configuration, we concluded that the CBP tag also did not change the properties of the HbpR protein to activate the PhbpC promoter. Furthermore, the CBP-HbpR fraction bound the hbpRC and hbpAD intergenic regions specifically, indicating that the CBP tag in the N-terminal part of HbpR did not disturb DNA binding. This notion is consistent with the finding that the N-terminal portions of the XylR and DmpR proteins did not affect DNA binding properties (19). In this respect, the 4-kDa CBP tag behaves in a manner similar to that of the 2-kDa His tag, which recently was used for purification of the XylR/DmpR-type transcription activator TouR (3).
In the absence of an effector and ATP, HbpR binds simultaneously to both pairs of UASs, a behavior similar to that of DmpR (34) but different from that of a constitutively active XylR variant devoid of the A domain (20). Furthermore, GMSAs with fragments with deletions of either UAS C-1 or UAS C-2 showed that one UAS was not sufficient for HbpR binding, not even in the presence of ATP and 2-HBP. This finding is consistent with initial observations made for the binding of NtrC from Salmonella enterica serovar Typhimurium (22) to the enhancer (UAS) in the glnA promoter (4, 24), although more recent studies demonstrated the binding of NtrC to an (artificial) single UAS (26, 31). The absence of binding to a single UAS suggests that HbpR binding is cooperative; i.e., the presence of the second UAS is needed to obtain full occupancy of both binding sites. Since we only observed one type of protein-DNA complex in GMSAs and DNA binding only with fragments containing both UASs, we presume that the complexes in GMSA consisted of two dimers of HbpR occupying both UASs, as in the case of NtrC (26). Therefore, we were unable to identify which of the UASs is bound by HbpR with a greater affinity.
The cooperativity of HbpR binding was not affected by an additional 10 bp between the two binding sites. Inserting 10 bp would result in the two binding sites being separated although maintained on the same side of the helix. Apparently, however, the HbpR dimers binding to this region could still produce the normal protein-protein interactions required for cooperativity. In fact, the insertion of 10 bp resulted in a configuration of both UASs similar to the spacing between the two palindromes in the hbpD promoter. Therefore, wider spacing between the UASs cannot explain the weaker affinity of HbpR for PhbpD than for PhbpC. A 5-bp insertion, on the other hand, would diminish the binding of HbpR, probably by positioning the UASs on opposite sides of the DNA helix. In addition, for XylR and DmpR it was shown that offsetting the distal UAS relative to the proximal one lowered promoter activity (20, 34).
Essential components in the complete activation cycle of NtrC-type proteins are the activation of the protein and the formation of an oligomeric complex at the UASs, most likely consisting of a tetramer of dimers (26, 31). The binding affinity of phosphorylated NtrC for the enhancer is slightly greater than that for unphosphorylated NtrC (4, 26), a fact which could explain why activated HbpR (in the presence of 2-HBP and ATP) binds the UAS DNA fragment more efficiently than inactive HbpR. The observed apparent change in the binding affinity of activated CBP-HbpR for the UASs (i.e., in the presence of 2-HBP and ATP) is the first time that this finding has been reported for a protein of the XylR/DmpR subclass. Studies with XylR have mostly been hindered by the inability to purify the intact protein. For this reason, a constitutively active form of XylR devoid of its N-terminal effector binding domain has been used because it could be purified. However, some of the results with respect to the cooperativity of DNA binding by XylR therefore may have been slightly different from those obtained with the complete protein (7). For example, DctD variants devoid of the N-terminal portion are constitutively active, like XylR but different from NtrC (16). However, the magnitude of the cooperative binding of DctD to its UASs changed when the N-terminal portion was deleted (30). For the wild-type DctD protein, the intrinsic affinity was 20-fold lower for the distal binding site than for the proximal binding site; however, for the truncated, constitutively active protein, DctDΔ1-142 (with a deletion of the N-terminal portion), the affinity difference was larger (30). These findings demonstrate that deletion of a portion of the protein not directly involved in DNA binding can have an influence on its DNA binding properties. For NtrC, a different phenomenon was observed. A constitutively active mutant form of NtrC exists, although it has a single amino acid change (i.e., S160F). The NtrC-S160F protein had the same binding affinity for its UASs as the NtrC protein but demonstrated greater cooperativity, apparently due to new protein-protein contacts (31). Since DNA binding studies are possible with an intact HbpR protein, the HbpR system may be important in refining the model of activation by XylR/DmpR-type transcription activators.
The full activation cycle for NtrC-type and XylR/DmpR-type transcription activators probably is as follows. In solution, the proteins exist as dimers, as proven for NtrC (26); under nonactivating conditions, the proteins bind as a pair of dimers to their enhancer (UASs) (26). In this form, NtrC has low ATPase activity (35), and it is assumed that no proper contacts can be formed between NtrC and σ54 RNA polymerase complexed to the promoter. Upon phosphorylation, NtrC changes affinity for its binding site and forms a larger complex at the UASs (most likely octamers) (26, 31). NtrC subunits inside this complex may have increased ATPase activity, and the complex itself will interact with the σ54 RNA polymerase-promoter complex, resulting in transcriptional initiation. For XylR, it has been proposed that in the presence of ATP, the protein-DNA structures seem to constantly assemble and disassemble between octamers and two dimer pairs (7). This scenario could explain why no higher-order structures of activated HbpR and the UAS C-1 and C-2 fragments were visible in GMSAs, as the equilibrium was mostly on the side of the dimer pair interaction.
REFERENCES
- 1.Abril, M. A., M. Buck, and J. L. Ramos. 1991. Activation of the Pseudomonas TOL plasmid upper pathway operon. J. Biol. Chem. 266:15832-15838. [PubMed] [Google Scholar]
- 2.Abril, M. A., and J. L. Ramos. 1993. Physical organization of the upper pathway operon promoter of the Pseudomonas TOL plasmid. Sequence and positional requirements for XylR-dependent activation of transcription. Mol. Gen. Genet. 239:281-288. [DOI] [PubMed] [Google Scholar]
- 3.Arenghi, F. L., P. Barbieri, G. Bertoni, and V. de Lorenzo. 2001. New insights into the activation of o-xylene biodegradation in Pseudomonas stutzeri OX1 by pathway substrates. EMBO Rep. 2:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, P., and L. J. Reitzer. 1995. Active contribution of two domains to cooperative DNA binding of the enhancer-binding protein nitrogen regulator I (NtrC) of Escherichia coli: stimulation by phosphorylation and the binding of ATP. J. Bacteriol. 177:2490-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lorenzo, V., M. Herrero, M. Metzke, and K. N. Timmis. 1991. An upstream XylR- and IHF-induced nucleoprotein complex regulates the σ54-dependent P u promoter of TOL plasmid. EMBO J. 10:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon, R. 1986. The xylABC promoter from the Pseudomonas putida TOL plasmid is activated by nitrogen regulatory genes in Escherichia coli. Mol. Gen. Genet. 203:129-136. [DOI] [PubMed] [Google Scholar]
- 7.Garmendia, J., and V. de Lorenzo. 2000. Visualization of DNA-protein intermediates during activation of the P u promoter of the TOL plasmid of Pseudomonas putida. Microbiology 146:2555-2563. [DOI] [PubMed] [Google Scholar]
- 8.Gomada, M., S. Inouye, H. Imaishi, A. Nakazawa, and T. Nakazawa. 1992. Analysis of an upstream regulatory sequence required for activation of the regulatory gene xylS in xylene metabolism directed by the TOL plasmid of Pseudomonas putida. Mol. Gen. Genet. 233:419-426. [DOI] [PubMed] [Google Scholar]
- 9.Inouye, S., M. Gomada, U. M. X. Sangodkar, A. Nakazawa, and T. Nakazawa. 1990. Upstream regulatory sequence for transcriptional activator XylR in the first operon of xylene metabolism on the TOL plasmid. J. Mol. Biol. 216:251-260. [DOI] [PubMed] [Google Scholar]
- 10.Inouye, S., A. Nakazawa, and T. Nakazawa. 1988. Nucleotide sequence of the regulatory gene xylR of the TOL plasmid from Pseudomonas putida. Gene 66:301-306. [DOI] [PubMed] [Google Scholar]
- 11.Jaspers, M. C., A. Schmid, M. H. Sturme, D. A. Goslings, H. P. Kohler, and J. R. Van Der Meer. 2001. Transcriptional organization and dynamic expression of the hbpCAD genes, which encode the first three enzymes for 2-hydroxybiphenyl degradation in Pseudomonas azelaica HBP1. J. Bacteriol. 183:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaspers, M. C., M. Sturme, and J. R. van Der Meer. 2001. Unusual location of two nearby pairs of upstream activating sequences for HbpR, the main regulatory protein for the 2-hydroxybiphenyl degradation pathway of ‘Pseudomonas azelaica’ HBP1. Microbiology 147:2183-2194. [DOI] [PubMed] [Google Scholar]
- 13.Jaspers, M. C., W. A. Suske, A. Schmid, D. A. Goslings, H. P. Kohler, and J. R. van der Meer. 2000. HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J. Bacteriol. 182:405-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohler, H. P. E., D. Kohler-Staub, and D. D. Focht. 1988. Degradation of 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1. Appl. Environ. Microbiol. 54:2683-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohler, H. P. E., A. Schmid, and M. van der Maarel. 1993. Metabolism of 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1: production and consumption of 2,2′,3-trihydroxybiphenyl. J. Bacteriol. 175:1621-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. H., D. Scholl, B. T. Nixon, and T. R. Hoover. 1994. Constitutive ATP hydrolysis and transcription activation by a stable, truncated form of Rhizobium meliloti DCTD, a σ54-dependent transcriptional activator. J. Biol. Chem. 269:20401-20409. [PubMed] [Google Scholar]
- 17.Morett, E., and L. Segovia. 1993. The σ54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez-Martín, J., and V. de Lorenzo. 1996. ATP binding to the σ54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell 86:331-339. [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Martín, J., and V. de Lorenzo. 1996. In vitro activities of an N-terminal truncated form of XylR, a σ54-dependent transcriptional activator of Pseudomonas putida. J. Mol. Biol. 258:575-587. [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Martín, J., and V. de Lorenzo. 1996. Physical and functional analysis of the prokaryotic enhancer of the σ54-promoters of the TOL plasmid of Pseudomonas putida. J. Mol. Biol. 258:562-574. [DOI] [PubMed] [Google Scholar]
- 21.Pérez-Martín, J., F. Rojo, and V. de Lorenzo. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58:268-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porter, S. C., A. K. North, A. B. Wedel, and S. Kustu. 1993. Oligomerization of NtrC at the glnA enhancer is required for transcriptional activation. Genes Dev. 7:2258-2273. [DOI] [PubMed] [Google Scholar]
- 23.Ravatn, R., S. Studer, A. J. B. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reitzer, L. J., and B. Magasanik. 1986. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell 45:785-792. [DOI] [PubMed] [Google Scholar]
- 25.Rippe, K. 2000. Simultaneous binding of two DNA duplexes to the NtrC-enhancer complex studied by two-color fluorescence cross-correlation spectroscopy. Biochemistry 39:2131-2139. [DOI] [PubMed] [Google Scholar]
- 26.Rippe, K., N. Mucke, and A. Schulz. 1998. Association states of the transcription activator protein NtrC from E. coli determined by analytical ultracentrifugation. J. Mol. Biol. 278:915-933. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid, A. 1997. Ph.D. thesis. Universität Stuttgart, Stuttgart, Germany.
- 30.Scholl, D., and B. T. Nixon. 1996. Cooperative binding of DctD to the dctA upstream activation sequence of Rhizobium meliloti is enhanced in a constitutively active truncated mutant. J. Biol. Chem. 271:26435-26442. [DOI] [PubMed] [Google Scholar]
- 31.Sevenich, F. W., J. Langowski, V. Weiss, and K. Rippe. 1998. DNA binding and oligomerization of NtrC studied by fluorescence anisotropy and fluorescence correlation spectroscopy. Nucleic Acids Res. 26:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shingler, V. 1996. Signal sensing by σ54-dependent regulators: derepression as a control mechanism. Mol. Microbiol. 19:409-416. [DOI] [PubMed] [Google Scholar]
- 33.Sticher, P., M. C. M. Jaspers, K. Stemmler, H. Harms, A. J. B. Zehnder, and J. R. van der Meer. 1997. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl. Environ. Microbiol. 63:4053-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sze, C. C., A. D. Laurie, and V. Shingler. 2001. In vivo and in vitro effects of integration host factor at the DmpR-regulated σ54-dependent Po promoter. J. Bacteriol. 183:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss, D. S., J. Batut, K. E. Klose, J. Keener, and S. Kustu. 1991. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell 67:155-167. [DOI] [PubMed] [Google Scholar]
- 36.Wyborski, D. L., J. C. Bauer, C. F. Zheng, K. Felts, and P. Vaillancourt. 1999. An Escherichia coli expression vector that allows recovery of proteins with native N-termini from purified calmodulin-binding peptide fusions. Protein Expr. Purif. 16:1-10. [DOI] [PubMed] [Google Scholar]
- 37.Wyman, C., I. Rombel, A. K. North, C. Bustamante, and S. Kustu. 1997. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science 275:1658-1661. [DOI] [PubMed] [Google Scholar]