Abstract
The halophilic bacterium Halomonas elongata synthesizes as its main compatible solute the aspartate derivative ectoine. We constructed a deletion mutant of H. elongata, KB1, defective in ectoine synthesis and tolerating elevated salt concentrations only in the presence of external compatible solutes. The dependency of KB1 on solute uptake for growth in high-salt medium was exploited to select insertion mutants unable to accumulate external solutes via osmoregulated transporters. One insertion mutant out of 7,200 failed to accumulate the osmoprotectants ectoine and hydroxyectoine. Genetic analysis of the insertion site proved that the mutation affected an open reading frame (ORF) of 1,281 bp (teaC). The nucleotide sequence upstream of teaC was determined, and two further ORFs of 603 bp (teaB) and 1,023 bp (teaA) were identified. Deletion of teaA and teaB proved that all three genes are mandatory for ectoine uptake. Sequence comparison showed significant identity of TeaA, TeaB, and TeaC to the transport proteins of the recently identified tripartite ATP-independent periplasmic transporter family (TRAP-T). The affinity of the cells for ectoines was determined (Ks = 21.7 μM), suggesting that the transporter TeaABC exhibits high affinity for ectoines. An elevation of the external osmolarity resulted in a strong increase in ectoine uptake via TeaABC, demonstrating that this transporter is osmoregulated. Deletion of teaC and teaBC in the wild-type strain led to mutants which excreted significant amounts of ectoine into the medium when cultivated at high salt concentrations. Therefore, the physiological role of TeaABC may be primarily to recover ectoine leaking through the cytoplasmic membrane.
Halophilic organisms living in saline environments such as salt lakes, coastal lagoons, and man-made salterns are challenged by two stress factors, the high inorganic ion concentration and the low water potential. A nonadapted organism exposed to such an environment must cope with its cytoplasmic water having a higher water potential than the water of the surrounding environment. Water always flows from a high to a low potential until the potential gradient is abolished. Thus, the cytoplasm, which is surrounded by a membrane freely permeable to water, will lose its free cytoplasmic water, resulting in cell shrinkage (4). The loss of water will cause cessation of growth, possibly due to molecular crowding, and thus reduced diffusion rates of proteins and metabolites.
Halophilic prokaryotes have developed two principal mechanisms to lower the potential of cytoplasmic water, avoiding the loss of water from the cell and achieving a cytoplasm of osmotic strength similar to that of the surrounding medium. These two mechanisms, which allow osmotic adaptation, are the salt-in-cytoplasm mechanism and the organic-osmolyte mechanism.
The salt-in-cytoplasm mechanism is considered the typical archaeal strategy of osmoadaptation (e.g., halobacteria). However, members of the Bacteria domain such as Salinibacter ruber and a specialized group of gram-positive anaerobic halophilic bacteria (Halanaerobiales) are now known to employ this strategy as well (37, 43). Organisms following this strategy adjust the water potential of the cell by raising the salt (KCl) concentration in the cytoplasm and must adapt the interior protein chemistry to a high salt concentration (6, 27). As a consequence, organisms using the salt-in-cytoplasm strategy display a relatively narrow adaptation and their growth is restricted to high saline environments.
The organic-osmolyte mechanism is widespread among Bacteria and also present in methanogenic Archaea (11, 26, 36, 54). In response to an osmotic stress these organisms mainly accumulate osmoprotective organic compounds such as amino acids and amino acid derivatives (11). These nonionic, highly water-soluble compounds do not disturb the metabolism even at high cytoplasmic concentrations and are therefore aptly named compatible solutes (2). Halophilic cells using compatible solutes can basically preserve the same enzymatic machinery as nonhalophiles and are more flexible, displaying tolerance to high and low salt concentrations.
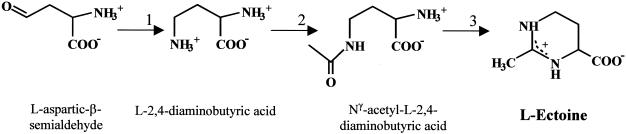
Many halophilic bacteria are able to synthesize their own compatible solutes such as glycine betaine, which is the typical solute of phototrophic bacteria (12, 30), and ectoine (Fig. 1), which is predominantly synthesized by chemoheterotrophic halophiles (48). However, halophilic microorganisms do not rely entirely upon de novo synthesis of solutes; they are also able to take up compatible solutes from the surrounding medium, which is a far more economical way to accumulate osmoprotectants (36).
FIG. 1.
Biosynthetic pathway of the compatible solute ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidine carboxylic acid) based on enzymological and genetic studies (14, 29, 40). 1, l-2,4-Diaminobutyric acid transaminase (encoded by ectB); 2, l-2,4-diaminobutyric acid Nγ-acetyltransferase (encoded by ectA); 3, ectoine synthase (encoded by ectC).
Halophilic bacteria taking up compatible solutes from the surrounding medium must be equipped with osmoregulated transporters facilitating this process. Such transporters have been studied, but only in nonhalophilic bacteria such as Escherichia coli, Corynebacterium glutamicum, Bacillus subtilis, and some halotolerant microorganisms (20, 24, 31, 39, 44). These organisms, which demonstrate limited abilities to cope with osmotic stress in comparison to real halophiles, possess either high-affinity binding protein-dependent ABC transporters or secondary transporters consisting of a single transmembrane protein.
To gain a deeper insight into the aspects of osmoregulated solute transport of truly halophilic Bacteria, we focused our interest on the extremophilic bacterium Halomonas elongata, a proteobacterium of the gamma subdivision (55). H. elongata displays a broad salt tolerance and synthesizes ectoine (Fig. 1) and hydroxyectoine as compatible solutes (56).
In this study, we identified by transposon mutagenesis a new type of osmoregulated solute transporter in bacteria. The transport system accepts ectoine and hydroxyectoine as substrates and is responsible for the uptake of ectoines leaking through the cytoplasmic membrane. We therefore propose that the system be designated TeaABC, transporter for ectoine accumulation ABC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains, vectors, and recombinant plasmids used for this study are listed in Table 1. H. elongata strains were grown aerobically at 30°C on MM63 medium (28) with glucose as a carbon source and in various NaCl concentrations. For physiological characterization H. elongata was grown in 100 ml of saline MM63 liquid medium contained in 250-ml flasks with side arms for turbidity measurements.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Sourceb or reference |
|---|---|---|
| H. elongata | ||
| DSM 2581T | Type strain | DSMZ |
| KB1 | ΔectA Ect− | This study |
| AFE35 | ΔectA teaC::Tn1732 Kmr Ect−; osmoregulatory ectoine uptake deficiency | This study |
| KB1-2 | ΔectA ΔteaB Ect−; osmoregulatory ectoine uptake deficiency | This study |
| KB1-3 | ΔectA ΔteaA Ect−; osmoregulatory ectoine uptake deficiency | This study |
| KB1-4 | ΔectA ΔteaC Ect−; osmoregulatory ectoine uptake deficiency | This study |
| KB1-5 | ΔectA Δ(orf1::Ω) Smr Spr Ect− | This study |
| KB2 | ΔteaC; osmoregulatory ectoine uptake deficiency, ectoine excretion | This study |
| KB2-1 | ΔteaCB; osmoregulatory ectoine uptake deficiency, ectoine excretion | This study |
| E. coli | ||
| K-12 | Wild type | DSMZ |
| DH5α | F− φ80dlacZDM15 Δ(lacZYA-argF)U169 recA1 hsdR17(rK− mK+) supE44 Δ−thi-1 gyrA relA1 | 17 |
| S17-1 | thi pro hsdR hsdM+recA Tpr Smr | 49 |
| SM10 | Kmr Thi Pro His | 49 |
| Plasmids | ||
| pSUP102-Gm::Tn1732 | Kmr Cmr | U. Priefer, University of Bielefeld |
| pK18mobsacB | Kmrmob sacB | 47 |
| pKGB1 | 1,043-bp PCR fragment for ectA deletion containing 510 bp upstream and 533 bp downstream of ectA, Kmr | This study |
| pKGB2 | 1,346-bp PCR fragment for teaB deletion containing 671 bp upstream and 675 bp downstream of teaB, Kmr | This study |
| pKGB3 | 1,863-bp PCR fragment for teaA deletion containing 940 bp upstream and 923 bp downstream of teaA | This study |
| pAVB1 | 2,197-bp PCR fragment for teaC deletion containing 1,090 bp upstream and 1,107 bp downstream of teaC, Kmr | This study |
| pKGB4 | 1,025-bp PCR fragment for orf1 deletion containing 512 bp upstream and 513 bp downstream of orf1 with Ω cassette insertion, Spr Smr Kmr | This study |
| pKGB6 | 1,045-bp PCR fragment for teaBC deletion containing 493 bp upstream and 552 bp downstream of teaBC | This study |
Abbreviations: Cm, chloramphenicol; Km, kanamycin; Sm, streptomycin; Tp, trimethoprim; Ect, ectoine.
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany.
E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB) (33) medium. Antibiotics were used at the following concentrations: kanamycin, 50 μg ml−1; streptomycin, 200 μg ml−1; and spectinomycin, 100 μg ml−1.
DNA isolation and manipulation.
Total DNA from H. elongata was isolated as described previously (22). Routine manipulation of DNA, plasmid isolation, construction of recombinant plasmids, electrophoresis of DNA, and transformation were carried out according to standard procedures (45). DNA sequencing, based on the method of Sanger et al. (46), was carried out by SequiServe (Vaterstetten, Germany). Southern hybridization experiments were performed after transfer of restriction fragments from 0.8% agarose gels to nylon membranes with a nonradioactive DNA labeling and detection kit under conditions specified by the manufacturer (Roche, Mannheim, Germany).
Transposon mutagenesis and selection for transport-deficient mutants.
Conjugation and Tn1732 mutagenesis were carried out by a modified method based on the technique described by Kunte and Galinski (22). After overnight filter mating of E. coli S17-1/pSUP102-Gm::Tn1732 and H. elongata KB1 on nutrient broth medium at 30°C, transconjugants of H. elongata were selected on MM63 medium (28) containing kanamycin (200 μg ml−1) and 0.34 M NaCl. After a 3-day incubation period, transconjugants were selected and placed onto a master plate of the same composition. Mutants with a deficiency in osmoregulatory solute uptake were then selected on saline MM63 medium (1.37 M NaCl) containing either ectoine (250 μM) or glycine-betaine (250 μM). Mutants that failed to grow on either medium were further analyzed by high-pressure liquid chromatography (HPLC) and uptake experiments.
Generation of deletion mutants.
DNA sequences upstream and downstream from the desired gene were joined together by applying the splicing-by-overlap-extension (SOE) PCR technique (18). The resulting PCR fragments were ligated into the shuttle vector pK18 mobsacB (47) and transferred into H. elongata by E. coli S17-1-mediated conjugation. Deletion mutants arising after double crossover were then selected on LBG medium containing 22% (wt/vol) sucrose at 37°C. The deletion sites were verified by PCR and DNA sequencing techniques.
Analytical methods.
For identification and quantification of intracellular compatible solutes, cells were harvested, freeze-dried, and extracted with methanol-chloroform-water as described by Galinski and Herzog (13). Cellular extracts as well as cultural medium were either analyzed by isocratic HPLC on an NH2 column (10) using a UV detector (220 nm) or by FMOC-HPLC as described by Kunte et al. (23).
Computer methods.
Protein and translated nucleotide databases were screened to find proteins similar to TeaABC, using the FASTA (38) and BLAST (1) programs. Multiple sequence alignments were constructed on an Apple Macintosh computer using the Clustal X program (51), in which the N-terminal leader sequences of TeaA and DctP were artificially removed in order to align the sequences correctly. The N-terminal leader sequence of TeaA was identified by using the SignalP program (34). Hydropathy profiles of proteins were constructed by the method of Kyte and Doolittle (25). Potential membrane-spanning units were identified by using the Dense Alignment Surface (DAS) program, available through the Transmembrane Prediction Server at the Stockholm Bioinformatics Center (www.sbc.su.se/∼mikkos/DAS/) (5).
Nucleotide sequence accession number.
The nucleotide sequence of teaABC, including orf1, was submitted to GenBank and assigned accession no. AY06146.
RESULTS
Generation of the ectA deletion mutant KB1 and selection for transposon mutants of KB1 defective in ectoine and hydroxyectoine uptake.
In order to identify transposon mutants with a defect in osmoregulatory uptake of osmoprotectants, a strain of H. elongata dependent on solute uptake from the medium for growth was constructed. To produce this strain, the ability of H. elongata to synthesize the compatible solute ectoine was eliminated. This was done by deleting the gene ectA, which encodes the l-2,4-diaminobutyric acid acetyltransferase, resulting in an interruption of the biosynthetic pathway of the ectoines (Fig. 1). The DNA regions upstream and downstream of ectA were joined together by applying the SOE-PCR technique. The resulting ΔectA fragment was cloned into the shuttle vector pK18 mobsacB and transferred into H. elongata by conjugation (22). The ΔectA mutants, arising after double crossover, were then selected on sucrose medium. Selected ΔectA mutant KB1 could only tolerate more than 0.86 M NaCl when compatible solutes (e.g., glycine-betaine or ectoines) were supplied in the medium (data not shown).
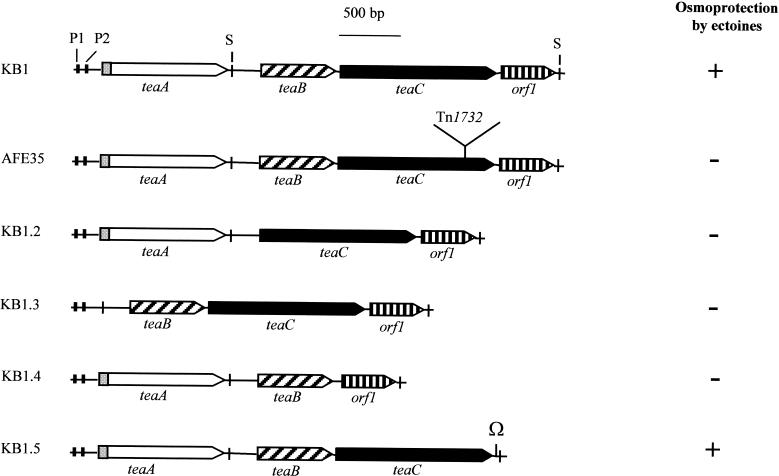
Tn1732 mutagensis was carried out with H. elongata KB1. The resulting 7,200 mutants were screened on mineral salt medium (1.38 M NaCl) containing either glycine betaine or ectoine-compatible solutes. One transposon mutant, H. elongata AFE35 (Fig. 2), no longer protected by exogenously provided ectoines, failed to grow on high-salt medium (>0.86 M NaCl). However, the mutation had no effect on the accumulation of glycine betaine, and AFE35 had the same salt tolerance in the presence of glycine betaine as H. elongata KB1 and the wild type of H. elongata.
FIG. 2.
Gene organization at the teaABC locus of H. elongata, location of Tn1732 insertion, deletion and marker replacement mutations, and osmoprotection analysis. The teaC gene in KB1 was identified by insertion mutagenesis (Tn1732). The DNA loci orf1 and teaAB were identified by ligase-mediated PCR and inactivated by deletion and marker replacement mutation (Ω), respectively. The relevance of teaABC and orf1 for osmoprotection by ectoine was verified by monitoring the growth of all mutants on high-salinity minimal plates (MM63 with 1.37 M NaCl) in the presence of 250 μM ectoine. Growth (growth, +; no growth, −) was scored after 3 days of incubation at 30°C. P1 and P2, potential promoter sequences; S, potential stem-loop sequences.
Osmoregulated uptake of ectoine and hydroxyectoine is impaired by the transposon insertion in H. elongata AFE35.
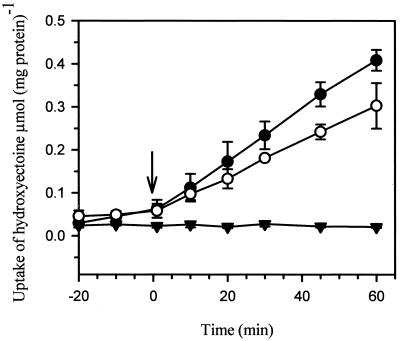
To test whether the osmoregulated uptake of ectoines was affected by the transposon insertion, H. elongata KB1 and the transposon mutant AFE35 were subjected to an osmotic upshift from 170 mM NaCl to 680 mM NaCl in the presence of ectoine and hydroxyectoine, respectively. While the osmotic upshock strongly induced the uptake of ectoines in H. elongata KB1, no osmoregulatory transport activities were detectable in the insertion mutant H. elongata AFE35 (Fig. 3). Cells of strain KB1 pretreated with 200 μg of chloramphenicol ml−1 were still able to respond to osmotic upshock and accumulated hydroxyectoine in the cytoplasm. However, in the presence of chloramphenicol, the final cytoplasmic ectoine content was about 25% lower than in untreated cells (Fig. 3).
FIG. 3.
Hydroxyectoine uptake mediated by the osmoregulated TeaABC transporter. Cells of H. elongata KB1 (ΔectA) and strain AFE35 (ΔectA teaC::Tn1732) were grown in saline minimal medium (MM63 with 170 mM NaCl). Hydroxyectoine (1 mM) was added to the medium 30 min prior to osmotic upshock. During exponential growth, KB1 and AFE35 were exposed to osmotic stress by increasing the salt concentration to 680 mM NaCl (↓). The uptake of hydroxyectoine was monitored for 60 min by analyzing the cytoplasm of KB1 and AFE by HPLC with a UV detector. At minimum, all experiments were carried out three times. Symbols: •, KB1; ▾, AFE35; ○, KB1 treated with chloramphenicol (200 μg ml−1)10 min before the cells were subjected to osmotic upshock at time zero.
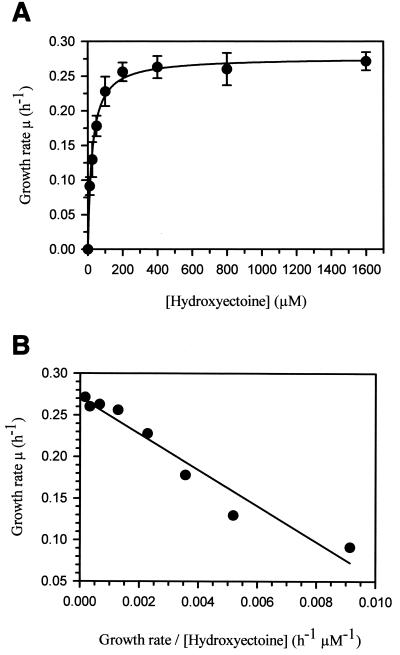
Since growth of H. elongata KB1 in high-saline minimal medium is dependent on the presence of external compatible solutes, lowering the ectoine concentration in the medium caused a decrease in growth rate, which enabled the affinity of the cells (Ks) for ectoines to be determined. The growth rates under ectoine-limited conditions were fitted by nonlinear regression, and the model that best described the correlation was the Monod model (Fig. 4). The plot of growth rate against growth rate/hydroxyectoine concentration (Eadie-Hofstee plot) showed that the cells had a saturation constant (Ks) of 21.7 μM for hydroxyectoine and 27.2 μM for ectoine, suggesting that the osmoregulated uptake of ectoines is mediated by a high-affinity transport system.
FIG. 4.
Affinity of whole cells for hydroxyectoine was determined in growth experiments with H. elongata KB1 under hydroxyectoine limitation. (A) Specific growth rates were determined from exponential-phase growth of hydroxyectoine-limited cultures in 1 M NaCl minimal medium (MM63). Lowering the solute concentration from 1,600 μM to 10 μM led to a decreasing growth rate of strain KB1. No growth was detected in hydroxyectoine-free medium. The growth rate data were fitted by nonlinear regression and described best by the Monod model. (B) The plot of growth rate against growth rate/hydroxyectoine concentration (Eadie-Hofstee plot) showed that the cells have a saturation constant of 21.7 μM. Growth rates were determined by a minimum of four independent experiments.
Tn1732 inserted into a DNA locus encoding a new type of osmoregulated transport system.
A 10-kb fragment containing the transposon Tn1732 was isolated from a partial gene bank of H. elongata AFE35, and 1,400 bp flanking the transposon insertion site were sequenced. For subsequent sequence analysis, it was taken into account that Tn1732 generates 5-bp direct repeats of the target DNA (53). Applying a ligase-mediated PCR technique, 2.3 and 1.2 kb of the regions adjacent to the transposon insertion site were isolated and their DNA sequences were determined.
Sequence analysis revealed that Tn1732 inserted into an open reading frame (ORF) of 1,281 bp, which we refer to as teaC (Fig. 2). This ORF encodes a 427-residue protein with a molecular mass of 44,925 Da. Two ORFs of 603 and 1,023 bp, designated teaB and teaA, respectively, were identified upstream of teaC. The ORFs teaB and teaA code for potential proteins with molecular masses of 22,168 and 38,240 Da, respectively. One ORF of 441 bp was found immediately downstream of teaC, called orf1, encoding a potential 147-residue protein with a mass of 15,741 Da. All four ORFs are preceded by potential ribosome-binding sites. In addition, the region upstream of teaA contains two potential σ70-dependent promoter sequences (Fig. 2). Following teaA and orf1, putative stem-loop structures were detected.
The deduced amino acid sequences of teaA, teaB, teaC, and orf1 were compared with sequences available from the databases. A comparison of the putative proteins TeaA, TeaB, and TeaC revealed a high degree of identity to tripartite ATP-independent periplasmic transporters (TRAP-T). TRAP-T-systems consist of three nonhomologous proteins: a large transmembrane protein, a small transmembrane protein, and a periplasmic substrate-binding protein (SBP). The putative TeaC protein shows 33.5% identical amino acids with the large transmembrane protein DctM of the DctPQM transporter specific for C4-dicarboxylate uptake in Rhodobacter capsulatus. This transporter was the first TRAP-T system to be described phenotypically and at the molecular level (9). The highest comparison scores were obtained with the putative TRAP-T proteins of Bacillus halodurans (GenBank accession no. AP001518) (50). More than 55% of the amino acids of the putative large membrane protein from B. halodurans were identical to and over 77% were similar to the amino acids of TeaC.
The small transmembrane proteins of TRAP-T are known to show little amino acid sequence similarity (36). However, TeaB showed a high amino acid sequence identity to the small transmembrane proteins of the TRAP-T family (35.1% of the amino acids were identical to the potential small transmembrane protein of B. halodurans). Both of the potential transmembrane proteins, TeaC and TeaB, are very hydrophobic, consisting of 61% (TeaB) and 73% (TeaC) apolar residues. The hydropathy profile of TeaB showed four hydrophobic regions of 19 to 23 amino acids which are potential membrane-spanning α-helices. The C and N termini of TeaB are predicted to face the cytoplasm. The same topology was recently proven to be correct for the small transmembrane protein DctQ of R. capsulatus by protein fusion analysis (57).
The hydropathy profile of TeaC revealed nine regions of hydrophobicity comprising 12 membrane-spanning units with a central hydrophilic loop, as indicated by the DAS transmembrane prediction program. The putative TeaA protein is 33.5% identical in amino acid sequence to the SBP of the potential TRAP system found in B. halodurans and shows 24.1% identical amino acids with the TRAP system of R. capsulatus. The gene teaA encodes a putative hydrophilic polypeptide which contains an N-terminal signal sequence of 25 residues, indicating that the TeaA protein is exported via the Sec pathway. The content of acetic amino acids is considerably high (19.3%), which is typical for proteins of halophilic bacteria in contact with a saline environment.
As indicated by the potential stem-loop sequences (Fig. 2), orf1, which is located 32 bp downstream of teaC, is thought to be transcribed along with teaBC. The potential 15.5-kDa protein encoded by this open reading frame shows similarities to the universal stress protein UspA found in E. coli (35) and UspA homologues identified in other prokaryotes.
teaA, teaB, and teaC are essential for ectoine transport; TeaABC mediates the uptake of ectoines leaking through the cytoplasmic membrane.
In order to determine the role of orf 1, teaA, teaB, and teaC in the uptake of ectoines in H. elongata, in-frame null mutations (ΔteaA, ΔteaB, and ΔteaC) and a marker replacement mutation (Δorf1) were constructed in strain KB1 using the SOE-PCR technique (Fig. 2). The replacement of orf1 by an Ω cassette (41) had no negative effects on osmoregulated ectoine uptake or on the growth at high salinities on minimal medium containing ectoine. In contrast, the deletion of teaA, teaB, or teaC and the transposon insertion in teaC abolished the ability of H. elongata to accumulate ectoine from the medium and proved that all three genes are required to express a functional ectoine transport system.
Hagemann et al. (16) reported that the glucosylglycerol transporter in the marine cyanobacterium Synechocystis sp. strain PCC 6803 is responsible for salvaging glucosylglycerol leaking into the medium. To prove whether TeaABC has a similar physiological function, we deleted teaC and teaBC in the wild-type strain of H. elongata. The teaC deletion mutant, named KB2, and the ΔteaBC mutant, called KB2-1, showed growth rates at different salt concentrations similar to those of the wild type and accumulated similar ectoine levels by de novo synthesis within the cytoplasm (Table 2; data not shown for KB2-1).
TABLE 2.
Growth rate and ectoine accumulation in cytoplasm and medium by H. elongata DSM 2581T (wild type) compared to teaC deletion mutant KB2
| NaCl concn (M) | Growth rate (h−1)
|
Ectoine content
|
||||
|---|---|---|---|---|---|---|
| Wild type | KB2 (ΔteaC) | Cytoplasm [μmol (mg of protein)−1]
|
Medium (μM)
|
|||
| Wild type | KB2 | Wild type | KB2 | |||
| 0.51 | 0.371 | 0.342 | 0.89 | 0.82 | —a | 214 |
| 1.03 | 0.243 | 0.276 | 2.34 | 2.29 | — | 288 |
| 1.71 | 0.212 | 0.227 | 3.54 | 3.77 | — | 370 |
—, not detectable by HPLC with a UV detector.
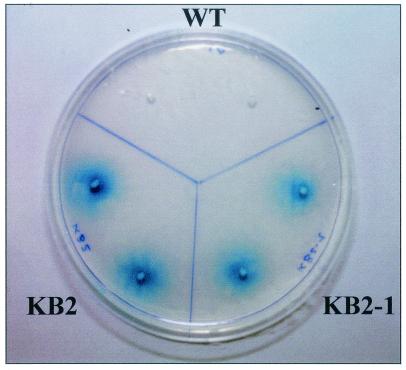
To verify the loss of the solute ectoine to the external medium, single colonies of KB2 and KB2-1 were transferred onto an agar plate, where 100 μl of an overnight culture of E. coli K-12 was spread out as an indicator organism. Since E. coli can tolerate up to 900 mM NaCl on minimal medium (MM63) only in the presence of external osmoprotectants (15), the growth of E. coli next to a colony of H. elongata would identify a mutant losing ectoine. No growth of E. coli was identified next to H. elongata DSM 2581T, whereas growth of the indicator organism was strongly supported in the surroundings of KB2-1 and KB2 due to ectoine leakage into the medium (Fig. 5). KB2, KB2-1, and the wild type of H. elongata were also grown in liquid mineral salt medium at different salt concentrations (MM63, 0.51, 1.03, and 1.71 M NaCl), and medium from exponentially growing cells was analyzed by HPLC. Both KB2 and KB2-1 lost ectoine, which accumulated to 0.370 mM in medium containing 1.71 M NaCl, while no ectoine was detectable in the medium of the wild type (Table 2).
FIG. 5.
Assay to detect compatible solute excretion by microorganisms, using E. coli K-12 as an indicator organism. An overnight culture of E. coli K-12 (100 μl) was spread onto agar medium (MM63) containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 900 mM NaCl, which inhibits the growth of E. coli in the absence of external compatible solutes. H. elongata DSM 2581T (wild type [WT]), strain H. elongata KB2 (ΔteaC), and strain KB2-1 (ΔteaBC) were transferred onto the assay plate and incubated for 48 h at 37°C. Growth of E. coli K-12 next to ectoine-excreting colonies is indicated by X-Gal, which is converted into a blue dye by the β-galactosidase activity of growing E. coli cells.
DISCUSSION
Until recently, the osmoregulated transport of compatible solutes in bacteria has been investigated mainly in nonhalophiles, in which several primary and secondary systems were described at the molecular and physiological level (21). In chemoheterotrophic halophiles, only one glycine-betaine transporter of unknown design was analyzed phenotypically (3). The data presented in this paper describe, for the first time, an osmoregulated compatible solute transport system at the molecular level and its physiological relevance for osmoadaptation in a halophilic bacterium.
We have identified a new type of osmoregulatory uptake system in the halophilic proteobacterium H. elongata, called TeaABC. TeaABC shows a high degree of sequence and structural similarities to uptake systems of the TRAP transporter family and is the first osmoregulated transporter of this type. TRAP transporters were found for the first time in Rhodobacter sphaeroides (19) and Rhodobacter capsulatus (9), where they have been shown to catalyze the nonosmoregulatory uptake of glutamate and C4-dicarboxylates, respectively. TRAP transporters appear to be widespread in prokaryotes, since homologues of the TRAP DctPQM system of Rhodobacter have been identified in numerous gram-negative and gram-positive Bacteria as well as in Archaea, based on results from database comparisons (9, 42).
Although the transport of substrate is mediated by a substrate-binding protein (SBP), TeaABC and other TRAP systems lack the typical sequences of the ATP-binding site described for primary SBP-dependent ABC transporters (Walker motif GxGKT and Rx4-12h4D). As shown by Jacobs et al. (19) and Forward et al. (9), transport activity of TRAP systems is not linked to ATP hydrolysis but is coupled to the cotransport of protons (or Na+ ions). The unique organization of the TRAP-T transmembrane domain, consisting of two proteins, is explained by the need for a membrane-based partner protein for the periplasmic SBP. According to the model proposed by Rabus et al. (42) and Driessen et al. (8), the larger membrane protein is thought to catalyze the actual transport reaction and is responsible for the energy coupling to the proton motive force, whereas the small membrane protein is not involved directly in the transport reaction but might be needed to interact with the SBP. This is plausible, since secondary transporters known to date do not contain SBP and are not designed to cooperate with periplasmic proteins.
TeaABC was proven to be an osmoregulated transporter catalyzing the uptake of ectoine and hydroxyectoine in response to osmotic upshock. In osmotic upshock experiments, chloramphenicol-treated cells of KB1 showed lower uptake activity for hydroxyectoine than cells from chloramphenicol-free medium, indicating that in addition to the regulation of TeaABC activity, the transcriptional overexpression also contributes to the osmoregulatory response. However, yet to be determined is whether this regulation pattern is present if cells of H. elongata are exposed to osmotic stress at higher salinity, where teaABC might be already fully induced.
TeaABC appears to be the only osmoregulated transporter of importance in H. elongata, since uptake of ectoines through other osmoregulated transporters is not detectable. Whether TeaABC accepts other nonectoine compounds has yet to be resolved. In contrast to many other osmoregulated compatible solute transporters, the physiological role of TeaABC is not restricted to the accumulation of external solutes in response to osmotic stress. Our studies revealed that TeaABC is also involved in maintaining a constant cytoplasmic ectoine level in the absence of external solutes by recovering synthesized ectoine leaking through the cell membrane. TeaABC might be designed primarily as a salvage system for ectoine lost from the cell, since the natural habitats of halophiles most likely do not contain sufficient ectoines to increase the salt tolerance significantly through uptake of external ectoine. Similar observations have been recorded for a halotolerant cyanobacterium which synthesizes glucosylglycerol as its main compatible solute (16, 32). The mutation of ggtA, a gene encoding a subunit of an ABC transport system mediating the uptake of glucosylglycerol in Synechocystis sp. strain PCC 6803, created a mutant leaky for glucosylglycerol.
The mechanism of compatible solute excretion is still unknown. It can be ruled out that the deficiency in one of the membrane components of the transporter (ΔteaC in KB2) caused the leakage (e.g., by leading to the misfolding of the remaining protein, TeaB). This was proven by deletion of both genes coding for the membrane-based proteins TeaB and TeaC, which created a mutant (KB2-1) that still lost ectoine to the medium. Touzé et al. recently identified a gene in Erwinia chrysanthemi named bspA, which was described as a regulator for a putative glycine betaine export channel (52). Mutation of bspA created a leaky mutant losing accumulated compatible solute glycine betaine to the medium. Such channels could explain how polar solutes (glucosylglycerol) and zwitterionic compounds (glycine betaine and ectoine) cross the cytoplasmic membrane. These results also indicate that the loss of osmoprotectants is not an uncontrolled process at all. Instead, export and recovery of compatible solutes such as ectoine might be part of an elegant mechanism to regulate the cytoplasmic solute concentration.
Osmoregulated compatible solute transporters such as TeaABC must be integrated (directly and indirectly) in the regulation of the cell's compatible solute synthetic pathways, since the uptake of external solutes results in an immediate decrease in the concentration of compatible solutes synthesized by the cell (7). In order to allow the ectoine pathway to be downregulated when solutes are transported from the medium, TeaABC must be linked to the synthesis of ectoine as well. This implies that any ectoine, regardless of its origin, even ectoine lost from the cell, will have this negative regulatory effect on the synthesis of ectoine. It is therefore possible that ectoine could serve as a signal for the regulation of its own synthesis. Increasing the ectoine concentration by de novo synthesis will lead to water influx and an increase in turgor pressure of salt-stressed cells. Assuming that the efflux of ectoine via an export channel is triggered by a signal such as membrane stretch, ectoine will be released to the periplasm if the turgor pressure reaches a certain threshold. Transport of exported ectoine back into the cytoplasm by the activated TeaABC system will downregulate ectoine synthesis and eventually stop the loss of ectoine.
The proposed regulation mechanism would allow the halophilic cell to maintain a compatible solute concentration, compensating for the external osmolality over a broad range of medium salinities. This model is also supported by the data obtained with mutants KB2 and KB2-1. Both strains not only leak ectoine but also overproduce ectoine, maintaining a cytoplasmic ectoine level identical to that of the wild type. The overproduction can be explained by missing regulation of the ectoine pathway through the TeaABC transporter. The way in which osmoregulated transporters are linked to the cell's metabolism is still to be resolved.
Acknowledgments
We thank Jörn Kalinowski (University of Bielefeld) for plasmid pK18 mobsacB. We also thank the Deutsche Studienstiftung for providing K.G. with a fellowship. We are grateful to Sharon Taylor for critical reading of the manuscript and to Birgit Amendt for valuable technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (Tr 133/27-1).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäfer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, A. D. 1976. Microbial water stress. Bacteriol. Rev. 40:803-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canovas, D., C. Vargas, L. N. Csonka, A. Ventosa, and J. J. Nieto. 1996. Osmoprotectants in Halomonas elongata: high-affinity betaine transport system and choline-betaine pathway. J. Bacteriol. 178:7221-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cayley, S., B. A. Lewis, and M. T. Record, Jr. 1992. Origins of the osmoprotective properties of betaine and proline in Escherichia coli K-12. J. Bacteriol. 174:1586-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 6.Dennis, P. P., and L. C. Shimmin. 1997. Evolutionary divergence and salinity-mediated selection in halophilic archaea. Microbiol. Mol. Biol. Rev. 61:90-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinnbier, U., E. Limpinsel, R. Schmid, and E. P. Bakker. 1988. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 150:348-357. [DOI] [PubMed] [Google Scholar]
- 8.Driessen, A. J. M., B. P. Rosen, and W. N. Konings. 2000. Diversity of transport mechanisms: common structural principles. Trends Biochem. Sci. 25:397-401. [DOI] [PubMed] [Google Scholar]
- 9.Forward, J. A., M. C. Behrendt, N. R. Wyborn, R. Cross, and D. J. Kelly. 1997. TRAP transporter: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J. Bacteriol. 179:5482-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frings, E., H. J. Kunte, and E. A. Galinski. 1993. Compatible solutes in representatives of the genera Brevibacterium and Corynebacterium: occurrence of tetrahydropyrimidine and glutamine. FEMS Microbiol. Lett. 109:25-32. [Google Scholar]
- 11.Galinski, E. A. 1995. Osmoadaptation in Bacteria. Adv. Microb. Physiol. 37:273-328. [PubMed] [Google Scholar]
- 12.Galinski, E. A., and H. G. Trüper. 1982. Betaine, a compatible solute in the extremely halophilic phototrophic bacterium Ectothiorhodospira halochloris. FEMS Microbiol. Lett. 13:357-360. [Google Scholar]
- 13.Galinski, E. A., and R. M. Herzog. 1990. The role of trehalose as a substitute for nitrogen-containing compatible solutes (Ectothiorhodospira halochloris). Arch. Microbiol. 153:607-613. [Google Scholar]
- 14.Göller, K., A. Ofer, and E. A. Galinski. 1998. Construction and characterization of a NaCl-sensitive mutant of Halomonas elongata impaired in ectoine synthesis. FEMS Microbiol. Lett. 161:293-300. [DOI] [PubMed] [Google Scholar]
- 15.Guesbet, G. M., M. Jebbar, R. Talibart, T. Bernard, and C. Blanco. 1994. Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiology 140:2415-2422. [DOI] [PubMed] [Google Scholar]
- 16.Hagemann, M., S. Richter, and S. Mikkat. 1996. The ggtA gene encodes a subunit of the transport system for the osmoprotective compound glucosylglycerol in Synechocystis sp. strain PCC 6803. J. Bacteriol. 179:714-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 18.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs, M. H. J., T. van der Heide, A. J. M. Driessen, and W. N. Konings. 1996. Glutamate transport in Rhodobacter sphaeroides is mediated by a novel binding protein-dependent secondary transport system. Proc. Natl. Acad. Sci. USA 93:12786-12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kappes, R. M., B. Kempf, and E. Bremer. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 22.Kunte, H. J., and E. A. Galinski. 1995. Transposon mutagenesis in halophilic eubacteria: conjugal transfer and insertion of transposon Tn5 and Tn1732 in Halomonas elongata. FEMS Microbiol. Lett. 128:293-299. [DOI] [PubMed] [Google Scholar]
- 23.Kunte, H. J., E. A. Galinski, and H. G. Trüper. 1993. A modified FMOC-method for the detection of amino acid-type osmolytes and tetrahydropyrimidines (ectoines). J. Microbiol. Methods 17:129-136. [Google Scholar]
- 24.Kunte, H. J., R. A. Crane, D. E. Culham, D. Richmond, and J. M. Wood. 1999. Protein ProQ influences osmotic activation of compatible solute transporter ProP in Escherichia coli K-12. J. Bacteriol. 181:1537-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 26.Lai, M.-C., D.-R. Yang, and M.-J. Chuang. 1999. Regulatory factors associated with synthesis of the osmolyte glycine betaine in the halophilic methanoarchaeon Methanohalophilus portucalensis. Appl. Environ. Microbiol. 65:828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanyi, J. K. 1974. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev. 38:272-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen, P. I., L. K. Sydne, B. Landfald, and A. R. Strøm. 1987. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: betaines, glutamic acid, and trehalose. Arch. Microbiol. 147:1-7. [DOI] [PubMed] [Google Scholar]
- 29.Louis, P., and E. A. Galinski. 1997. Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology 143:1141-1149. [DOI] [PubMed] [Google Scholar]
- 30.Mackay, M. A., R. S. Norton, and L. J. Borowitzka. 1984. Organic osmoregulatory solutes in cyanobacteria. J. Gen. Microbiol. 130:2177-2191. [Google Scholar]
- 31.MacMillan, S. V., D. A. Alexander, D. E. Culham, H. J. Kunte, E. V. Marshall, D. Rochon, and J. M. Wood. 1999. The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim. Biophys. Acta 1420:30-44. [DOI] [PubMed] [Google Scholar]
- 32.Mikkat, S., M. Hagemann, and A. Schoor. 1996. Active transport of glucosylglycerol is involved in salt adaptation of the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 142:1725-1732. [DOI] [PubMed] [Google Scholar]
- 33.Miller, H. J. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Nielsen, H., J. Engelbrecht, S. Brunak, and G. van Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 35.Nyström, N., and F. C. Neidhardt. 1992. Cloning, mapping, and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 6:3187-3198. [DOI] [PubMed] [Google Scholar]
- 36.Oren, A. 1999. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63:334-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oren, A., and L. Mana. 1February2002, posting date. Amino acid composition of bulk protein and salt relationships of selected enzymes of Salinibacter ruber, an extremely halophilic bacterium. Extremophiles [Online.] http://link.springer.de/link/service/journals/00792/contents/01/00241/. [DOI] [PubMed]
- 38.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peter, H., B. Weil, A. Burkovski, R. Krämer, and S. Morbach. 1998. Corynebacterium glutamicum is equipped with four secondary carriers for compatible solutes: identification, sequencing and characterization of the proline/ectoine uptake system, ProP, and the ectoine/proline/glycine betaine carrier, EctP. J. Bacteriol. 180:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters, P., E. A. Galinski, and H. G. Trüper. 1990. The biosynthesis of ectoine. FEMS Microbiol. Lett. 71:157-162. [Google Scholar]
- 41.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 42.Rabus, R., D. L. Jack, D. J. Kelly, and M. H. Saier, Jr. 1999. TRAP transporters: an ancient family of extracytoplasmic solute-receptor-dependent secondary transporters. Microbiology 145:3431-3445. [DOI] [PubMed] [Google Scholar]
- 43.Rengpipat, S., S. E. Lowe, and J. G. Zeikus. 1988. Effect of extreme salt concentrations on the physiology and biochemistry of Halobacteroides acetoethylicus. J. Bacteriol. 170:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rübenhagen, R., H. Rönsch, H. Jung, R. Krämer, and S. Morbach. 2000. Osmosensor and osmoregulator properties of the betaine carrier BetP from the Corynebacterium glutamicum in proteoliposomes. J. Biol. Chem. 275:735-741. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 48.Severin, J., A. Wohlfarth, and E. A. Galinski. 1992. The predominant role of the recently discovered tetrahydropyrimidines for the osmoadaptation of halophilic eubacteria. J. Gen. Microbiol. 138:1629-1638. [Google Scholar]
- 49.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 50.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, Y. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Touzé, T., G. Gouesbet, C. Boiangiu, M. Jebbar, S. Bonnassie, and C. Blanco. 2001. Glycine betaine loses its osmoprotective activity in a bspA strain of Erwinia chrysanthemi. Mol. Microbiol. 42:87-99. [DOI] [PubMed] [Google Scholar]
- 53.Ubben, D., and R. Schmitt. 1986. Tn 1721 derivatives for transposon mutagenesis, restriction mapping and nucleotide sequence analysis. Gene 41:145-152. [DOI] [PubMed] [Google Scholar]
- 54.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of aerobic moderately halophilic bacteria. Microbiol. Mol. Biol. Rev. 62:504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vreeland R. J., C. D. Litchfield, E. L. Martin, and E. Elliot. 1980. Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int. J. Syst. Bacteriol. 30:485-495. [Google Scholar]
- 56.Wohlfarth, A., J. Severin, and E. A. Galinski. 1990. The spectrum of compatible solutes in heterotrophic halophilic eubacteria of the family Halomonadaceae. J. Gen. Microbiol. 136:705-712. [Google Scholar]
- 57.Wyborn, N. R., J. Alderson, S. C. Andrews, and D. J. Kelly. 2001. Topological analysis of DctQ, the small integral membrane protein of the C4-dicarboxylate TRAP transporter of Rhodobacter capsulatus. FEMS Microbiol. Lett. 194:13-17. [DOI] [PubMed] [Google Scholar]