Abstract
For Escherichia coli, growth in the absence of ammonia is termed nitrogen limited and results in the induction of genes that assimilate other nitrogen sources, a response mediated by σ54 and nitrogen regulator I (NRI, also called NtrC). The astCADBE operon, which is required for growth with arginine as the sole nitrogen source, is moderately expressed during general nitrogen limitation and maximally expressed in the presence of arginine. The operon is also induced in stationary phase. Primer extension analysis of E. coli revealed the presence of a σ54-dependent promoter utilized in exponential phase during nitrogen limitation and a σS-dependent promoter active during stationary phase. We used an ast-lacZ fusion to show that arginine stimulates expression, that ArgR, the arginine repressor, enhances expression from both promoters but is not essential, and that transcription by the two forms of the RNA polymerase is competitive and mutually exclusive. We demonstrated the binding of RNA polymerase holoenzymes, NRI, and ArgR to the promoter region in vitro. We also reconstituted transcription from both promoters with purified components, which confirmed the accessory role of ArgR for the σ54-dependent promoter. Thus, the ast operon exhibits nitrogen source-specific induction that is unique for an NRI-dependent gene. The transcriptional regulation of the ast operon in E. coli differs from that in Salmonella enterica serovar Typhimurium, in which ArgR is required for ast operon expression.
Escherichia coli can utilize several compounds as the sole source of nitrogen in defined minimal media (33). Growth without ammonia, the compound that gives the fastest growth rate, induces proteins that transport and catabolize other nitrogen sources and assimilate the resulting nitrogen (25, 33). Alternative sigma factor σ54 and σ54-dependent activator nitrogen regulator I (NRI, also called NtrC) mediate this nitrogen-regulated (Ntr) response (25, 33).
Most Ntr genes that have been studied thus far are expressed in nitrogen-limiting (ammonia-lacking) media, regardless of the alternate nitrogen source present. Exceptions are some genes regulated by the σ70-dependent activator, Nac, itself the product of an Ntr gene (4). Such nitrogen source-specific regulation of Nac-dependent genes can be the result of a specific regulator that responds to a particular nitrogen source (4; S. Ruback and L. Reitzer unpublished results). Such regulation is common for σ70-dependent promoters but is unusual for σ54-dependent promoters. An exception appears to be the astCADBE operon. The genes of this operon encode the enzymes of the arginine succinyltransferase (AST) pathway and are required for growth of E. coli with arginine as the nitrogen source (40). Gene array analysis showed that expression of the ast operon is NRI dependent and Nac independent (52). Although arginine is not required for expression, arginine stimulates maximal expression (40). Expression of the ast operon in Salmonella enterica serovar Typhimurium implicated the arginine repressor protein, ArgR (23). ArgR is a homohexameric protein that, when complexed with arginine, represses arginine biosynthetic genes by binding at sites overlapping their promoters (24). Similar regulation of the E. coli ast operon could explain the arginine-specific induction.
In addition to Ntr control, the ast operon is induced by stationary phase, conditioned media, and carbon starvation (2, 5). Such regulation is physiologically advantageous since the resulting arginine catabolism produces citric acid cycle intermediates. Stationary-phase sigma factor σS has been implicated in this induction (15, 23).
We undertook a detailed in vivo and in vitro analysis of astCADBE expression and regulation. We verified the presence of two promoters, presented evidence that competition between these promoters exists, and obtained genetic and biochemical evidence for a nonessential accessory role of ArgR in the NRI-dependent expression of the σ54-dependent promoter. We also show that the regulation of the ast operon in E. coli differs significantly from that in S. enterica serovar Typhimurium.
(This work is a partial fulfillment of the requirement for a Ph.D. degree at the University of Texas at Dallas, Richardson, Tex., for A. Kiupakis.)
MATERIALS AND METHODS
Strains and plasmids.
All strains used for assays in this study are derivatives of E. coli K-12 strain W3110 (41). Strains and plasmids used are listed in Table 1, and oligonucleotide primers are listed in Table 2.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| W3110 (wild type) | lacL8 lacIq | Laboratory strain |
| AK15 | W3110 trp::putPA1303::[Kanr-astC′-′lac] | This study |
| AK16 | W3110 trp::putPA1303::[Kanr-astC′-′lac] rpoS::tet | This study |
| AK17 | W3110 trp::putPA1303::[Kanr-astC′-′lac] himD::cam | This study |
| AK18 | W3110 trp::putPA1303::[Kanr-astC′-′lac] argR::cam | This study |
| AK21 | W3110 trp::putPA1303::[Kanr-astC′-′lac] Δcrp Strr | This study |
| AK22 | W3110 rpoN::FRT-Kanr-FRT | This study |
| AK23 | W3110 ΔrpoN | This study |
| AK24 | W3110 trp::putPA1303::[Kanr-astC′-′lac] ΔrpoN | This study |
| AK26 | W3110 trp::putPA1303::[Kanr-astC′-′lac] ΔrpoN rpoS::tet | This study |
| CA8445 | Δcrp Strr | 37 |
| K5746 | Overproduces both IHF subunits | 30 |
| TE2680 | Δ(lac)X74 recD1903::Tn10 trpDC700::putPA1303::[Kans-Camr-′lac] | 13 |
| K4633 | recD::tet | D. Friedman (University of Michigan) |
| BW12848 | himD::cam | B. Wanner (Purdue University) |
| UM315 | rpoS::tet | Laboratory strain originally obtained from P. C. Loewen |
| Plasmids | ||
| pRS551 | Ampr Kanr-′lac | 42 |
| pTE103 | Ampr | 14 |
| pAK10 | AmprastCp | This study |
| pUTmini-Tn5 Cm | Ampr MCS-cam-MCS | 10 |
| pQErpoS30 | Ampr ptac::His6-rpoS | 3 |
| pTH7 | Ampr ptac::rpoN | 19 |
| pXZCRP | Amprcrp | 51 |
TABLE 2.
Oligonucleotide primers
| Primer | Sequence |
|---|---|
| XTH1 | GGCGA ATTCG GTGAG GTCTG GCGCG CAGGC CG |
| SOT3 | GGCGA ATTCT CATCC ATTCA TCAAA GTTTT CACGC G |
| SOT4 | GGATC CTGAG ACATA GCGAC CTCTA C |
| argR5 | CCAGG ATCCG GAGTC GCATC TTCAC C |
| argR6 | CCAGA ATTCT ATGCA AACAG TCAGC CC |
| argR7 | GCTCA AGCTT AATCT CTGCC CCGTC G |
| argR8 | CCACG GTACC AGCAT TTCAC GCATA TCC |
| himD1 | GTAAT TCTCT GACTC TTCGG |
| himD2 | CCCGA GGCAT ATTCA GAAG |
| CRP1 | CAGAG GATAA CCGCG C |
| CRP2 | ACGCG CCACT CCGAC G |
| RPOS1 | CCGTA AACCC GCTGC G |
| RPOS2 | TTGCG TGGTA TCTTC CGG |
| rpoN(f)-P4 | TGCGA CGTTT TAGCA GGAGA GTACG ATTCT GAACA TGAAG ATTCC GGGGA TCCGT CGACC |
| rpoN(r)-P1 | TGTTG AGCTG CATAG TGTCT TCCTT ATCGG TTGGG TCAAA GTGTA GGCTG GAGCT GCTTC G |
| rpoN(f)1 | AGCGT CTATA CCTTG GGG |
| rpoN(r)1 | TCTCG ACGTT ATTTC CGG |
The ast promoter was isolated from genomic DNA by PCR with primers XTH1 and SOT3 and cloned into the EcoRI site of pUC18, giving plasmid pUC-astp, and the sequence of the insert was verified. This insert was cloned into the EcoRI site of pTE103 to give plasmid pAK10, which was used for in vitro transcription. A 410-bp EcoRI/ClaI fragment from pUC-astp was blunted and cloned into the HincII site of pUC18 in both orientations to give plasmids pUC-astp(+) and pUC-astp(−). EcoRI/PstI fragments from these plasmids were used for DNase I footprinting.
An ast-lacZ fusion was constructed by cloning the insert of pUC-astp into the EcoRI site of pRS551. An 8.5-kb XhoI/SacII fragment of the resulting plasmid was transformed into strain TE2680 to place the ast-lacZ fusion into the trp operon of E. coli. The fusion was moved into W3110 by P1 transduction of kanamycin resistance to give strain AK15.
We constructed an rpoN derivative of W3110 by P1 transduction of the rpoN208::Tn 10 allele from strain YMC18 (46), but the resulting strain did not exhibit the correct phenotype, e.g., it grew with alanine or arginine as the sole nitrogen source (this has been observed before [15]). We decided, therefore, to construct an in-frame deletion of the rpoN gene using the method of Datsenko and Wanner (9). We transformed a PCR product obtained from plasmid pKD13 with primers rpoN(f)-P4 and rpoN(r)-P1 into W3110/pKD46. We removed the Kanr cassette from the resulting insertion-deletion strain (AK22) by FLP recombination to give strain AK23. We isolated the rpoN allele of AK23 by PCR amplification with primers rpoN(f)1 and rpoN(r)1. We cloned the resulting fragment and sequenced the rpoN region to verify that the in-frame deletion removed the entire rpoN open reading frame (ORF) except of the first two codons and the stop codon. The resulting 30-amino-acid product of the ORF shows no homology to anything in the databases. The ast-lacZ fusion from AK15 was moved into AK23 by P1 transduction to give strain AK24.
To disrupt argR, two genomic DNA PCR products, obtained with primer pairs argR5 and argR6 and argR7 and argR8, and a 3.6-kb EcoRI fragment from pUTmini-Tn5 Cm carrying the Camr gene were cloned consecutively into pBlueScript (Stratagene) between the BamHI and EcoRI sites, between the HindIII and KpnI sites, and into the EcoRI site, respectively. The resulting argR::cam allele was recombined onto the chromosome by transformation of a 4.65-kb SacI-KpnI fragment into strain K4633 and then moved by P1 transduction into AK15 to give strain AK18. The disruption, which removes the entire argR ORF, was verified by genomic PCR using primers argR5 and argR8.
The rpoS, himD, and crp deletions were moved into AK15 by P1 transduction from strains UM315, BW12848, and CA8445, respectively, to give strains AK16, AK17, and AK20. The rpoS::tet allele of UM315 was also moved into AK24 to give AK26. The disruptions in these strains were verified by genomic PCR with primer pairs RPOS1 and RPOS2, himD1 and himD2, and CRP1 and CRP2.
Cell growth.
Minimal media contained W salts (36), 0.02% thiamine, 0.4% (wt/vol) glucose or glycerol, and a nitrogen source (0.2%) (glutamine-arginine media contained 0.2% glutamine and 0.1% arginine). Overnight cultures (2 or 3 ml in various media) with the appropriate antibiotic were harvested, washed with 1 ml of 150 mM NaCl, resuspended in 0.5 ml of 150 mM NaCl, and used to inoculate cultures without antibiotics for assays. Unless otherwise specified, antibiotic concentrations were 100 μg/ml for ampicillin, 25 μg/ml for kanamycin, 15 μg/ml for chloramphenicol, 30 μg/ml for streptomycin, and 20 μg/ml for tetracycline. All cultures for assays were grown at 30°C.
RNA extraction.
Ten-milliliter cultures were grown to a density of ∼80 Klett units (filter 42) for exponential-phase assays or grown to stationary phase and harvested the next morning. The flask was moved to −80°C until the culture became slushy, and the cells were harvested by centrifugation. The cell pellets were used immediately or frozen at −80°C for no longer than 2 days. RNA was extracted as described previously (1) with the following modification: after addition of the acidic phenol (pH 4.2; Sigma) the solution was incubated at 60°C for 10 min with vortexing every 2.5 min. The RNA was quantitated by measuring A280.
Primer extension analysis.
Primer extension experiments were carried out essentially as described previously (38), except that 10 or 15 μg of total RNA was dried in a SpeedVac and resuspended in 5 to 10 μl of the 32P-labeled primer (17 to 34 pmol) and the solution was evaporated in a SpeedVac until just dry before resuspension in the hybridization solution. The reaction mixture did not contain actinomycin. After the reaction 10 μl of sequencing loading dye (80% formamide, 1 mM EDTA [pH 8.0], 0.2% [wt/vol] xylene cyanol, 0.2% [wt/vol] bromophenol blue) was added to the reaction tubes, which were placed on ice until they were heated and the contents were loaded on a sequencing gel.
β-Galactosidase assays.
Cells were grown in the various media containing 0.01% tryptophan. For exponential-phase measurements, 10-ml cultures were grown to a density of 90 to 120 Klett units. Stationary-phase measurements were from 2-ml cultures grown for 36 to 48 h. Cells were harvested by centrifugation and washed with 1 ml of 150 mM NaCl, and cell pellets were frozen for no longer than 2 days. Cells were thawed on ice, resuspended in 0.5 ml of 50 mM NaPO4, pH 7.5-1 mM β-mercaptoethanol, and lysed by sonication. After centrifugation at 4°C, the supernatants were used for β-galactosidase assays as described previously (28). The protein concentrations of the extracts were determined with bovine serum albumin as a standard (22).
Proteins.
For purification of σ54, W3110 carrying plasmid pTH7 was grown to late exponential phase in 6 liters of Luria broth (LB) with 0.5 mg of ampicillin/ml, incubated with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h, harvested by centrifugation, and washed with 500 ml of 150 mM NaCl, and the cell pellet was stored at −80°C. All subsequent steps were done at 4°C. Cells were thawed, resuspended in 60 ml of buffer A (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 10% glycerol) with 1 mM phenylmethylsulfonyl fluoride and lysed by sonication. Streptomycin sulfate was added to the clarified lysate to 30 mg/ml, the lysate was stirred for 30 min, and the insoluble material was removed by centrifugation. The 40 to 65% (NH4)2SO4 precipitate was resuspended in 3 ml and dialyzed twice against 1 liter of buffer A, loaded on a Mono-Q column (Pharmacia), and eluted with a 100 to 550 mM KCl gradient. RpoN fractions (0.30 to 0.45 mM KCl) were pooled, precipitated with 70% ammonium sulfate, resuspended in 0.2 ml of buffer B (20 mM NaPO4 [pH 7.0], 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol), dialyzed against the same buffer, concentrated on a Centricon (Amicon), and loaded on a Sephadex column (Pharmacia). Fractions containing σ54 were again pooled and precipitated, resuspended in buffer C (20 mM NaPO4 [pH 7.0], 1 mM EDTA, 1 mM DTT, 10% glycerol) containing 1 M (NH4)2SO4, and loaded on a phenyl-Sepharose column (Pharmacia). RpoN eluted close to the end of a 1 to 0 M (NH4)2SO4 gradient. The protein was precipitated with ammonium sulfate, resuspended, dialyzed in buffer A, concentrated, and stored at −80°C with 40% glycerol. The final preparation appeared to be over 90% pure by visual inspection after gel electrophoresis.
RpoS was purified as described previously (3) from W3110 carrying plasmid pQErpoS30 except that the cells were lysed by sonication, RpoS was precipitated from the clarified cell lysate with 40% (NH4)2SO4, chromatography was done in batch in buffers containing 600 mM KCl, and the binding buffer contained 10 mM imidazole.
The cyclic AMP (cAMP) receptor protein (CRP) was purified by modifications of previously described procedures (6, 12, 17). A 1-liter culture of strain CA8445/pXZCRP was grown in LB with ampicillin at 37°C overnight, harvested, washed with 0.5 liter of 150 mM NaCl, and resuspended in 30 ml of buffer D (20 mM KPO4 [pH 7.7], 40 mM KCl, 1 mM EDTA, 1 mM DTT, 10% glycerol) with 10 mM phenylmethylsulfonyl fluoride. After sonication, 0.5% Polymin P was added to the clarified supernatant and the mixture was stirred for 10 min and then centrifuged. The 42 to 60% (NH4)2SO4 pellet of the Polymin P supernatant was resuspended in buffer D, pH 6.7, dialyzed overnight against 1 liter of the same buffer, and loaded on a 7-ml BIO-REX 70 column (Bio-Rad), and CRP was eluted with a 100 to 700 mM KCl gradient. CRP-containing fractions were pooled, concentrated, and dialyzed twice against 1 liter of buffer TG (50 mM Tris-HCl [pH 8.15], 1 mM EDTA, 1 mM DTT, 10% glycerol). The sample was loaded by gravity on a 1.5-ml DEAE-Spherilose column (ISCO), and CRP was collected in the flowthrough, concentrated, and stored at −80°C. When we used the purified CRP for in vitro transcription, we discovered that it contained an RNase activity. To remove this activity, our CRP preparation was subjected to gel filtration through a Sephadex column (Pharmacia), which resulted in the separation of two RNase activities, one of which coeluted with CRP. Fractions containing CRP were pooled, concentrated by ammonium sulfate precipitation, resuspended in buffer E (20 mM Tris-HCl [pH 8.8], 150 mM KCl, 1 mM EDTA, 1 mM DTT, 10% glycerol), dialyzed against the same buffer, and loaded on a Mono-Q column (Pharmacia), and CRP was collected in the flowthrough. The final preparation was >90% pure, did not contain any detectable RNase activity, and was active in a mobility shift assay using a 40-bp oligonucleotide containing the CRP consensus binding sequence.
NRI (29) and integration host factor (IHF) (30) were purified as described previously, and ArgR was a generous gift from Greg VanDuyne.
DNase I footprinting.
DNase I footprinting assays were performed as described previously (7). Footprinting with ArgR was done in the presence of 5 mM arginine.
Mobility shift assays.
These assays were done in 25 μl containing 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM DTT, 5 mM arginine, 100 μg of acetylated bovine serum albumin/ml, 0.6 μg of sonicated salmon sperm DNA/ml, 10% glycerol, 0.1 nM 32P-labeled oligonucleotide, and various concentrations of the ArgR protein. The tubes were incubated for 20 min at room temperature and then loaded immediately on a 4% (49:1) polyacrylamide gel containing 40 mM Tris-HCl, pH 8.0, 10 mM MgCl2, and 5 mM arginine in both the gel and the running buffer. The gel was run at 8 V/cm for 1.5 h, followed by 4 V/cm for 3.5 h.
In vitro transcription.
Single-round in vitro transcription reaction mixtures contained, in a final volume of 25 μl, transcription buffer (50 mM Tris-HCl [pH 7.5], 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.1 mM EDTA), 5 mM arginine (when indicated in Fig. 10A and in all lanes in Fig. 10B), 5 mM cAMP (for Fig. 10B), 10 nM pAK10 as a template, 4 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 0.1 mM UTP, 1.25 μl of [α-32P]UTP (800 Ci/mmol, 10 mCi/ml; ICN), 150 μg of heparin, 100 nM core RNA polymerase (Epicenter Technologies), 300 nM σ54 or σS, 50 nM NRI dimer, 50 nM NRII dimer, 50 nM ArgR hexamer, 100 nM IHF, and various concentrations of CRP.
A mixture containing core polymerase, sigma factor, arginine, cAMP and 3 U of RNAGuard (Pharmacia) in transcription buffer was incubated for 5 min at 37°C. NRI, NRII, and ATP were added, and the mixture was incubated for another 10 min. The mixture was incubated for 10 more minutes after addition of ArgR and IHF in that order for reactions with σ54 or of ArgR and CRP simultaneously for reactions with σS. All components were in transcription buffer, and the final volume after these additions was 20.5 μl. Transcription was initiated by addition of 4.5 μl of a solution containing GTP, CTP, cold and hot UTP, and heparin in transcription buffer and was allowed to proceed for 15 min at 37°C, after which the reactions were stopped by addition of 25 μl of 50 mM EDTA-100 μg of bakers’ yeast tRNA/ml and the reaction mixtures were stored on ice. The reaction mixtures were extracted with 50 μl of acidic phenol (pH 4.2; Sigma), and 40 μl of the upper phase was added to 10 μl of sequencing loading dye. The tubes were heated to 90°C for 3 min and centrifuged briefly before the contents were loaded on a 5% acrylamide-8 M urea-0.5× Tris-borate-EDTA gel.
RESULTS
Expression of the ast genes is regulated at two promoters.
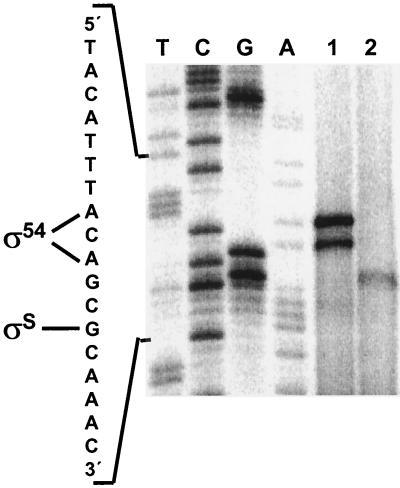
Direct assays of the AST pathway enzymes have shown that general nitrogen limitation, the specific nitrogen source, and the carbon source affect ast operon expression (40). Additionally, other reports published during the course of this work found that the growth phase also affects expression of the operon in E. coli (2, 5) and S. enterica serovar Typhimurium (23) and that σ54 and σS are both involved in this regulation. To analyze in more detail the regulation of the ast operon, we performed primer extension analysis on E. coli total RNA isolated from cells grown in different media and harvested at different phases of growth. As shown in Fig. 1, exponentially growing cells in glucose-arginine minimal medium expressed the ast operon from an upstream promoter (which will be shown to require σ54), while stationary-phase cells in complex media (LB) expressed the operon from a second promoter (which will be shown to require σS). Transcription from the σ54-dependent promoter initiated at two adenines found 2 bp apart in the promoter sequence. It is possible that the shorter primer extension product is a degradation product of the longer one. The transcriptional start site for the σS-dependent promoter was a guanine 5 bp downstream from the most upstream σ54-dependent start site.
FIG. 1.
Start site mapping of the ast promoter. Extension of primer SOT3 was performed with RNA isolated from W3110 cells growing exponentially on glucose-arginine minimal medium (lane 1) or in stationary phase after growth in LB (lane 2), and the reaction products were run next to a sequencing ladder. The promoter sequence and the start sites for the σ54 and σS promoters are shown.
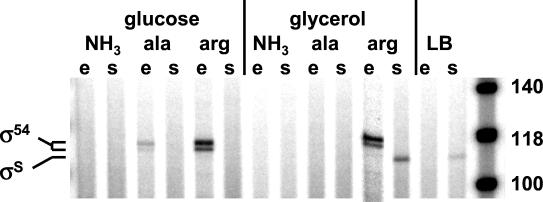
The proximity of the initiation sites raised the question of competition between the two holoenzymes for binding to the ast promoters. To investigate this issue, we examined promoter utilization in cells grown on a more diverse collection of media, including different carbon and nitrogen sources, and in cells in both phases of growth. Cells growing exponentially in nitrogen-limited minimal media, especially those with arginine as the sole nitrogen source, utilized the σ54-dependent promoter (Fig. 2). In contrast, stationary-phase cells grown in media with arginine but without glucose (glycerol-arginine minimal medium or LB) utilized the σS-dependent promoter. We never observed expression from both promoters simultaneously, and the case of glycerol-arginine medium is extreme--promoter utilization switched from the σ54-dependent promoter during exponential phase to the σS-dependent promoter in stationary phase. This suggests that expression from the two promoters is mutually exclusive and that the two forms of the polymerase compete for binding.
FIG. 2.
Promoter utilization. The products of primer extension reactions on RNA from cells grown under various conditions were run on a sequencing gel next to size markers (rightmost lane). Primer SOT4 was used. The carbon and nitrogen source is indicated, as is the phase of growth (e, exponential; s, stationary). The promoter from which the transcripts originate is noted. (The glycerol-arginine exponential-phase lane is from a different gel and is inserted here for presentation purposes).
In vivo analysis of astCADBE expression.
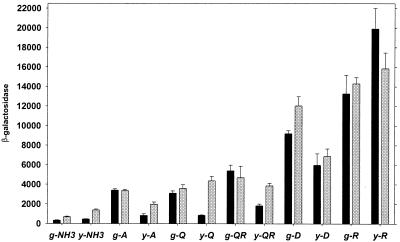
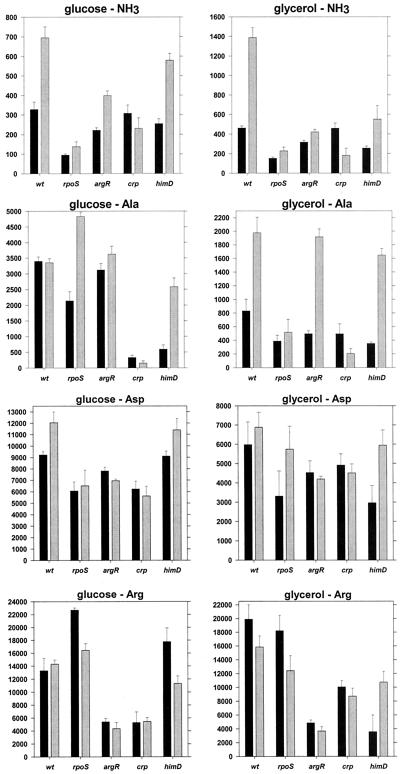
To quantitatively analyze transcriptional regulation of the ast operon, we constructed a strain with an ast promoter-lacZ fusion in the E. coli chromosome. The fusion did not disrupt the wild-type ast operon, which permits growth with arginine as a nitrogen source. We assayed β-galactosidase activity in cells grown in minimal media with different carbon and nitrogen sources and harvested during exponential or stationary phase. We found that the ast operon was highly inducible, with a 60-fold range of expression (Fig. 3). This could not be attributed to differences in the growth rate, which showed only a sixfold range for the different media (not shown). As expected, nitrogen limitation increased expression, and there appeared to be specific induction, since β-galactosidase levels were not the same for all growth-limiting nitrogen sources but were highest when arginine was the sole source of nitrogen, followed by aspartate. These results agree with direct enzyme assays (40), which validates use of the lacZ fusion for expression studies. In cells growing exponentially (Fig. 3) with a poor nitrogen source other than arginine, expression was higher with glucose as the carbon source than it was with glycerol, while the same was not always true in stationary phase (Fig. 3). Since expression under these conditions was σ54 dependent (Fig. 2), this observation suggests that transcription by σ54 is reduced in carbon-limited media. In general, activity was the same in exponentially growing and stationary-phase cells, although, as indicated above, this does not imply utilization of one promoter.
FIG. 3.
In vivo expression of the ast operon. Strain AK15 was grown on a variety of minimal media, and β-galactosidase levels in exponential (black bars) and stationary (gray bars) phases were measured. Units are nanomoles of product per minute per milligram of total soluble protein. Averages of at least three independent determinations and standard errors of the means are shown. Carbon sources: g, glucose; y, glycerol. Nitrogen sources are indicated with the one-letter amino acid designation.
Factors affecting expression of the ast operon.
Based on the expression analysis presented here a number of proteins could be involved in the transcriptional regulation of the astCADBE operon: the alternative sigma factors σ54 and σS, the σ54-dependent activator NRI, and CRP. Additionally, the arginine repressor, ArgR, has been shown to be involved in the transcriptional regulation of the ast operon in S. enterica serovar Typhimurium (23), and IHF is involved in the regulation of some NRI- and σ54-dependent promoters. To investigate the possible effect of each of these factors on the expression of the ast operon in E. coli, we constructed rpoN, rpoS, crp, argR, and himD (encoding one of the IHF subunits) mutants and an rpoN rpoS double mutant in a W3110 background and assayed the expression of β-galactosidase from cells with the ast-lacZ fusion in a variety of media. (The rpoN mutant is essentially a glutamine auxotroph in nitrogen-limited media and, therefore, could not be grown in many of the media used.)
(i) Loss of the sigma factors.
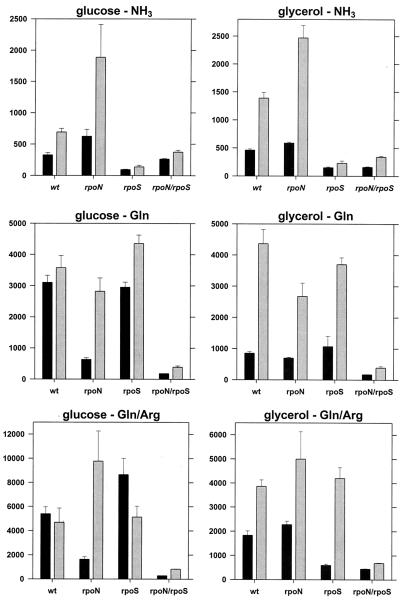
The rpoN rpoS double mutant had basal levels of transcription for cells in all media, which implies that only σ54 and σS participate in the expression of the ast operon in E. coli (Fig. 4).
FIG. 4.
Sigma factors and ast expression. Strains carrying the ast-lacZ fusion were grown on the indicated media and assayed for β-galactosidase activity in exponential (black bars) and stationary (gray bars) phases. Values are reported as in Fig. 3. Note the differences in the scaling of the y axes. wt, wild type.
The rpoN mutant (σ54 deficient) had lower β-galactosidase activity than a wild-type strain, but only during exponential growth in nitrogen-limited media (glucose as the carbon source and either glutamine or glutamine plus arginine as the nitrogen sources) (Fig. 4). With glycerol as the carbon source, this mutant had essentially the same β-galactosidase activity as the wild-type strain (less than 20% difference for all media), which implies that this activity requires σS (Fig. 4). The rpoN mutant had higher activity than the wild type in stationary phase with ammonia and either glucose or glycerol. This is consistent with the hypothesis of holoenzyme competition (Fig. 4). Interestingly, the rpoN and rpoN rpoS strains do not show the reduced expression in glycerol-containing media described previously (Fig. 3 and 4). This may suggest that σ54 mediates the glycerol effect.
Loss of σS was most noticeable for cells grown in nitrogen-rich (ammonia-containing) media, regardless of the carbon source or growth phase (Fig. 4). Such growth results in minimal phosphorylation of NRI, which should prevent activation of the σ54-dependent promoter. For exponentially growing cells with glutamine, glutamine with arginine, alanine, aspartate, or arginine as the nitrogen source, deletion of rpoS had little effect on β-galactosidase activity (Fig. 4 and 5). These results were expected since NRI is phosphorylated, which should activate the σ54-dependent promoter. β-Galactosidase activities for the wild-type and rpoS strains harvested in stationary phase were similar (Fig. 5). This was unexpected since expression in W3110 should be primarily from the σS-dependent promoter (Fig. 2, glycerol-arginine medium). This may imply a switch in promoter utilization, from the σS-dependent promoter in the wild type to the σ54-dependent promoter in the rpoS mutant. However, the accumulation of β-galactosidase during exponential growth may account for most of the activity, and there may be little or no ast operon expression during stationary phase in the rpoS mutant.
FIG. 5.
Effects of regulatory mutants on ast expression. The presentation of the data is the same as that for Fig. 4. wt, wild type.
(ii) ArgR.
The argR deletion had a significant effect on transcription only under conditions that resulted in the highest levels of expression, i.e., with arginine as the sole nitrogen source (Fig. 5). Loss of ArgR appeared to prevent arginine-specific induction, which suggests that ArgR mediates this induction. Effectively, the argR deletion acted to limit the highest attainable level of expression to ∼6,000 nmol min−1 mg of protein−1. This was true for the exponential and stationary phases and suggests that, while not essential, ArgR enhances transcription at least from the σ54-dependent promoters and from the σS-dependent promoter to the extent that β-galactosidase activity reflects synthesis in stationary phase and not from previous accumulation during exponential growth.
(iii) CRP.
Deletion of crp lowered β-galactosidase activity, especially in stationary phase and in media in which there were low levels of expression (ammonia or alanine as the nitrogen source) (Fig. 5). The crp mutant, like the argR mutant, seemed to limit the highest level of expression that could be achieved. However, this may have been an indirect effect of the slower growth of this mutant in most media.
(iv) IHF.
Finally, the himD mutation did not have an appreciable effect on the β-galactosidase levels under most conditions (Fig. 5). Maximal activity was attainable (in glucose-arginine medium), and there was no discernible pattern in the cases where this mutation apparently impaired expression. As was the case with crp, the himD strain grew slower in most media, further complicating the interpretation of our results. The data presented here do not support an essential role for IHF in the regulation of the ast operon in E. coli.
Protein-DNA interactions at the ast promoter in vitro.
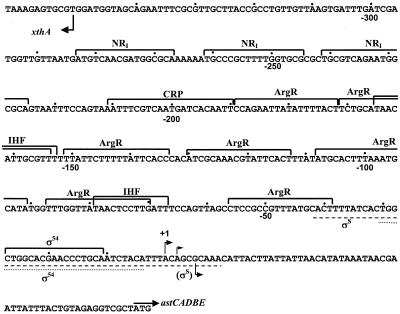
Computer analysis and visual inspection of the promoter sequence revealed putative regulatory elements throughout the promoter region (Fig. 6). A σ54 site near the transcription start site and two closely spaced NRI-binding sites, centered at −233 and −253, were easily recognizable. There are seven potential ArgR-binding sites between the σ54 and the NRI sites and also a potential CRP-binding site just downstream from the enhancer sites. Two potential IHF sites, overlapping ArgR sites, were also discernible. To confirm the results concerning the factors that control astCADBE expression and to determine whether the potential binding sites actually bind proteins, we analyzed the interaction of purified proteins with the ast promoter region in vitro with DNase I footprinting.
FIG. 6.
The ast promoter of E. coli. The sequence of the ast promoter is shown together with the potential binding sites of various factors. Bent arrows, transcription start sites; straight arrow, start codon of astC. Dots mark every 10th nucleotide from the most upstream transcription initiation site (+1). The lines under the sequence indicate the extents of the footprints for the σS holoenzyme (dashed line) and the σN holoenzyme (dotted line).
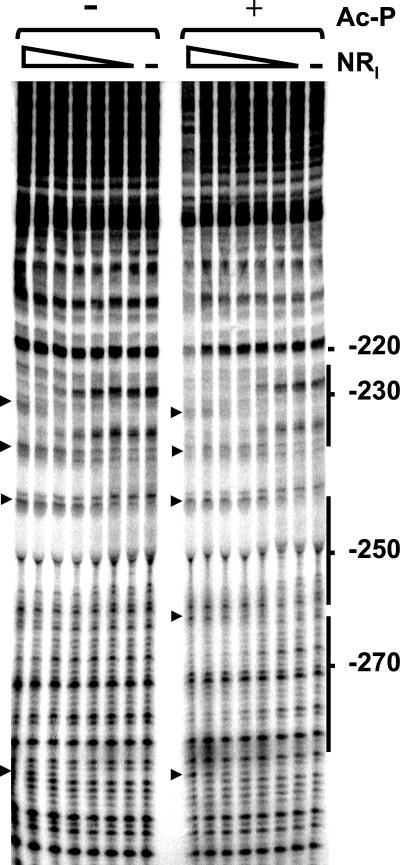
(i) NRI.
NRI bound at the two predicted sites and, at higher concentrations, at an additional site centered at −275 (Fig. 7). A number of hypersensitive sites were seen, usually at the ends of the binding sites; these hypersensitive sites are indicative of DNA bending by NRI and have been observed at other NRI-dependent promoters (see, for example, Fig. 2 of reference 7). Quantitative analysis of the gel showed that the concentration of NRI required for half-maximal occupancy of the downstream site decreases from 35 to 13 nM upon phosphorylation by acetyl-phosphate. This concentration is 5 to 10 times higher than that reported for the glnA promoter of E. coli (7, 50), which means that the enhancer of the ast operon is weaker than the glnALG enhancer.
FIG. 7.
Footprinting with NRI. DNase I footprinting was performed on a fragment carrying the ast promoter region. The concentrations of the NRI dimer used are 0, 4, 8, 16, 31, 62.5, 125, and 250 nM. Reactions were done in the presence or absence of acetyl-phosphate (Ac-P) as indicated. The position on the ast promoter sequence is also indicated. Solid lines, NRI-binding sites; arrowheads, hypersensitive sites.
(ii) ArgR.
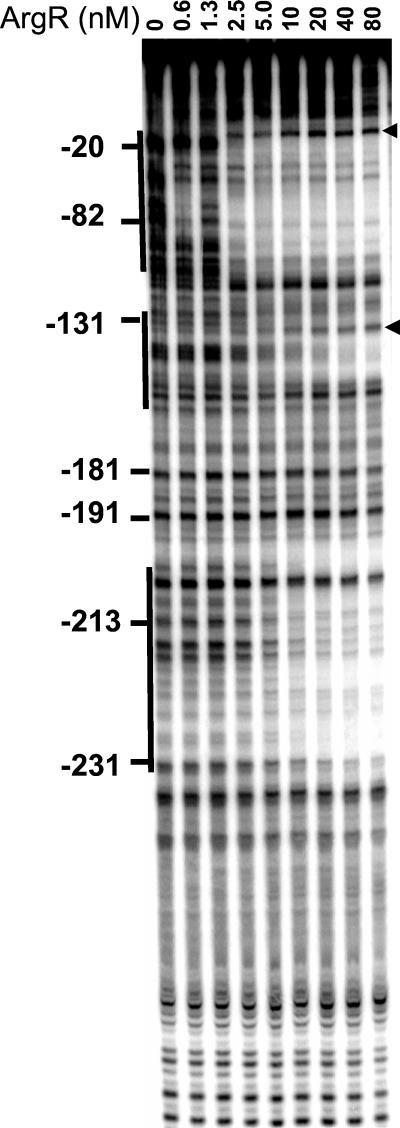
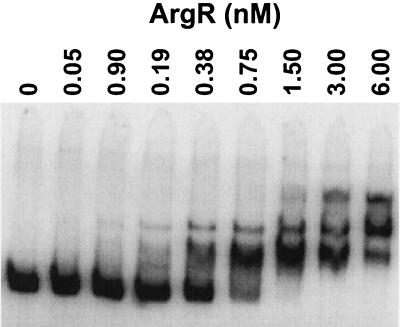
The arginine repressor in its free form did not bind to the promoter (data not shown). ArgR with 5 mM arginine bound to an extended region from −20 to −120 (Fig. 8). At higher protein concentrations additional footprints were observed in regions −130 to −150 and −200 to −230. As with NRI, sites of increased sensitivity to digestion denoted the bending caused by ArgR binding to the promoter. The size of the footprint in region −20 to −120 suggests that four ArgR-binding sites in the ArgR-promoter complex are occupied (Fig. 6). Assuming that one ArgR hexamer binds to two sites, as it does in other promoters (24), the binding of more than one ArgR hexamer would be required to give the observed footprint. An electrophoretic mobility shift assay showed multiple slowly migrating complexes (Fig. 9), suggesting that two or three ArgR hexamers might bind to the ast promoter simultaneously.
FIG. 8.
Footprinting with ArgR. DNase I footprinting was performed on a fragment carrying the ast promoter with the indicated concentration of ArgR monomer and 5 mM arginine. The position on the ast promoter is shown. Solid lines, regions of protection; arrowheads, hypersensitive sites.
FIG. 9.
Electrophoretic mobility shift assay with ArgR. A DNA fragment carrying the ast promoter region was incubated with the indicated concentration of ArgR monomer and 5 mM arginine prior to electrophoresis.
(iii) IHF.
IHF protected two sites around −80 and −160 at concentrations higher than 20 nM (data not shown). These footprints overlap the ArgR-binding sites.
(iv) CRP.
We could not detect specific binding of purified CRP to the ast promoter by DNase I footprinting with or without cAMP (not shown). We also did not observe binding in mobility shift assays, except at very high concentrations of CRP. With a 200-fold excess of CRP to DNA, we saw an abrupt transition from no binding to a shift where almost all the probe failed to enter the gel. We suspect that this is nonspecific binding. Results with cells containing the ast-lacZ fusion suggested that CRP does not obviously contribute to ast expression in E. coli. This, together with the failure to observe specific binding, suggests that CRP does not directly control the ast operon. These results differ from genetic and biochemical results obtained with S. enterica serovar Typhimurium that suggest a direct role for CRP (23).
(v) RNA polymerase.
DNase I footprinting showed that purified σ54 or σS together with core RNA polymerase bound the region around the sites of transcriptional initiation. The extent of the holoenzyme footprints is shown in Fig. 6.
Transcription from the ast promoters with purified components.
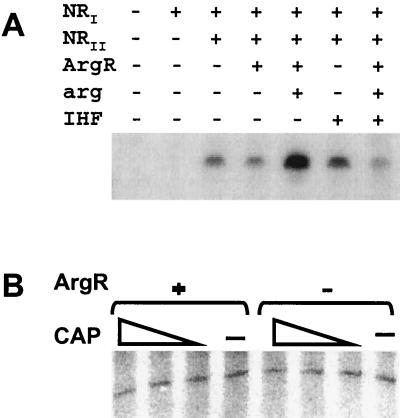
The combination of in vivo and in vitro data presented thus far argues that the expression of the ast operon is regulated at two promoters. To further verify this conclusion, we analyzed transcription from these promoters with purified proteins. As shown in Fig. 10A, expression from the σ54 promoter was dependent on the presence of NRI-phosphate. This transcription did not require the ArgR-arginine complex, although this complex enhanced transcription. IHF did not stimulate transcription and appeared to inhibit it when ArgR was also present.
FIG. 10.
In vitro transcription from the ast promoters. Single-round transcription experiments with purified components were performed from a supercoiled template with σ54 holoenzyme (A) or σS holoenzyme (B) and the indicated additions. The concentrations of CRP dimer used were 0, 200, 400, and 800 nM. The expected ∼420-nucleotide transcript was obtained in both cases.
Transcription from the σS-dependent ast promoter required only σS holoenzyme (Fig. 10B). ArgR and CRP, alone or in combination, did not affect transcription (Fig. 10B). IHF also failed to affect transcription (not shown).
DISCUSSION
σ54 and σS are the major participants in ast operon expression.
Mutants lacking both sigma factors have very low ast expression. There is some evidence for a σ70-dependent promoter based on sequence analysis (just downstream of the σ54 and σS promoters) (see Fig. 3 of reference 15) and for the dependence of expression on σ70 based on results from a coupled in vitro transcription-translation system in crude extracts with a plasmid carrying an ast-lacZ fusion (5). However, our results clearly show that, if this σ70-dependent promoter exists, it is not quantitatively important under the conditions we examined.
RNA polymerase holoenzyme competition.
An interesting aspect of the regulation of the ast operon is the apparent competition between Eσ54 and EσS. Several lines of evidence indicate that each holoenzyme inhibits transcription by the other one. First, the proximity of the transcriptional start sites might preclude simultaneous binding and initiation. Second, during our primer extension analysis we did not encounter a condition in which we could detect transcripts from both promoters, which would indicate that, in every condition, the dominant holoenzyme takes over transcription of the operon entirely. Third, in some cases removal of a sigma factor responsible for transcription in a particular growth medium did not result in a significant change in the expression of the operon (Fig. 2 and 5; the rpoS mutant grown in glycerol-arginine medium and harvested in stationary phase). This indicates that only one promoter is utilized because of competition and not because the conditions do not allow for the utilization of the other promoter. Finally, in conditions in which expression could be initiated by only one form of the polymerase (e.g., EσS in ammonia-containing media, which results in inactive Eσ54 since NRI is largely unphosphorylated), deletion of the other sigma factor increases expression (Fig. 4). This idea of competition between the promoters in the ast operon has been previously postulated (15).
Stationary-phase induction and CRP.
Our results show that the ast operon is induced in stationary phase, which confirms previous results. The ast operon was identified in a search for E. coli genes activated in response to conditioned medium (2) and another study looking for carbon starvation-induced genes (5). The apparent rationale for such control is the provision of citric acid cycle intermediates.
Stationary-phase expression initiates from the σS promoter, and we reconstituted σS-dependent transcription in vitro. The β-galactosidase results show that, as is the case in exponential phase, arginine stimulates expression but that exogenous arginine is not required for expression. This is most apparent for the rpoN mutant in glucose-containing media (compare β-galactosidase activity for cells grown in glucose-glutamine with that for cells grown in glucose-glutamine plus arginine in Fig. 4). This suggests that ArgR might also be involved in the regulation of the σS promoter, perhaps by facilitating an interaction between EσS and another DNA-bound transcription factor. Despite the effect of the carbon source on the expression of astCADBE, we could not demonstrate a clear involvement of CRP in vivo or in vitro. This is in contrast with what is found for S. enterica serovar Typhimurium, where CRP seems to be essential for stationary-phase expression (23), but is in agreement with results for E. coli arguing against a role of cAMP in the regulation of an ast-lacZ fusion (5). The basis for the difference in CRP responsiveness is not clear. An E. coli site with significant homology to the CRP consensus sequence can be found at the same position as the S. enterica serovar Typhimurium site. The sites in the two species differ by only 3 bp, with the S. enterica serovar Typhimurium site being closer to the consensus by only 1 bp. Unexpectedly though, while binding to the site on the S. enterica serovar Typhimurium promoter can be demonstrated in vitro (23), we could not obtain CRP binding to the E. coli promoter by mobility shift or DNase I footprinting.
Our results do not exclude the possibility that there is a binding site for an unidentified activator and that ArgR enhances its ability to activate transcription. It is known that stationary-phase induction of the ast operon is complex. In strain MC1061, this induction is mediated, at least in part, through indole (47). However, while indole production by W3110 has been reported previously (21), some W3110 strains contain an insertion of an IS 5 element in the tnaB gene of the tnaAB operon (20) (tnaA codes for tryptophanase, the indole-producing enzyme [43]), and our W3110 strain is indole negative based on a standard microbiological test (H. Kasbarian and L. Reitzer, unpublished results). We tested induction by indole in 0.5× LB medium (the Eσ54 promoter is inactive) and found that ast-lacZ expression in mid-exponential phase increased from 1,300 to 5,700 nmol min−1 mg of protein−1. Therefore, although our W3110 does not produce indole, it retains the factors necessary to respond to its presence. Nonetheless, there is an additional unidentified factor that activates the ast operon and other operons in stationary phase (47), and ArgR could potentially enhance its ability to stimulate transcription.
Carbon source control of ast expression.
There is lower ast operon expression in exponential phase with glycerol instead of glucose as the carbon source (Fig. 3). This effect appears to be both CRP and σ54 dependent (Fig. 4 and 5). It has been reported that CRP-cAMP can interact directly with σ54 and inhibit its ability to activate transcription (45, 48). This could explain the carbon source control.
General induction by nitrogen limitation.
The astCADBE operon in E. coli codes for the enzymes of the AST pathway, which are necessary for the utilization of arginine as a nitrogen source (40). The AST pathway produces the ammonia required for growth in nitrogen-limiting minimal media and also produces glutamate. The levels of the AST enzymes are increased in nitrogen-limiting conditions, as would be expected given the function of the AST pathway (40). This aspect of the regulation of the operon is mediated through σ54 and NRI. The involvement of NRI has been seen before through measurements of AST enzyme levels (40) and gene arrays (52). Here, we complement these results by reconstituting the activation of the ast promoter by NRI in vitro.
ArgR and arginine-specific enhancement during nitrogen-limited growth.
Most NRI-dependent Ntr genes are expressed in nitrogen-limiting conditions regardless of the specific nitrogen source. The ast operon is different: arginine as a nitrogen source enhances expression (40; this study). Based on the in vivo and in vitro results of our analysis, ArgR mediates this specific induction. The ArgR-dependent induction is not essential for expression, either in vivo or in vitro. For several reasons it seems that the role of ArgR is to facilitate the interaction between NRI and Eσ54. First, the NRI-binding sites are centered at −233 and −253 from the start site of transcription, a distance significantly greater than that in any other known NRI-dependent promoter. This distance might make direct contact between the promoter-bound polymerase and the enhancer-bound NRI less likely. Second, the ArgR-binding sites are positioned appropriately to facilitate an RNA polymerase-NRI interaction. Third, this proposal is consistent with the known properties of ArgR. The binding of an ArgR hexamer to two ArgR-binding sites causes the DNA to bend by 70° (44). If more than one hexamer binds to the ast promoter, as implied by the mobility shift and footprinting results, they could cause a bend closer to 180°, which would be comparable to the bend caused by IHF binding (34). The E. coli ast operon differs in its regulation from the S. enterica serovar Typhimurium operon, which appears to absolutely require ArgR for expression. It is not clear why this role of ArgR is essential in S. enterica serovar Typhimurium but not in E. coli.
It is not uncommon for a DNA-bending protein to promote an interaction between Eσ54 and an activator, and IHF is frequently the DNA-bending protein (8, 11, 18, 39, 49). However, our results do not suggest an effect of the himD mutation in the regulation of astCADBE in vivo. Furthermore, the IHF footprint was weak, and purified IHF had no positive effect on transcription in vitro. These results suggest that IHF does not positively contribute to ast operon expression. The IHF footprint could be an artifactual result of the AT richness of the ast promoter, which is a feature of both ArgR-binding sites and σ54-dependent promoters (IHF-binding sites are AT rich). It is also possible that, earlier in bacterial evolution, IHF was involved in ast operon expression and that ArgR preempted this function. In this case, a residual IHF-binding site might still remain.
Arginine-responsive transcription factors commonly participate in the regulation of arginine synthesis or degradation or both in many diverse bacterial species. The ArgR protein of Bacillus licheniformis activates the arcABCD gene cluster, which encodes the enzymes of the arginine deiminase pathway (26). The ArgR homologue of Bacillus subtilis, AhrC, activates the rocABC and rocDEF operons of arginine and ornithine catabolism by a mechanism which, apparently, involves direct protein-protein interaction with RocR, a σ54-dependent activator (16, 27). Overexpression of the argR gene of Streptomyces clavuligerus causes repression of an arginine biosynthetic enzyme and activation of a catabolic one (35). Finally, the ArgR of Pseudomonas aeruginosa, a transcriptional activator of the AraC/XylS family unrelated to the ArgR of the enteric bacteria and the bacilli, activates the arginine anabolic genes and represses the arginine catabolic genes (32).
The functions of nonspecific ArgR-independent induction and ArgR-dependent superinduction.
Regardless of the promoter and sigma factor used, there is ArgR-independent expression overlaid with ArgR-dependent expression (or superinduction). Therefore, the function of ArgR-dependent control must be considered within the context of general control mechanisms and their functions. It has been proposed that a major function of the σ54-dependent Ntr response is nitrogen scavenging (52). This is reasonable based on the nonspecific (nitrogen source-independent) induction of Ntr genes. Perhaps it is even surprising that all known genes induced by nitrogen limitation in E. coli do not require specific induction, even though specific regulators sometimes exist (e.g., for the ast and gab operons). Nonspecific induction might reflect the environment, i.e., nitrogen limitation may frequently involve a number of nitrogen sources at low concentration. However, a scavenging function does not account for why E. coli scavenges a limited number of nitrogen-containing compounds and why E. coli has so few σ54-dependent catabolic pathways for nitrogen source utilization. Very few nitrogen sources are degraded by σ54-dependent enzymes, and these include arginine, γ-aminobutyrate (via Nac) (52; S. Ruback, A. Kiupakis, and L. Reitzer, unpublished results), and cytosine (52). There is actually a good reason for a σ54-dependent pathway of arginine catabolism. It can be calculated from the composition of E. coli (31) that arginine in protein (281 μmol/g dry weight) constitutes 10.9% of the total cellular nitrogen (1,124 μmol of nitrogen in arginine compared to 10,283 μmol of nitrogen/g of E. coli), which is surprisingly large. (Guanine nucleotides comprise 11.1%, and adenine nucleotides comprise 9.2%, of total nitrogen, but no other single macromolecular monomer contains as much nitrogen.) Assuming that other organisms have similar nitrogen contents, then a major source of organic nitrogen from recycled organisms would be arginine. Therefore, an arginine catabolic pathway would be important for nitrogen acquisition, which accounts for general induction by nitrogen limitation. An ArgR-dependent superinduction would be reasonable if there were environments in which arginine was a major component. Some organisms, such as Saccharomyces cerevisiae, have vacuoles that contain arginine. The lysis of organisms such as this could create an arginine-rich milieu.
The same pattern of nonspecific induction followed by a possible ArgR-dependent superinduction may also exist for the σS-dependent promoter. The rationale for such regulation might be similar to that for the two layers of control for the σ54-dependent promoter. A nonspecific induction during stationary-phase metabolism would provide citric acid cycle intermediates. The superinduction would then occur only in arginine-rich environments, and this might require ArgR with an as yet unidentified activator.
Acknowledgments
This work was supported by grants MCB-9723003 and MCB-0077904 from the National Science Foundation.
We acknowledge Barry Wanner (Purdue University), Richard Ebright (Rutgers University), Alex Ninfa (University of Michigan, Ann Arbor), Hua Tsen and Stephen Levene (University of Texas at Dallas), and Regine Hengge-Aronis (Freie Universität Berlin) for strains and plasmids, and Greg Van Duyne (University of Pennsylvania) for his gift of ArgR protein. We gratefully acknowledge the help of Barbara Schneider.
REFERENCES
- 1.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. [PubMed] [Google Scholar]
- 2.Baca-DeLancey, R. R., M. M. South, X. Ding, and P. N. Rather. 1999. Escherichia coli genes regulated by cell-to-cell signaling. Proc. Natl. Acad. Sci. USA. 96:4610-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, G., E. Klauck, and R. Hengge-Aronis. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. USA 96:6439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, R. A. 1991. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol. Microbiol. 5:2575-2580. [DOI] [PubMed] [Google Scholar]
- 5.Blum, P. H., S. B. Jovanovich, M. P. McCann, J. E. Schultz, S. A. Lesley, R. R. Burgess, and A. Matin. 1990. Cloning and in vivo and in vitro regulation of cyclic AMP-dependent carbon starvation genes from Escherichia coli. J. Bacteriol. 172:3813-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boone, T., and G. Wilcox. 1978. A rapid high-yield purification procedure for the cyclic adenosine 3′,5′-monophosphate receptor protein from Escherichia coli. Biochim. Biophys. Acta 541:528-534. [DOI] [PubMed] [Google Scholar]
- 7.Chen, P., and L. J. Reitzer. 1995. Active contribution of two domains to cooperative DNA binding of the enhancer-binding protein nitrogen regulator I (NtrC) of Escherichia coli: stimulation by phosphorylation and the binding of ATP. J. Bacteriol. 177:2490-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claverie-Martin, F., and B. Magasanik. 1991. Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc. Natl. Acad. Sci. USA 88:1631-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn 5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dworkin, J., G. Jovanovic, and P. Model. 1997. Role of upstream activation sequences and integration host factor in transcriptional activation by the constitutively active prokaryotic enhancer-binding protein PspF. J. Mol. Biol. 273:377-388. [DOI] [PubMed] [Google Scholar]
- 12.Eilen, E., C. Pampeno, and J. S. Krakow. 1978. Production and properties of the α core derived from the cyclic adenosine monophosphate receptor protein of Escherichia coli. Biochemistry 17:2469-2473. [DOI] [PubMed] [Google Scholar]
- 13.Elliott, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, T., and E. P. Geiduschek. 1984. Defining a bacteriophage T4 late promoter: absence of a “−35”region. Cell 36:211-219. [DOI] [PubMed] [Google Scholar]
- 15.Fraley, C. D., J. H. Kim, M. P. McCann, and A. Matin. 1998. The Escherichia coli starvation gene cstC is involved in amino acid catabolism. J. Bacteriol. 180:4287-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardan, R., G. Rapoport, and M. Debarbouille. 1997. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol. Microbiol. 24:825-837. [DOI] [PubMed] [Google Scholar]
- 17.Heyduk, T., and J. C. Lee. 1989. Escherichia coli cAMP receptor protein: evidence for three protein conformational states with different promoter binding affinities. Biochemistry 28:6914-6924. [DOI] [PubMed] [Google Scholar]
- 18.Hoover, T. R., E. Santero, S. Porter, and S. Kustu. 1990. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell 63:11-22. [DOI] [PubMed] [Google Scholar]
- 19.Hunt, T. P., and B. Magasanik. 1985. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc. Natl. Acad. Sci. USA 82:8453-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamath, A. V., K. Gish, and C. Yanofsky. 1994. A copy of insertion element IS 5 is present within tnaB in the Kohara library of Escherichia coli W3110. J. Bacteriol. 176:1546-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kell, R. G. D. B. 1993. Rapid and quantitative analysis of bioprocesses using pyrolysis mass spectrometry and neural networks: application to indole production. Anal. Chim. Acta 279:17-26. [Google Scholar]
- 22.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 23.Lu, C. D., and A. T. Abdelal. 1999. Role of ArgR in activation of the ast operon, encoding enzymes of the arginine succinyltransferase pathway in Salmonella typhimurium. J. Bacteriol. 181:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magasanik, B. 1996. Regulation of nitrogen utilization, p. 1344-1356. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 26.Maghnouj, A., T. F. de Sousa Cabral, V. Stalon, and C. Vander Wauven. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J. Bacteriol. 180:6468-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, C. M., S. Baumberg, and P. G. Stockley. 1997. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol. Microbiol. 26:37-48. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Moore, J. B., S. P. Shiau, and L. J. Reitzer. 1993. Alterations of highly conserved residues in the regulatory domain of nitrogen regulator I (NtrC) of Escherichia coli. J. Bacteriol. 175:2692-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nash, H. A., C. A. Robertson, E. Flamm, R. A. Weisberg, and H. I. Miller. 1987. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J. Bacteriol. 169:4124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neidhardt, F. C., and H. E. Umbarger. 1996. Chemical composition of Escherichia coli, p. 13-16. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington D.C.
- 32.Park, S. M., C. D. Lu, and A. T. Abdelal. 1997. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 179:5300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reitzer, L. J. 1996. Sources of nitrogen and their utilization, p. 380-390. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 34.Rice, P. A., S. Yang, K. Mizuuchi, and H. A. Nash. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87:1295-1306. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Garcia, A., M. Ludovice, J. F. Martin, and P. Liras. 1997. Arginine boxes and the argR gene in Streptomyces clavuligerus: evidence for a clear regulation of the arginine pathway. Mol. Microbiol. 25:219-228. [DOI] [PubMed] [Google Scholar]
- 36.Rothstein, D. M., G. Pahel, B. Tyler, and B. Magasanik. 1980. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc. Natl. Acad. Sci. USA 77:7372-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabourin, D., and J. Beckwith. 1975. Deletion of the Escherichia coli crp gene. J. Bacteriol. 122:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Santero, E., T. R. Hoover, A. K. North, D. K. Berger, S. C. Porter, and S. Kustu. 1992. Role of integration host factor in stimulating transcription from the sigma 54-dependent nifH promoter. J. Mol. Biol. 227:602-620. [DOI] [PubMed] [Google Scholar]
- 40.Schneider, B. L., A. K. Kiupakis, and L. J. Reitzer. 1998. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 180:4278-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider, B. L., S. P. Shiau, and L. J. Reitzer. 1991. Role of multiple environmental stimuli in control of transcription from a nitrogen-regulated promoter in Escherichia coli with weak or no activator-binding sites. J. Bacteriol. 173:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 43.Snell, E. E. 1975. Tryptophanase: structure, catalytic activities, and mechanism of action. Adv. Enzymol. Relat. Areas Mol. Biol. 42:287-333. [DOI] [PubMed] [Google Scholar]
- 44.Tian, G., D. Lim, J. Carey, and W. K. Maas. 1992. Binding of the arginine repressor of Escherichia coli K12 to its operator sites. J. Mol. Biol. 226:387-397. [DOI] [PubMed] [Google Scholar]
- 45.Tian, Z. X., Q. S. Li, M. Buck, A. Kolb, and Y. P. Wang. 2001. The CRP-cAMP complex and downregulation of the glnAp2 promoter provides a novel regulatory linkage between carbon metabolism and nitrogen assimilation in Escherichia coli. Mol. Microbiol. 41:911-924. [DOI] [PubMed] [Google Scholar]
- 46.Ueno-Nishio, S., K. C. Backman, and B. Magasanik. 1983. Regulation at the glnL-operator-promoter of the complex glnALG operon of Escherichia coli. J. Bacteriol. 153:1247-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Y. P., A. Kolb, M. Buck, J. Wen, F. O'Gara, and H. Buc. 1998. CRP interacts with promoter-bound sigma54 RNA polymerase and blocks transcriptional activation of the dctA promoter. EMBO J. 17:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wassem, R., E. M. De Souza, M. G. Yates, F. D. Pedrosa, and M. Buck. 2000. Two roles for integration host factor at an enhancer-dependent nifA promoter. Mol. Microbiol. 35:756-764. [DOI] [PubMed] [Google Scholar]
- 50.Weiss, V., F. Claverie-Martin, and B. Magasanik. 1992. Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc. Natl. Acad. Sci. USA 89:5088-5092. (Erratum, 89:8856.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, X. P., A. Gunasekera, Y. W. Ebright, and R. H. Ebright. 1991. Derivatives of CAP having no solvent-accessible cysteine residues, or having a unique solvent-accessible cysteine residue at amino acid 2 of the helix-turn-helix motif. J. Biomol. Struct. Dyn. 9:463-473. [DOI] [PubMed] [Google Scholar]
- 52.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]