Abstract
Integrons capture gene cassettes by using a site-specific recombination mechanism. As only one class of integron and integron-determined site-specific recombination system has been studied in detail, the properties of a second class, the only known class 3 integron, were examined. The configuration of the three potentially definitive features of integrons, the intI3 gene, the adjacent attI3 recombination site, and the Pc promoter that directs transcription of the cassettes, was similar to that found in the corresponding region (5′ conserved segment) of class 1 integrons. The integron features are flanked by a copy of the terminal inverted repeat, IRi, from class 1 integrons on one side and a resolvase-encoding tniR gene on the other, suggesting that they are part of a transposable element related to Tn402 but with the integron module in the opposite orientation. The IntI3 integrase was active and able to recognize and recombine both known types of IntI-specific recombination sites, the attI3 site in the integron, and different cassette-associated 59-be (59-base element) sites. Both integration of circularized cassettes into the attI3 site and excision of integrated cassettes were also catalyzed by IntI3. The attI3 site was localized to a short region adjacent to the intI3 gene. Recombination between a 59-be and secondary sites was also catalyzed by IntI3, but at frequencies significantly lower than observed with IntI1, the class 1 integron integrase.
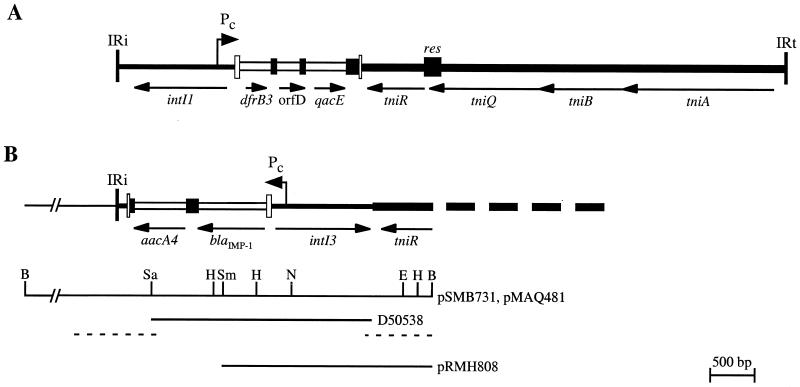
The role of integrons and gene cassettes in the spread of antibiotic resistance genes in gram-negative bacteria is well established (see references 21, 22, 48, and 49 for reviews). Integrons are able to capture genes that are part of gene cassettes (48) via a site-specific recombination event between two sites, one in the integron and one in the cassette (8-10). In class 1 integrons, which are the only well-characterized group, a specific recombination site, attI1, is located next to the intI1 gene and is recognized by the IntI1 integrase (27, 44, 50), and a promoter, Pc (formerly Pant or P1), which directs transcription of the cassette-borne genes, lies within the intI1 gene (11, 53). The configuration of these three features, shown in Fig. 1A, is now often viewed as definitive for all integrons, although this has never been shown experimentally for a second class. For class 1 integrons, intI1, attI1, and Pc are found within mobile elements that are transposons, e.g., Tn402 (47) (Fig. 1) or defective transposon derivatives (4, 25, 45, 46, 53).
FIG. 1.
Structure of (A) the class 1 integron Tn402 and (B) the class 3 integron. The 25-bp inverted repeats are indicated by tall bars labeled IRi and IRt. The 5′ conserved segment (5′-CS) of class 1 and the equivalent region in the class 3 integron are shown as medium lines, with the extent and direction of the intI gene indicated. Each gene cassette is shown as an open box that includes the gene and an adjacent filled box that represents the 59-be. The two parts of the attI sites flanking the cassettes are shown as open bars, and the positions and directions of the Pc promoters are indicated. Parts of the tni module are indicated by thick lines. For Tn402, the transposition genes, tni, and the res site are indicated. (B) The extent of the BamHI fragment of the class 3 integron cloned in pSMB731 and pMAQ481 is shown as a line below, with the positions of some of the restriction enzyme sites. The extent of the sequence in GenBank accession no. D50538 is indicated, and the additional regions sequenced in this work are shown by dashed lines. The fragment present in pRMH808 is shown below. Restriction enzyme sites: B, BamHI; E, EcoRI; H, HindIII; N, NcoI; Sa, SacI; Sm, SmaI.
To date, five distinct integron classes have been found associated with cassettes that contain antibiotic resistance genes (1, 23, 24, 29, 49, 53; accession no. AJ277063). For class 2, the intI2* gene, which includes a termination codon, and presumably also attI2 and Pc are found within transposons such as Tn 7 (23, 24, 55, 56). Classes 1 and 2 are most common in resistant bacteria, and the mobility of these integrons was undoubtedly important in facilitating their spread into many different bacterial species. It is possible that integrons that successfully carry resistance genes or other useful genes into new species will be translocatable, as is the case for classes 1 and 2, but to date only one example of each of the remaining three, class 3 (1), class 9 (29), and an unnumbered class (accession no. AJ277063), has been found.
However, the role of integron and gene cassette systems in the evolution of bacterial and plasmid genomes is now known to be much broader than their role in the dissemination of antibiotic resistance genes. An integron (class 4) was recently found in the small chromosome of several different Vibrio cholerae strains (6, 7, 37), and in the V. cholerae strain that has been sequenced, the integron contains an array of 179 cassettes, only a few of which, e.g., the catB9 gene cassette, contain genes that are likely to determine resistance to an antibiotic (28). Furthermore, different V. cholerae strains contain different cassette arrays (7). Other Vibrio species also contain a chromosomally located integron (7, 52). Some species of Pseudomonas, Xanthomonas, and various other bacteria whose genomes have been partially sequenced also contain integrons, as do unidentified bacteria from environmental soil samples (41, 52, 57). However, whether these integrons are located on plasmids or in the chromosome remains to be established.
The IntI proteins, IntI1, IntI2, IntI3, etc., encoded by different integron types, are 34 to 94% identical or 57 to 96% similar in pairwise comparisons and form a distinct family of the tyrosine recombinase superfamily (40, 41). Most of the genes that encode IntI-type integrases identified to date have been found adjacent to gene cassettes, indicating that they are part of an integron. Each distinct IntI (intI) type (>98% identical) thus defines an integron class which encompasses all integrons with the same intI gene but different cassette arrays (20, 48). The number assigned to each class corresponds to the number assigned to the intI gene and IntI integrase.
The IntI1 integrase has been shown to recognize and recombine both the attI1 site found in the integron and the 59-be (59-base element) sites found in gene cassettes (21) which have different organizations (12, 44, 54). IntI1 catalyzes integration of gene cassettes into attI1 (8) and excision of cassettes (9) to create free circular gene cassettes (10). Integrative and excisive recombination between attI1 and a 59-be and between two 59-be, and integrative recombination between two attI1 sites have also been demonstrated in an in vivo cointegration assay (13, 24, 27, 35, 36, 44, 50, 54). In the same assay, recombination between a 59-be and secondary recombination sites occurs at very low but nonetheless significant levels (16, 17, 50), and recombination between attI1 and secondary recombination sites is less frequent (13, 27, 44).
Though the three features found in class 1 integrons, an intI gene, an adjacent attI site, and a Pc promoter, have come to be viewed as the defining features of an integron (19, 20, 22, 40), in most cases only the intI gene has been located. The presence of an attI site and Pc promoter in addition to the intI gene is inferred, but this has not been established experimentally. The presence of gene cassettes implies that a functional attI site is present, and because the gene cassettes found in class 2 and class 3 integrons contain antibiotic resistance genes that are expressed, a Pc promoter is also presumed to be present. As these cassettes are identical to ones that are found in class 1 integrons, it is also possible to define the point at which cassettes are integrated and thus the crossover point at one end of the putative attI2 and attI3 sites. However, there is little obvious similarity between the sequences of attI1 and the regions in class 2 and class 3 integrons that are predicted to contain the attI2 and attI3 sites (26). Recently, the boundary between the region containing the intI4 gene and the attI4 site in the Vibrio cholerae genome and the first gene cassette has also been unambiguously defined (7), and the sequence of the attI4 region is also unrelated to the known attI sites. In other cases, the presumptive attI site is also minimally related to attI1 (40). However, in all of the attI sites, a simple site consisting of two inversely oriented putative IntI binding sites can be discerned (26, 40).
Here, we have sought to establish the generality of the arrangement of key integron features, intI gene, attI site, and Pc promoter, by examining in detail the properties of another class of integron. To establish experimentally if IntI that are not very closely related recognize the same sites, namely the 59-be in gene cassettes, and if cassettes are preferentially incorporated at the attI site, we also examined the reactions catalyzed by this further IntI-type integrase. We used the class 3 integron isolated by Arakawa et al. (1). The IntI3 integrase is 59% identical to IntI1 and has been shown to be active in preliminary experiments (26).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli UB5201 is F− pro met recA56 gyrA; E. coli UB1637 is F− his lys trp recA56 rpsL; and E. coli DH5α is supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1. Plasmids are described in Table 1. pSMB731, which contains a 9-kb BamHI fragment from Serratia marcescens cloned in the vector pMK16 (1), was used as the source of the class 3 integron. The 9-kb BamHI fragment from pSMB731 was recloned into the BamHI site of pACYC184 to generate pMAQ481, in which the direction of transcription of the cassette genes, blaIMP-1 and aacA4, is the same as that of the tetracycline resistance gene (tetA) of the vector. A 2.5-kb BamHI-SmaI fragment of pMAQ481 including the intI3 gene and part of blaIMP-1 was subcloned between the BamHI and NruI sites of pACYC184 to generate pRMH807. An EcoRV-XbaI fragment containing the tetA promoter was deleted from pRMH807 to form pRMH808, which was used as the substrate for insertion of aadA2 and aacA4 cassettes.
TABLE 1.
Plasmids
| Plasmid | Descriptiona | Relevant phenotype | Reference |
|---|---|---|---|
| R388 | 33-kb IncW plasmid containing In3 | Sur Tpr Tra+ IntI1+ | 2 |
| pACYC184 | Cloning vector | Cmr Tcr | 5 |
| pSMB731 | 9-kb BamHI fragment from S. marcescens AK7393 in pMK16; cassettes blaIMP-1-aacA4 | Apr Kmr IntI3+ | 1 |
| pMAQ28 | 400-bp Sau3A-HindIII fragment in BamHI-HindIII sites of pACYC184 (orientation 1); contains aadB/qacE 59-be | Cmr | 24 |
| pMAQ481 | BamHI fragment of pSMB731 in BamHI site of pACYC184 (orientation 2) | Apr Cmr Kmr IntI3+ | This study |
| pMAQ484 | PCR fragment (HS215/HS216) from pSMB731 cloned in pACYC184 (orientation 1); contains 131-bp att13/34-bp blaIMP-1 | Cmr | This study |
| pMAQ487 | PCR fragment (HS207/HS208) from pSMB731 cloned in pFLAG-MAC | Apr IntI3+ | 26 |
| pMAQ495 | R388 with aphA1 inserted in intI1 | Kmr Sur Tpr Tra+ | 12 |
| pMAQ545 | Religation of a HindIII fragment from cointegrate of pMAQ484 with pMAQ495 (orientation 1); contains 131-bp att13/198-bp qacE | Cmr | This study |
| pMAQ636 | AvaI-HindIII fragment in pACYC184 (orientation 1); contains aacA4/qacE 59-be | Cmr | This study |
| pRMH251 | 262-bp TaqI-HindIII fragment in pACYC184 (orientation 1); contains 64-bp attI1/198-bp qacE | Cmr | 50 |
| pRMH505 | intI1 gene from pSU2056 in pFLAG-MAC | Apr IntI1+ | 12 |
| pRMH555 | EcoRV fragment in pACYC184 (orientation 1); contains dfrA7/qacE 59-be | Cmr | 13 |
| pRMH807 | BamHI-SmaI fragment of pMAQ481 in BamHI-NruI sites of pACYC184; contains attI3/blaIMP-1 | Cmr IntI3+ | This study |
| pRMH808 | XbaI-EcoRV deletion of pRMH807 (removes tetA promoter) | Cmr IntI3+ | This study |
| pRMH813 | IntI3-mediated insertion of aadA2 cassette into pRMH808; cassette aadA2 (blaIMP-1) | Cmr Smr/Spr IntI3+ | This study |
| pRMH819 | IntI3-mediated insertion of aacA4 cassette into pRMH813; cassette aacA4-aadA2 (blaIMP-1) | Cmr Kmr Smr/Spr IntI3+ | This study |
| pRMH850 | Derived from pRMH505 by end-filling and religation of the EcoRI site | Apr IntI1+ | This study |
| pRMH851 | Derived from pMAQ487 by end-filling and religation of the EcoRI site | Apr IntI3+ | This study |
| pRMH863 | IntI3-mediated deletion of aacA4 cassette from pRMH819; cassette aadA2 (blaIMP-1) | Cmr Smr/Spr IntI3+ | This study |
| pRMH864 | IntI3-mediated deletion of aadA2 cassette from pRMH819; cassette aacA4 (blaIMP-1) | Cmr Kmr IntI3+ | This study |
| pRMH866 | NcoI-HindIII fragment in EcoRV-HindIII sites of pACYC184 (orientation 1); contains 284-bp attI3/198-bp qacE | Cmr | This study |
| pRMH891 | IntI3-mediated deletion of aacA4 and aadA2 cassettes from pRMH819 | Cmr IntI3+ | This study |
Orientation 1 refers to that orientation of the cloned fragment in the vector pACYC184, in which the direction of transcription from Pc is the same as that of the rep gene of pACYC184, while orientation 2 is the opposite (13).
pMAQ495 is a derivative of R388 in which the intI1 gene is interrupted by insertion of an aphA1 (kanamycin resistance) gene (12).
Culture conditions.
Bacteria were routinely cultured in Luria-Bertani (LB) medium or LB agar (38) supplemented as appropriate with ampicillin (100 μg/ml), chloramphenicol (Cm; 25 μg/ml), kanamycin (Km; 5 μg/ml), nalidixic acid (Nx; 25 μg/ml), spectinomycin (Sp; 25 μg/ml), streptomycin (25 μg/ml), sulfamethoxazole (Su; 25 μg/ml), or trimethoprim (Tp; 25 μg/ml). Antibiotics were obtained from Sigma.
Determination of transcription start point.
mRNA from DH5α/pMAQ481 was used as a template for extension of oligonucleotide RH230 using reverse transcriptase as previously described (11). The extension product was electrophoresed through a 6% denaturing polyacrylamide gel together with dideoxy sequencing tracks using pMAQ481 DNA as the template and RH230 as the primer.
Determination of antibiotic resistance level.
Stationary-phase cultures of E. coli UB5201 containing pRMH813 were diluted in saline (0.85% NaCl, wt/vol), and 100 to 300 cells were plated on LB agar containing different concentrations of streptomycin as described before (11). The concentration of streptomycin required for 50% loss of colony-forming ability (IC50) was determined as described previously (11). Values from four independent cultures were averaged.
Cloning and expression of integrase.
pMAQ487 contains the complete intI3 gene, amplified from pSMB731 using primers HS207 and HS208 and cloned between the EcoRI and BglII sites of pFLAG-MAC (International Biotechnologies) in frame with the Flag octapeptide (26). The Flag-IntI3 protein has an N-terminal extension of 17 amino acids. pRMH505 contains the complete intI1 gene cloned in pFLAG-MAC so that the IntI1 expressed is fused at the N terminus to the Flag octapeptide (12). pRMH850 and pRMH851 are derived from pRMH505 and pMAQ487, respectively, by cleavage and end-filling of the EcoRI site, followed by religation, thus disrupting the fusion to the Flag leader peptide and creating an N-terminal fusion to the 12-amino-acid peptide MTKSSFSRKFSC in the expressed integrases.
Conduction assays.
The ability of IntI1 or IntI3 to support integrative recombination was determined by measuring the frequency with which a pACYC184-based plasmid containing a cloned recombination site was transferred from the donor strain UB1637 (RecA− Smr) to the recipient UB5201 (RecA− Nxr) after fusion with pMAQ495, a derivative of the conjugative plasmid R388 (see above and Table 1) as described previously (36, 54). Integrase was supplied in trans using plasmid pRMH505, pRMH850, pMAQ487, or pRMH851 (see above and Table 1). Transconjugants were selected on agar containing trimethoprim and nalidixic acid and recombinants were selected on agar containing chloramphenicol and nalidixic acid. Cointegration frequency was expressed as the ratio of Cmr to Tpr transconjugants.
Cassette insertion and deletion assays.
Integration of gene cassettes was performed as described previously (8). Briefly, cassettes were constructed in vitro by digesting a plasmid containing a tandem duplication of a cassette with a restriction enzyme cleaving at a unique site in the cassette. The resulting linear cassette was isolated from an agarose gel using Geneclean (Bio 101, Inc.), circularized by ligation, and transformed by electroporation into DH5α harboring pACYC184 derivatives that contained cloned recombination sites, as well as pMAQ487 or pRMH851 to supply IntI3. Transformants were isolated on LB agar containing chloramphenicol and the antibiotics to which the resident cassette (if any) and the incoming cassette conferred resistance. Plasmid DNA was isolated from the resulting colonies and retransformed into DH5α. DNA from these secondary transformants was then checked for integration of the incoming cassette by restriction mapping.
Deletion of cassettes was assayed as described previously (9). DH5α was transformed with a pACYC184 derivative containing the aacA4 or aadA2 cassette, followed by pMAQ487 or pRMH851 (Table 1) to supply the appropriate integrase in trans, and then grown in LB medium containing ampicillin and chloramphenicol for approximately 50 generations. Plasmid DNA was retransformed into DH5α, and transformant colonies selected with chloramphenicol were screened for loss of the antibiotic resistance encoded by the cassette gene(s) by patching to L agar containing kanamycin for aacA4 or spectinomycin for aadA2. Deletions were confirmed by restriction mapping with NcoI.
DNA manipulations.
Plasmid DNA was isolated by the alkaline lysis method (3). Plasmid DNA for double-strand sequencing was purified by using Wizard columns (Promega) and annealed with the primer according to the method of Jones and Schofield (31). Sequencing was carried out using a Sequenase kit (United States Biochemical). Alternatively, automated sequencing was carried out at the Macquarie Sequencing Facility, Department of Biological Sciences, Macquarie University, Sydney, using an ABI Prism 377 DNA sequencer (PE Biosystems) and Big Dye terminator mixes.
Oligonucleotides (with positions in GenBank accession no. D50438) used for amplification of intI3 were HS207 (GGAATTCCTGCATGAACAGGTATAACGG, 1066 to 1046) and HS208 (CGGGATCCCGTCAGCCGGGCGACAAGTGCA, 23 to 52), and for amplification of attI3 they were HS215 (CGGGATCCCGCGTTATACCTGTTCATGCAT, 1049 to 1067) and HS216 (CGGGATCCCGTACAGATAACTTGCTCATAC, 1193 to 1212). RH230 (GAATACAGATAACTTGCTCATAC, 1215 to 1193; in blaIMP-1) was used for primer extension. The 5′ extensions containing restriction sites to facilitate cloning are shown in bold type. HS207 includes an EcoRI site, and HS208, HS215, and HS216 include a BamHI site (underlined).
RESULTS
Structure of class 3 integron.
Only a single example of a class 3 integron, isolated from a carbapenem-resistant Serratia marcescens strain, has been described (1). A gene encoding a putative integrase that is 59% identical (72% similar) to IntI1 was identified adjacent to the blaIMP-1 gene cassette (Fig. 1) that had previously been found in a class 1 integron (42). The blaIMP-1 gene is expressed, as the metallo-β-lactamase it encodes confers resistance to carbapenems (1, 30), and this implies that a Pc promoter is present. The reported sequence (1) (GenBank accession no. D50438) also includes part of an aacA4 cassette (aacA4 confers resistance to aminoglycosides) located downstream of the blaIMP-1 cassette. Here, the 9-kb BamHI fragment that includes this region was recloned into pACYC184 to permit detection of resistance to aminoglycosides, and the E. coli strain containing the resultant plasmid, pMAQ481 (Fig. 1B), was resistant to kanamycin.
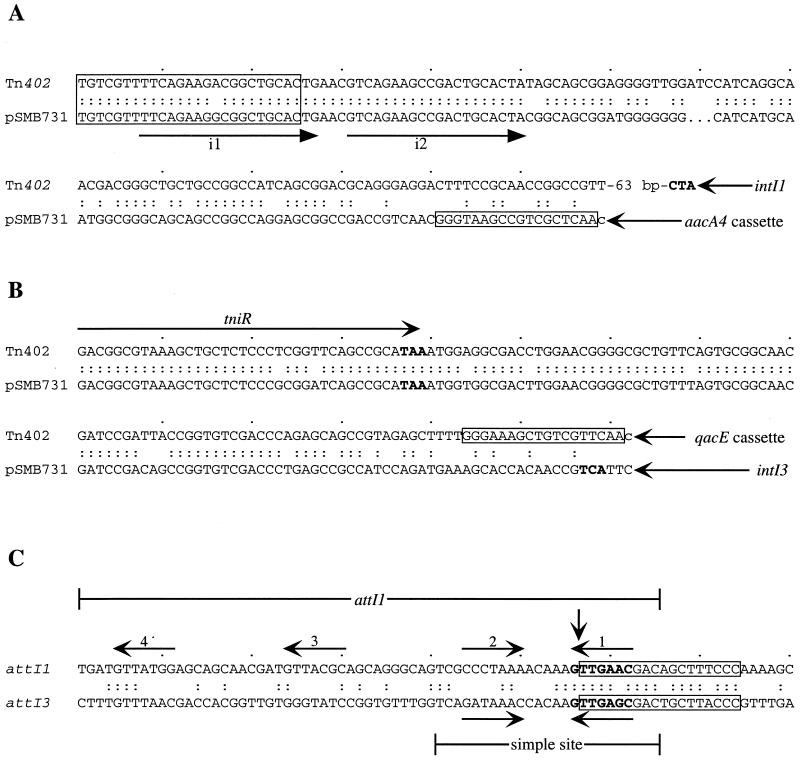
The sequence of the available BamHI fragment was extended in both directions (GenBank accession no. AF416297). The complete aacA4 cassette was present, and the sequence immediately beyond it, which should comprise either the start of a further gene cassette or the remainder of the attI3 site, was related to a short region of Tn402 that abuts the qacE cassette, which is the last cassette in Tn402 (Fig. 2). As Tn402 is a class 1 integron, this region comprises the remainder of the original attI1 site that is separated away when a cassette is incorporated into attI1 (Fig. 1A). Thereafter, a region of about 110 bp was clearly related to the IRi end of class 1 integrons (Fig. 2A). This segment includes both of the binding domains for the TniA transposase recently identified in binding studies (33).
FIG. 2.
Alignments of homologous sequences from Tn402 and the class 3 integron. Identical nucleotides are indicated by colons. The locations and orientations of various genes are shown by arrows, with stop codons in bold type. (A) IRi and adjacent regions. The 25-bp IRi of both sequences is boxed. The arrows beneath the sequences labeled i1 and i2 show the extent of a 19-bp repeat bound by tniA (33). The 18-bp region boxed only in the pSMB731 sequence is aligned with the equivalent region from Tn402 in panel C. (B) tniR genes and adjacent regions. The 18-bp region boxed only in the Tn402 sequence is aligned with the equivalent region from pSMB731 in panel C. (C) attI regions. The cassette sequences have been removed to join the regions present in the original attI sites. The vertical arrow represents the recombination crossover point at which cassettes are integrated. The extents of attI1 (44) and of the simple site are indicated. The core sites that form the central region of the IntI binding sites are shown by horizontal arrows.
The sequence downstream of the intI3 gene contained most of a tniR gene (the codons for the first 12 amino acids of the product are missing, as they lie beyond the BamHI site) whose product is 95% identical to TniR of Tn402 (Fig. 1). The similarity between the Tn402 and pSMB731 sequences extends beyond the end of the tniR genes to close to the end of the qacE cassette or intI3 gene (Fig. 2B). The intI3 gene thus lies in a similar position to intI1 within a similar transposon backbone, but the intI genes are in opposite orientations with respect to this backbone (Fig. 1).
Location of Pc promoter.
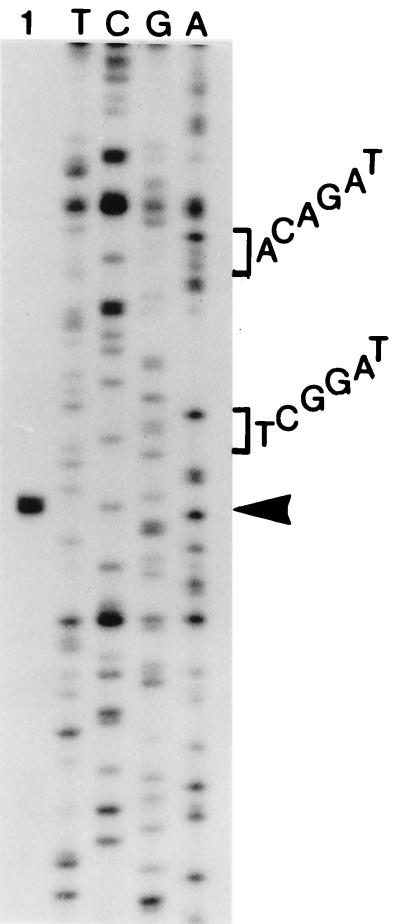
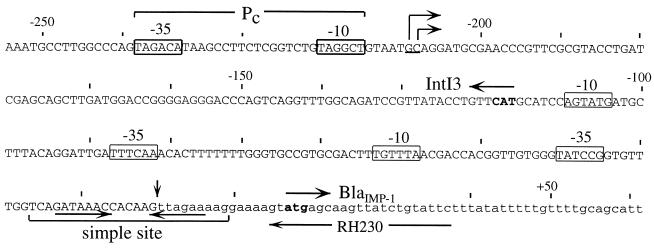
The start point for transcription of the cassette genes was mapped by primer extension. RNA from cells containing pMAQ481 (Fig. 1B) was used as the template, and the primer is complementary to a sequence just inside the blaIMP-1 gene. Two extension products of approximately equal intensity that differed in size by one base were detected (Fig. 3). The mRNA start points lay 208 and 209 bases to the left of the boundary of the blaIMP-1 cassette, at a G and a C residue (underlined in Fig. 4), and the most likely promoter sequence is TAGACA-N17-TAGGCT (boxed in Fig. 4). Thus, Pc is located just inside the intI3 gene, and its position in the coding region is precisely equivalent to that of Pc in class 1 integrons.
FIG. 3.
Transcription start point for the class 3 Pc. RNA from cells containing pMAQ481 was used as the template for the extension of primer RH230, which is complementary to the sequence just inside the blaIMP-1 gene. The extension products (lane 1) were separated by electrophoresis on a denaturing acrylamide gel, together with dideoxy sequencing reactions as markers (lanes T, C, G, and A), generated by extension of RH230 with pMAQ481 DNA as the template. The position of the transcription start point is marked by an arrowhead, and the sequences of the Pc −35 and −10 regions are indicated to the right.
FIG. 4.
Promoter and attI3 region of the class 3 integron. The pair of nucleotides representing the 5′ end of the Pc mRNA extension product (Fig. 3) are underlined, and the inferred start points for transcription are marked with bent arrows. The position of Pc is indicated, with −35 and −10 regions boxed. The −10 and −35 sequences for the two possible Pint promoters for transcription of the intI3 gene are also boxed. The sequence of the beginning of the blaIMP-1 cassette is shown in lowercase letters, and the position of primer RH230 is indicated by a horizontal arrow. Start codons for the intI3 and blaIMP-1 genes are in bold type, and the directions are marked with arrows above the sequence. The extent of the attI3 simple site (inferred by analogy with attI1) includes a pair of 7-bp consensus sequences in inverted orientation, marked with arrows, and a vertical arrow marks the recombination crossover point.
To estimate the strength of the class 3 Pc promoter, the level of resistance to streptomycin conferred by the aadA2 gene when the aadA2 cassette is in first position was measured as described previously (11). Using E. coli UB5201 harboring pRMH813, which contains a cloned fragment of the class 3 integron with the aadA2 cassette inserted in the first position (see below and Fig. 5A), the concentration of streptomycin required to reduce the plating efficiency to 50% (IC50) was 340 μg/ml. This lies within the range of the class 1 Pc, for which a number of variants that differ in strength over a 20- to 30-fold range (IC50 of streptomycin, 65 to 1,200 μg/ml) have been identified (11, 34).
FIG. 5.
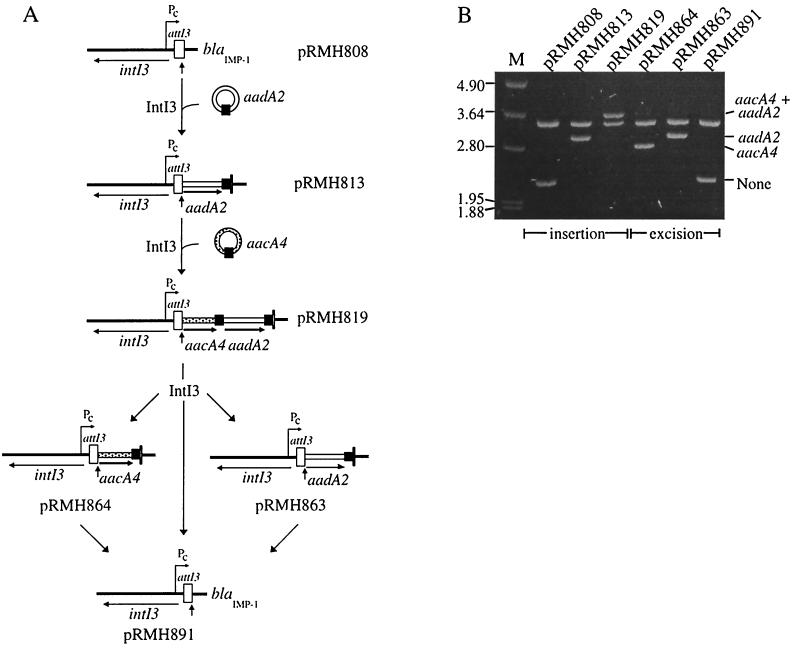
Insertion and excision of gene cassettes. (A) IntI3-catalyzed site-specific recombination between the 59-be in the circular cassette and the attI3/blaIMP-1 recombination site in pRMH808 incorporates the cassette into the integron. IntI3-dependent recombination also excises the cassettes, independently or together. The position of the Pc promoter and the extent of the coding regions of the intI3, aacA4, and aadA2 genes are marked by arrows. Other features are as shown in Fig. 1. (B) Analysis of the products of cassette insertion and excision. Plasmids generated by the IntI3-mediated events depicted in panel A were digested with NcoI, and fragments were separated by electrophoresis in a 0.8% agarose gel. The integron insert region is included in one of the NcoI fragments, the length of which indicates the presence of the cassettes shown to the right. Size markers (in kilobases) are shown to the left.
Activity of IntI3 and attI3.
To determine whether the IntI3 protein is active, the intI3 gene was initially cloned in the expression vector pFLAG-MAC under the control of the tac promoter so that the IntI3 protein is fused in frame to a 17-amino-acid peptide that includes the Flag octapeptide (26). The resulting plasmid (pMAQ487) and a further plasmid, pRMH851, in which the Flag leader peptide is replaced by a different 12-amino-acid peptide (see Materials and Methods) were used as the source of IntI3 in a standard conduction assay (13, 35, 36, 50, 54). The assay measures the frequency of formation of cointegrates between a pACYC184 derivative containing a cloned recombination site (59-be or attI) and a conjugative plasmid related to R388. To eliminate the possibility of recombination due to IntI1, the intI1 gene in R388 was disrupted by insertion of a kanamycin resistance gene, and this derivative of R388, pMAQ495, was used. R388 and pMAQ495 contain a class 1 integron with two inserted cassettes, dfrB2 and orfA, and thus have three composite recombination sites, attI1/dfrB2 and the dfrB2/orfA and orfA/qacE 59-be, hereafter referred to as attI1, dfrB2 59-be, and orfA 59-be, respectively. However, if the cloned recombination site is in the configuration used here, the predominant event has been shown to be recombination between the cloned site and the orfA 59-be (13).
Flag-IntI3 catalyzed cointegration of pMAQ495 with plasmids containing either a 59-be (pMAQ28, pRMH555, and pMAQ636) or the region predicted to include the attI3 site (pMAQ484, pMAQ545, and pRMH866) (Table 2). For comparison, assays for the equivalent reactions catalyzed by equivalent versions of IntI1 are also shown in Table 2. IntI3 recombines both the 59-be and attI3 sites with the orfA 59-be in R388, albeit less efficiently than equivalent constructs of IntI1 recombined the 59-be or attI1 with the orfA 59-be. Restriction mapping of 12 cointegrates isolated from the crosses involving pMAQ28 or pMAQ545 and catalyzed by IntI3 (pRMH851) confirmed that in all cases, recombination had occurred between the aadB/qacE 59-be or the attI3 site and the orfA 59-be in pMAQ495. These experiments also localize the region required for full attI3 site activity to 131 bp to the left of the crossover point.
TABLE 2.
Activity of IntI3 recombinase
| Recombination site | Fragment lengtha (nt) | Plasmidb | Mean cointegration frequency (no. of assays)
|
||||
|---|---|---|---|---|---|---|---|
| pMAQ487 (Flag-IntI3) | pRMH851 (IntI3) | pRMH505 (Flag-IntI1) | pRMH850 (IntI1) | No integrase | |||
| aadB/qacE | 202/198 | pMAQ28 | 1.5 × 10−4 (10) | —c | 1.3 × 10−3 (6) | 1.8 × 10−3 (4) | 9.5 × 10−8 (3) |
| dfrA7/qacE | 221/755 | pRMH555 | 7.0 × 10−4 (6) | — | — | — | — |
| aacA4/qacE | 228/198 | pMAQ636 | 3.9 × 10−3 (6) | — | — | — | — |
| attI3/qacE | 284/198 | pRMH866 | — | 2.2 × 10−3 (5) | — | — | — |
| attI3/qacE | 131/198 | pMAQ545 | 7.1 × 10−4 (6) | 1.4 × 10−3 (5) | — | — | 2.1 × 10−7 (3) |
| attI3/blaIMP-1 | 131/34 | pMAQ484 | 8.1 × 10−4 (5) | 8.4 × 10−4 (4) | — | — | — |
| attI1/qacE | 64/198 | pRMH251 | — | — | 1.1 × 10−2 (7) | 8.6 × 10−3 (6) | 3.4 × 10−7 (4) |
| None | pACYC184 | — | 5.0 × 10−7 (6) | — | 1.3 × 10−5 (6) | — | |
Number of nucleotides (nt) in the cloned fragment to the left/right of the recombination crossover point.
Cells also contained pMAQ495.
—, not determined.
IntI3 catalyzes cassette insertion at attI3.
To demonstrate that gene cassettes can be inserted at attI3 by IntI3-mediated recombination, circular aadA2 cassettes, constructed in vitro as described previously (8), were transformed into DH5α harboring pRMH851 to supply IntI3 and pRMH808 (Fig. 1B), which contains the attI3 recombination site. Approximately 100 colonies appeared after overnight incubation of transformants on medium containing spectinomycin (Spr, specified by the aadA2 gene) and chloramphenicol (Cmr, specified by the vector). Even though the intI3 gene is present on pRMH808, it does not appear to be significantly expressed, as Spr Cmr transformants were not recovered unless pRMH851 was present to supply IntI3. Integration of the aadA2 cassette was also observed when pRMH851 was replaced by pMAQ487, which produces IntI3 with the Flag peptide fused to the N terminus.
Plasmid DNA was isolated from one clone and retransformed into DH5α to segregate parental from recombinant plasmids (8). Plasmid DNA from six of the resulting secondary transformants was mapped using the restriction enzyme NcoI to confirm that integration of an aadA2 cassette at attI3 had occurred. In each case, the size of the insert region had increased by 0.9 kb (Fig. 5B), which is the size of the aadA2 cassette, and sequencing of the boundaries confirmed that the aadA2 cassette had been precisely inserted into the attI3/blaIMP-1 site (Fig. 6). The sequence of the recombinant junction is consistent with the crossover's having occurred at the expected position within the GTTAGA of the core site of attI3/blaIMP-1 and the GTTAGA at the 3′ end of the aadA2/aadA2 59-be (Fig. 6).
FIG. 6.
Sequences of the boundaries of inserted cassettes. The sequence surrounding the recombination point in the 59-be of each circular cassette (lowercase) and the attI3 site (uppercase) as well as the sequence of each boundary of the newly inserted cassette is shown. The bases to which the recombination crossover can be confined are underlined or overlined, and the sequence of the core IntI binding site is in bold type.
For class 1 integrons that contain a gene cassette and thus have both an attI1 and a 59-be site available, IntI1 preferentially integrates cassettes at attI1 (8). To determine whether this preference also applies for attI3 and IntI3, circular aacA4 cassettes (637 bp) were constructed and transformed into pRMH813 (see Fig. 5A), which contains two sites, attI3/aadA2 and the aadA2/blaIMP-1 59-be. Plasmid DNA prepared from seven independent transformants selected on medium containing kanamycin and spectinomycin was retransformed into DH5α, and restriction mapping of one transformant from each revealed that every isolate contained an aacA4 cassette inserted at the attI3/aadA2 junction (Fig. 5B). This was confirmed by sequencing for one isolate (Fig. 6). Thus, IntI3-mediated insertion of a cassette into the class 3 integron also occurs preferentially at the attI3 site.
IntI3-catalyzed excision of one or both cassettes from pRMH819, assayed as previously described (9), was also observed (Fig. 5).
IntI3 catalyzes recombination between 59-be and secondary sites.
To determine whether IntI3 can catalyze recombination at secondary or nonspecific sites, recombination of the vector pACYC184 with pMAQ495 was examined. A low frequency of recombination (1.7 × 10−6) that is 10-fold lower than that for the equivalent reaction mediated by IntI1 (1.3 × 10−5) was observed (Table 2). The resulting cointegrates were examined by restriction analysis and found to have been formed by recombination either between the dfrB2 59-be or the orfA 59-be in pMAQ495 and several different sites in pACYC184. Thus, like IntI1, IntI3 is capable of catalyzing a low frequency of recombination between a 59-be and a secondary site. However, a notable difference is that for IntI3, several of these reactions involved the dfrB2 59-be rather than the orfA 59-be, which is exclusively involved when the recombinase is IntI1 (16, 50). Sequencing revealed that the secondary site was predominantly GTT (Table 3). This is again different from IntI1, where GTT and GAT triplets are used equally efficiently as secondary sites. Two unusual sites, GTA and GCT, were also found.
TABLE 3.
Nucleotide sequences surrounding secondary recombination sites in pACYC184
| pACYC184 sequencea | Positionb (strand) | pMAQ495 recombination site | No. of independent isolates |
|---|---|---|---|
| CCTTAGCTCCTGAAAATCTC GAT AACTCAAAAAATACGCCCGG | 248 (+) | orfA | 1 |
| CTACCGGGCGTATTTTTTGA GTT ATCGAGATTTTCAGGAGCTA | 253 (−) | dfrB2 | 1 |
| AATGAGACGTTGATCGGCAC GTA AGAGGTTCCAACTTTCACCA | 312 (−) | orfA | 1 |
| ATCATACACTAAATCAGTAA GTT GGCAGCATCACCCGACGCAC | 457 (−) | dfrB2 | 1 |
| TTCTATCAGCTGTCCCTCCT GTT CAGCTACTGACGGGGTGGTG | 529 (+) | dfrB2 | 1 |
| CGCGGCCCTCTCACTTCCCT GTT AAGTATCTTCCTGGCATCTT | 816 (−) | orfA | 1 |
| CGCGGCCCTCTCACTTCCCT GTT AAGTATCTTCCTGGCATCTT | 816 (−) | dfrB2 | 2 |
| CGATAAGCTTTAATGCGGTA GTT TATCACAGTTAAATTGCTAA | 1539 (+) | orfA | 1 |
| TAATGCGGTAGTTTATCACA GTT AAATTGCTAACGCAGTCAGG | 1549 (+) | orfA | 2 |
| AGGATGACGATGAGCGCATT GTT AGATTTCATACACGGTGCCT | 1591 (−) | orfA | 2 |
| AGCCGCGAGCGATCCTTGAA GCT GTCCCTGATGGTCGTCATCT | 2585 (−) | orfA | 1 |
| ATAAAAACGGTTAGCGCTTC GTT AATACAGATGTAGGTGTTCC | 3497 (−) | orfA | 1 |
| TTGGTGCCCTTAAACGCCTG GTT GCTACGCCTGAATAAGTGAT | 3767 (−) | dfrB2 | 1 |
Nucleotides in bold type are those to which recombination crossover can be confined.
Numbers indicate the position of the G residue in the central triplet in the pACYC184 sequence (51) (modified according to accession no. X06403). The T at position 3766 is replaced by a TT in the pACYC184 used here. + indicates the strand shown in the published sequence, and − indicates the complementary strand.
DISCUSSION
Analysis of the single known representative of a class 3 integron revealed that the integron module, consisting of the intI3 gene, attI3 site, and Pc promoter, is configured in the same way as the class 1 integron module, with attI3 upstream of intI3 and Pc within the intI3 gene. This is consistent with the use of these three features as definitive for integrons. However, it cannot be assumed that a promoter is present in chromosomally located integrons, and experimental data are needed.
Based on the presence of IRi and a tniR gene in the region sequenced here, the class 3 integron module is also likely to be part of a transposon. The backbone is very clearly related to that of Tn402, which is the only known class 1 integron that contains a complete transposition module (47). Additional support for this conclusion comes from our finding that the IRi end is adjacent to a gene encoding a putative resolvase that is over 85% identical to the ParA resolvase of plasmid RP1 (GenBank accession no. L27758) (43). Class 1 integrons and related transposons that have similar IR and transposition genes are known to target the res sites associated with resolvase genes (32, 39). An interesting feature of this putative transposon is that though the intI3/attI3/Pc module has been incorporated into the backbone in the opposite orientation to the intI1/attI1/Pc module, both integron modules are in a similar location.
The fact that all three integron classes (1, 2, and 3) isolated from clinical antibiotic-resistant bacterial strains are both transposons and integrons is likely to be a testament to the utility of translocation in bacterial responses to strong selective pressure. These transposons may well have captured the intI/attI module from the chromosome of a bacterium that possesses an integron. If this is the case, it seems likely that further integron modules will be found in plasmids or transposons, and a further example has been reported recently (accession no. AJ277063) in a plasmid from Vibrio salmonicida. The close relationship of the IntI protein to that of the other Vibrio species (61 to 73% identity) suggests that it may even represent a duplication of the V. salmonicida chromosomal integron.
Most of the reactions previously shown to be catalyzed by the IntI1 integrase were also catalyzed by IntI3. Only recombination of attI3 with attI3 was not examined here. Furthermore, a new cassette was preferentially incorporated at the attI site, which is also the case for IntI1. Despite showing only 59% identity, IntI1 and IntI3 can recognize the same 59-be sites, allowing cassettes to move from one integron class to another. This feature is obviously important if cassettes from the pools found in chromosomally located integrons are to be available to other bacterial species. However, it is in contrast to the stringent target site specificities of the only other pair of related tyrosine-type recombinases that have been examined in detail. The integrases specified by phages λ and HK022 are 70% identical but are able to recombine only their cognate att sites, even though the att sites are quite similar (14, 15).
The most obvious difference between the class 1 and class 3 site-specific recombination systems is in the sequence of the attI sites (Fig. 2C), though they may have similar structures. The attI1 site is made up of 56 bp of the 5′-conserved segment and a further 9 or 10 bp from the adjacent module (44, 50) and contains four IntI1 binding sites (12, 18). The attI3 site was localized here to within a region of 131 bp from the conserved region, and it is presumed that a further 9 or 10 bp from the adjacent module are also needed. The attI1 site (Fig. 2C) consists of a simple site within which the recombination crossover occurs and two further directly oriented IntI1 binding sites that act to enhance the recombination efficiency (26, 44). IntI1 binds to these accessory sites more strongly than it binds to the simple site (12, 18). Whether attI3 shares these features with attI1 remains to be established, and further studies are in progress to more accurately define the attI3 site.
The experiments reported here did not examine the ability of IntI3 to recognize the attI1 site as a recombination substrate and recombine it with a 59-be type site or vice versa. Granted that the attI sites show only very limited sequence similarity, it is possible that each of them is only recognized by the cognate IntI. Our preliminary data (26) indicate that this may be so, and more detailed experiments are currently under way to examine this question.
Acknowledgments
We thank Y. Arakawa for supplying pSMB731 and Rachelle Ellis and Debbie Watson for technical assistance.
REFERENCES
- 1.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avila, P., and F. de la Cruz. 1988. Physical and genetic map of the IncW plasmid R388. Plasmid 20:155-157. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, C. A., L. Purins, P. Kaewrakon, and P. A. Manning. 1997. VCR repetitive sequence elements in the Vibrio cholerae chromosome constitute a mega-integron. Mol. Microbiol. 26:1137-1143. [DOI] [PubMed] [Google Scholar]
- 7.Clark, C. A., L. Purins, P. Kaewrakon, T. Focareta, and P. A. Manning. 2000. The Vibrio cholerae O1 chromosomal integron. Microbiology 146:2605-2612. [DOI] [PubMed] [Google Scholar]
- 8.Collis, C. M., G. Grammaticopoulos, J. Briton, H. W. Stokes, and R. M. Hall. 1993. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9:41-52. [DOI] [PubMed] [Google Scholar]
- 9.Collis, C. M., and R. M. Hall. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collis, C. M., and R. M. Hall. 1992. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 6:2875-2885. [DOI] [PubMed] [Google Scholar]
- 11.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collis, C. M., M.-J. Kim, H. W. Stokes, and R. M. Hall. 1998. Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol. Microbiol. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 13.Collis, C. M., G. D. Recchia, M. J. Kim, H. W. Stokes, and R. M. Hall. 2001. Efficiency of recombination reactions catalyzed by the class 1 integron integrase IntI1. J. Bacteriol. 183:2535-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorgai, L., E. Yagil, and R. A. Weisberg. 1995. Identifying determinants of recombination specificity: construction and characterization of mutant bacteriophage integrases. J. Mol. Biol. 252:178-188. [DOI] [PubMed] [Google Scholar]
- 15.Dorgai, L., S. Sloan, and R. A. Weisberg. 1998. Recognition of core binding sites by bacteriophage integrases. J. Mol. Biol. 277:1059-1070. [DOI] [PubMed] [Google Scholar]
- 16.Francia, M. V., F. de la Cruz, and M. García Lobo. 1993. Secondary sites for integration mediated by the Tn 21 integrase. Mol. Microbiol. 10:823-828. [DOI] [PubMed] [Google Scholar]
- 17.Francia, M. V., P. Avila, F. de la Cruz, and M. García Lobo. 1997. A hot spot in plasmid F for site-specific recombination mediated by Tn21 integron integrase. J. Bacteriol. 179:4419-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gravel, A., B. Fournier, and P. H. Roy. 1998. DNA complexes obtained with the integron integrase IntI1 at the attI1 site. Nucleic Acids Res. 26:4347-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, R. M. 1997. Mobile gene cassettes and integrons: moving antibiotic resistance genes in gram-negative bacteria, p. 192-205. In D. J. Chadwick and J. Goode (ed.), Ciba Foundation Symposium 207. Antibiotic resistance: origins, evolution, selection and spread. John Wiley and Sons, Chichester, England. [DOI] [PubMed]
- 20.Hall, R. M. 2001. Integrons, p. 1041-1045. In S. Brenner and J. H. Miller (ed.), Encyclopaedia of genetics. Academic Press, San Diego, Calif.
- 21.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 22.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updates 1:109-119. [DOI] [PubMed] [Google Scholar]
- 23.Hall, R. M., and C. Vockler. 1987. The region of the IncN plasmid R46 coding for resistance to β-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn 21-like transposons. Nucleic Acids Res. 15:7491-7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination crossover point. Mol. Microbiol. 5:1941-1959. [DOI] [PubMed] [Google Scholar]
- 25.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall, R. M., C. M. Collis, M. J. Kim, S. R. Partridge, G. D. Recchia, and H. W. Stokes. 1999. Mobile gene cassettes in evolution. Ann. N. Y. Acad. Sci. 87:68-80. [DOI] [PubMed] [Google Scholar]
- 27.Hansson, K., O. Sköld, and L. Sundström. 1997. Non-palindromic attI sites of integrons are capable of site-specific recombination with one another and with secondary targets. Mol. Microbiol. 26:441-453. [DOI] [PubMed] [Google Scholar]
- 28.Heidelberg, J. F., J. A. Elsen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragol, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito, H., Y. Arakawa, S. Ohsuka, R. Wacharotayankun, N. Kato, and M. Ohta. 1995. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob. Agents Chemother. 39:824-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, D. S. C., and J. P. Schofield. 1990. A rapid method for isolating high quality plasmid DNA suitable for DNA sequencing. Nucleic Acids Res. 18:7463-7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamali-Moghaddam, M., and L. Sundström. 2000. Transposon targeting determined by resolvase. FEMS Microbiol. Lett. 186:55-59. [DOI] [PubMed] [Google Scholar]
- 33.Kamali-Moghaddam, M., and L. Sundström. 2001. Arrayed transposase-binding sequences on the ends of transposon Tn5090/Tn402. Nucleic Acids Res. 29:1005-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lévesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 35.Martinez, E., and F. de la Cruz. 1988. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol. Gen. Genet. 211:320-325. [DOI] [PubMed] [Google Scholar]
- 36.Martinez, E., and F. de la Cruz. 1990. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 9:1275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazel, D., B. Dychinco, V. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Minakhina, S., G. Kholodii, S. Mindlin, O. Yurleva, and V. Nikiforov. 1999. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol. 33:1059-1068. [DOI] [PubMed] [Google Scholar]
- 40.Nield, B. S., A. J. Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. H. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 41.Nunes-Düby, S. E., H. J. Kwon, R. S. Tirumalai, T. E. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 44.Partridge, S. R., G. D. Recchia, C. Scaramuzzi, C. M. Collis, H. W. Stokes, and R. M. Hall. 2000. Definition of the attI1 site of class 1 integrons. Microbiology 146:2855-2864. [DOI] [PubMed] [Google Scholar]
- 45.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rådström, P., O. Skold, G. Swedberg, J. Flensburg, P. H. Roy, and L. Sundström. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 176:3257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 49.Recchia, G. D., and R. M. Hall. 1997. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 389:389-394. [DOI] [PubMed] [Google Scholar]
- 50.Recchia, G. D., H. W. Stokes, and R. M. Hall. 1994. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res. 22:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose, R. E. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rowe-Magnus, D. A., A.-M. Guerot, P. Ploncard, B. Dychinco, J. Davies, and D. Mazel. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl. Acad. Sci. USA 98:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 54.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 55.Sundström, L., P. H. Roy, and O. Sköld. 1991. Site-specific insertion of three structural gene cassettes in transposon Tn7. J. Bacteriol. 173:3025-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tietze, E., J. Brevet, and H. Tschäpe. 1987. Relationships among the streptothricin resistance transposons Tn1825 and Tn1826 and the trimethoprim resistance transposon Tn7. Plasmid 18:246-249. [DOI] [PubMed] [Google Scholar]
- 57.Vaisvila, R., R. D. Morgan, J. Posfai, and E. Raleigh. 2001. Discovery and distribution of super-integrons among pseudomonads. Mol. Microbiol. 42:587-601. [DOI] [PubMed] [Google Scholar]