Abstract
The cyanobacterium Synechocystis sp. strain PCC 6803 is able to acclimate to levels of salinity ranging from freshwater to twice the seawater concentrations of salt by accumulating the compatible solute glucosylglycerol (GG). Expression of the ggpS gene coding for the key enzyme (glucosylglycerol-phosphate synthase) in GG synthesis was examined in detail. Under control conditions, the GgpS protein is stable, so that weak constitutive transcription of the ggpS gene resulted in a significant protein content. However, the enzyme activity was biochemically switched off, and no GG was detectable. After a salt shock, an immediate increase in mRNA content proportional to the salt content occurred, while the GgpS protein and GG contents rose in a linear manner. Furthermore, the stability of the ggpS mRNA increased transiently. In salt-acclimated cells expression of the ggpS gene, the GgpS protein content, and the amount of accumulated GG depended linearly on the external salt concentration. Mapping of the 5′ end of the ggpS transcript revealed a long nontranslated 5′ sequence and a putative typical cyanobacterial promoter, which did not show any obvious salt-regulatory element. The alternative σ factor σF was found to be involved in salt-dependent regulation of ggpS, since in a σF mutant induction of this gene was strongly reduced. The present study demonstrated that in addition to biochemical regulation of GgpS activity, alterations of ggpS expression are involved in regulation of GG synthesis in Synechocystis sp. strain PCC 6803. A model showing the interaction of the two regulatory levels is presented.
The strategies used by bacteria to survive in environments with high and changing salinities have received much attention in the last few years. The general physiological response after an upshift of the salt concentration in a medium includes three phases. First, inorganic ions (usually Na+ and Cl−) enter the cell after the turgor collapse resulting from the large difference in water potentials across the cytoplasmic membrane. Second, Na+ is exchanged for K+, which saves the cell metabolism from the toxic influence of high Na+ concentrations. Stabilization of turgor by accumulation of ions as a long-term acclimation strategy is possible only in halophilic archaea and some halophilic bacteria. Therefore, in the third phase so-called compatible solutes are usually accumulated, which allows exclusion of the inorganic ions without a further change of turgor (1).
For some model organisms, like the gram-negative enteric bacterium Escherichia coli, the gram-positive soil bacterium Bacillus subtilis, and the cyanobacterial freshwater isolate Synechocystis sp. strain PCC 6803, the acclimation processes have been investigated in detail. In minimal media these strains produce by de novo synthesis amounts of the compatible solutes trehalose (E. coli) (11), proline (B. subtilis) (26), and glucosylglycerol (GG) (Synechocystis) that are proportional to the stress (20, 5). Mutants defective in trehalose, proline, or GG production exhibit a growth defect in hyperosmotic media but are able to take up these or other osmolytes from the medium to survive (1, 14, 19). In E. coli the stress-related trehalose synthesis system consists of two enzymes. A trehalose-6-phosphate synthase catalyzes the enzymatic condensation of UDP-glucose and glucose 6-phosphate, and the subsequent fast dephosphorylation of the intermediate trehalose 6-phosphate is mediated by trehalose-6-phosphate phosphatase. These enzymes are encoded by the genes otsA and otsB, respectively, which are transcribed together in the otsBA operon (11, 12). The proline synthesis in B. subtilis after an osmotic upshift starts from glutamate with two enzymes not involved in anabolic synthesis of proline. The genes that encode these enzymes, proH and proJ, are organized in an operon whose expression is salt induced (1). The synthesis of GG in Synechocystis sp. strain PCC 6803 is performed by a two-step reaction, in which enzymatic condensation of ADP-glucose and glycerol 3-phosphate by glucosylglycerol-phosphate synthase (GgpS) is followed by dephosphorylation of the intermediate by glucosylglycerol-phosphate phosphatase (GgpP) (5). The genes that encode the enzymes, ggpS and stpA, respectively, are not organized in an operon (7, 17).
Successful salt acclimation involves tight regulation of many processes, not only the processes involved in the production of compatible solutes. The regulation can be based on modulation of gene expression or the activities of existing proteins. Often these two mechanisms are combined. Clear induction of the otsBA operon in E. coli has been found after salt shock and in cells entering the stationary phase, which is dependent on the activity of the alternative σ factor RpoS (9). Additionally, the activities of the trehalose-synthesizing enzymes are directly stimulated by enhanced salt concentrations (3). Furthermore, the high-affinity uptake systems for the compatible solutes ProU and ProP in E. coli are activated after salt stress in a direct manner combined with 3-fold induction and 100-fold induction, respectively, of gene expression as measured by using promoter fusions with lacZ (18). Regulators have also been identified for expression of the glycine betaine uptake system OpuE induced by salt and during the stationary phase in B. subtilis (24). Two promoters and sigma factors are involved. Promoter P1 mediates strong induction directly after a salt shock and is σB dependent. σB also regulates the general stress response of B. subtilis (8). The P2 promoter is σA dependent and mediates only weak induction directly after an upshock but was found to be essential for opuE expression in cells acclimated to higher salt concentrations.
In Synechocystis sp. strain PCC 6803 GG production is activated primarily by a biochemical, salt-dependent mechanism (5). Here we show that transcription of the ggpS gene encoding the key enzyme in GG synthesis is clearly induced after an osmotic upshift. However, in shocked cells no direct correlation between mRNA content and the translated GgpS protein was observed. In contrast, in cells completely acclimated to different salt concentrations the ggpS mRNA content, the GgpS protein content, and the amount of accumulated GG are determined by the external salt concentration in a linear relationship. Two distinct mechanisms, biochemical modulation of enzyme activity and increased gene expression, are obviously involved in the regulation of GG production in Synechocystis sp. strain PCC 6803 that is proportional to stress.
MATERIALS AND METHODS
Strains and culture conditions.
A derivative of Synechocystis sp. strain PCC 6803 with enhanced transforming capacity (referred to below as Synechocystis) was obtained from S. Shestakov (Moscow State University, Moscow, Russia) and was used in all experiments. Synechocystis belongs to the group of moderately halotolerant cyanobacteria that resist up to 1.2 M NaCl by accumulating GG as the main compatible solute and traces of sucrose (20). Axenic cells were cultivated in batch cultures at 30°C with bubbling with air enriched with CO2 (5%, vol/vol) and continuous illumination (170 μmol m−2 s−1; Osram L58W/32 Lumilux de luxe) by using a KNO3-containing standard medium with 10 mM Na+ (17). Different salt concentrations were obtained by addition of appropriate amounts of NaCl. In order to obtain cells completely acclimated to a certain salt concentration, cells were precultured for at least 5 days in medium containing NaCl at this concentration.
RNA techniques.
For RNA extraction 7 ml of cells was harvested by centrifugation (4,000 × g, 5 min, 2°C), immediately frozen, and stored at −80°C. RNA was isolated by using a HIGH PURE RNA isolation kit (Roche Diagnostics) after preextraction with hot phenol and chloroform. The methods used for separation of RNA, blotting, and hybridization were described in detail previously (7). Gene-specific DNA probes for Northern blot experiments were obtained after PCR amplification of the coding sequences of the ggpS and psbC genes (sll1566 and sll0851) (13) by using the primers ggpS5′ (CGG GAT CCA TGA ATT CAT CAT CCC TTG TGA TCC), ggpS3′ (CGG GAT CCC TAC ATT TGG GGG GGC TGT CCC) (17), psbD-fw (CCA TGA CTA TTG CAG TCG GA), and psbC_rev (CCA AAG TCT CAA TCT AGT CG) (boldface type indicates BamHI recognition sites, and underlining indicates start and stop codons). A 16S ribosomal DNA fragment was amplified by using primers 16SrDNA27f (AGA GTT TGA TCM TGG CTC AG) and 16SrDNA1525 (AAG GAG GTG WTC CAR CC) (16) binding to the 5′ and 3′ ends of the coding sequences, respectively. The DNA was labeled with [α-32P]dATP (Amersham Buchler) by using a random prime labeling kit (MBI Fermentas). The rRNA bands in ethidium bromide-stained gels were used to determine fragment sizes (23). Hybridization signals were recorded and quantified by using a phosphorimager (BAS1000; Fuji). In order to quantify the data and correct errors in gel loading, all calculations were made on the basis of hybridization signals obtained after a radiolabeled 16S ribosomal DNA probe was applied to the same filters.
Primer extension experiments were performed by using a Super Script II kit (Invitrogen/Life Technologies). One microgram of total RNA isolated from cells in control cultures or 2 h after a salt shock with 684 mM NaCl and 2 pmol of primer PE_3321 (AGT CCA CAG ACA TCG CGT TC) were used for reverse transcription according to the manufacturer's instructions. The same primer with an IRD800 label was used in a sequencing reaction (Thermo Sequenase fluorescently labeled primer cycle sequencing kit with 7-deaza-dGTP; Amersham), and all products were separated and analyzed in a polyacrylamide gel by using an automated sequencer (LI-COR Bioscience) according to the manufacturer's instructions. As the template for the sequencing reaction, a plasmid was used which was obtained after cloning of a PCR fragment containing the complete coding region and 4,929 bp upstream of the ggpS gene into pGEM-T Vector System I (Promega). For the PCR, primers hyp_slr1670_5′ (CCT AGC ATT CCT TCT TCG) and ggpS3′ were used and chromosomal DNA of Synechocystis was used as the template.
The half-life of mRNA was analyzed by incubation of Synechocystis cells grown under control or salt shock (684 mM NaCl) conditions with rifampin (final concentration, 150 μg ml−1; Sigma) as a transcriptional inhibitor. At different time points samples were taken from the cultures and treated with rifampin for 5, 10, 15, 20, or 25 min under the same light and temperature conditions. Cells were harvested, and total RNA was isolated. Specific mRNA contents were estimated by Northern blot analysis. The signal intensities were plotted versus time, and the half-life was calculated. In the same manner the half-life of mRNAs was determined by using cells completely acclimated to 684 mM NaCl.
Protein techniques.
The amount of the GgpS protein was estimated performing immunoblot experiments with an GgpS-specific antibody raised in a rabbit (Eurogentec) against the purified protein obtained after overexpression in E. coli (17). Total soluble protein was extracted by sonication and separated by polyacrylamide gel electrophoresis as described by Hagemann et al. (7). Binding of the antibody was detected by using an enhanced chemiluminescence kit (Amersham). The signals were detected with X-ray film and were quantified by video densitometric evaluation by using the Bioprofil 1D software package (Vilbert Lourmat).
The half-life of the GgpS protein was analyzed by incubation of cells with lincomycin (final concentration, 200 μg ml−; Sigma) as a specific inhibitor of translation. Soluble proteins were isolated from cells grown under control conditions and from cells completely acclimated to 684 mM NaCl in the presence or absence of lincomycin. The specific GgpS protein content was estimated before and 3, 6, 12, 18, and 24 h after addition of lincomycin. The signal intensities were plotted versus time, and the half-life was calculated.
Physiological characterization.
Shock treatment experiments were performed with batch cultures after separate additions of NaCl, sucrose, and sorbitol to compare salt and nonionic pure osmotic stress. In acclimation experiments cells were used after precultivation for 5 days at the desired NaCl concentration with daily medium changes. The content of the low-molecular-mass carbohydrates was analyzed by high-performance liquid chromatography (7). Growth and cell density were monitored by determining the absorption at 750 nm (A750) of diluted cyanobacterial suspensions with a double-beam UV/VIS spectrophotometer (U2000; Hitachi). Photosystem II fluorescence with and without 10 μM 3-(3,4-dichlorophenyl)-1,1-dimethylurea was recorded by using a microplate fluorescence reader (Lambda Fluoro 320; Bio-Tek Instruments, MWG Biotech) after excitation at 440 nm and emission at 680 nm, respectively.
Below, means and standard deviations from three independent experiments are given or the results of one typical experiment are shown.
RESULTS
Expression of ggpS is induced after salt shock and in salt-acclimated cells.
In order to investigate the regulation of ggpS gene expression in detail, first mRNA, protein, and GG levels were measured in cells shocked with 684 mM NaCl (about 50% of the maximal tolerance level). While in control cells almost no ggpS mRNA was detected, ggpS mRNA appeared immediately after salt was added (Fig. 1A). A main transcript consisting of about 2,000 nucleotides was detected. After an approximately 25-fold increase in ggpS mRNA content during the first 2 h, a new steady-state level was reached around 6 h after the shock, which was still about five times higher than the level in control cells. The ggpS mRNA content was also analyzed after nonionic pure osmotic shocks were applied by using sucrose or sorbitol. In both cases an increase in the transcript level was observed. The kinetics of ggpS mRNA accumulation were comparable in experiments in which NaCl shock was used and in experiments in which nonionic osmotic shock was used (data not shown).
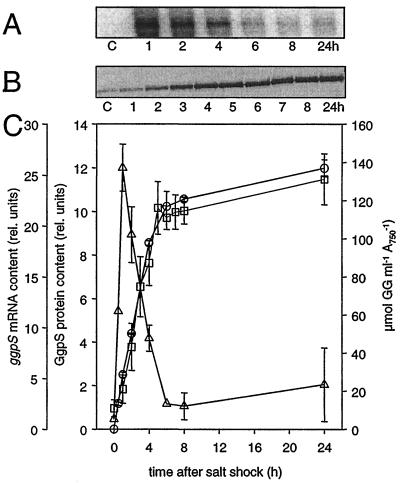
FIG. 1.
Cellular levels of ggpS mRNA, GgpS protein, and GG in salt-shocked Synechocystis cells. Changes in the cellular contents of ggpS mRNA, GgpS protein, and GG in Synechocystis cells shocked by 684 mM NaCl at time zero were determined. (A and B) Signals for the ggpS mRNA obtained by Northern blot analysis (A) and for the GgpS protein obtained by immunoblot analysis using a specific antibody (B). Lane C, control. (C) Densitometric estimation of the relative mRNA and protein steady-state levels and the absolute amount of GG. Symbols: ▵, ggpS mRNA; □, GgpS protein; ○, GG. rel. units, relative units.
In contrast to the ggpS transcript level, a basal level of GgpS protein was present in control cells, and the level increased linearly for at least 8 h after a salt shock with 684 mM NaCl (Fig. 1B). The different kinetics of ggpS mRNA and GgpS protein accumulation show clearly that the initial transient increase in mRNA content is not directly translated into the GgpS protein, which indicates that after salt shock transcription and translation are not completely linked. GG accumulation after addition of NaCl started immediately (Fig. 1C), because the synthesizing enzymes are preformed and are activated directly by a biochemical mechanism (5). However, a comparison of GgpS protein and GG contents in salt-shocked Synechocystis cells showed that there was a direct and strong correlation of the two parameters within the first 24 h after salt shock.
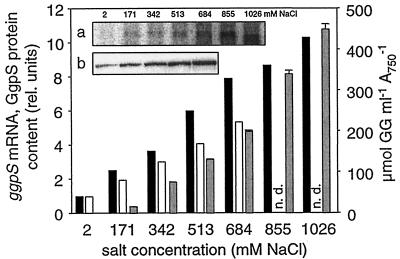
In further experiments, the amounts of ggpS mRNA, GgpS protein, and accumulated GG were compared in Synechocystis cells completely acclimated for 5 days to different salt concentrations. In contrast to the situation immediately after salt shock, in these acclimated cells a linear dependence of all three parameters (ggpS mRNA, GgpS protein, GG) on the external salt concentration was found, and in control cells, despite the detectable amounts of ggpS mRNA and protein, no GG was detectable (Fig. 2), showing the importance of the biochemical regulation.
FIG. 2.
Cellular levels of ggpS mRNA, GgpS protein, and GG in completely salt-acclimated Synechocystis cells. Changes in the ggpS mRNA (solid bars) and GgpS protein (open bars) steady-state contents and the amounts of accumulated GG (gray bars) in Synechocystis cells completely acclimated to different salt concentrations were determined. rel. units, relative units; n.d., not determined. (a) Signals for the ggpS mRNA obtained by Northern blot analysis; (b) signals for the GgpS protein obtained by immunoblot analysis using a specific antibody.
The increase in the ggpS transcript level is the result of de novo synthesis of ggpS mRNA.
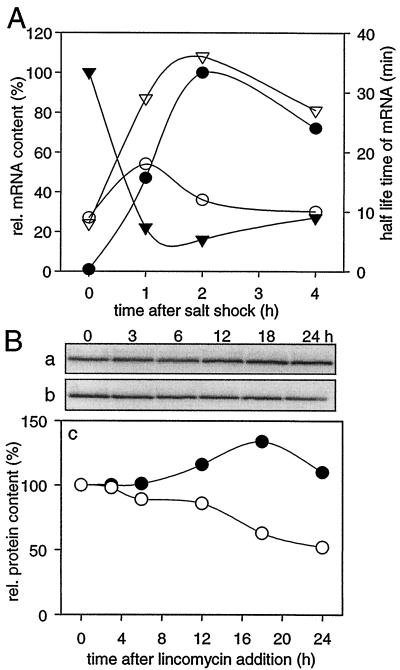
In order to exclude the possibility that the rise in ggpS mRNA content resulted from specific stabilization of this transcript, we analyzed the half-life of ggpS mRNA after a salt shock with 684 mM NaCl using rifampin-treated cells. As a control, the half-life of the psbC mRNA was estimated (Fig. 3A). In cells not treated with rifampin, the typical increase in the ggpS transcript level was observed after the salt shock, while a strong decrease in the psbC transcript level was found under these conditions. However, for both transcripts an increase in the half-life was measured; the half-life of the ggpS mRNA increased from 9 to 18 min, and the half-life of the psbC mRNA increased from 8 to 36 min. In spite of the greater stability of the psbC mRNA (whose half-life was two times longer than that of the ggpS transcript), the amount of psbC mRNA quickly decreased after the salt shock. Therefore, it can be concluded that the increase in the ggpS mRNA level resulted from de novo synthesis, while the decrease in the psbC mRNA content indicated an almost complete stop of transcription of this mRNA. In cells completely acclimated to 684 mM NaCl, the half-lives of the ggpS and psbC transcripts were found to be about 7 and 25 min, respectively (data not shown).
FIG. 3.
Influence of salt treatment on the half-lives of mRNA (A) and the GgpS protein (B). (A) Analysis of the levels (solid symbols) and half-lives in the presence of rifampin (open symbols) of the ggpS (circles) and psbC (triangles) mRNA after salt shock consisting of 684 mM NaCl. The mRNA content is expressed in relative units (the maximal steady-state contents were defined as 100%; the signal intensity of the ggpS maximum was about 60% of the signal intensity of the psbC maximum). (B) Estimation of the half-life of the GgpS protein in the presence of lincomycin (added at zero time) in cells grown under control conditions (•) and in cells completely acclimated to 684 mM NaCl (○). Changes in GgpS-specific signals determined by imunoblotting using a specific antibody are shown for lincomycin-treated control cells (a) and salt-acclimated cells (b). Densitometric evaluation of these signals revealed the relative protein content (c) (the level in cells before lincomycin addition was defined as 100%).
Additionally, the stability of the GgpS protein was analyzed in cells grown under control conditions or acclimated to 684 mM NaCl by measuring its half-life in the presence of lincomycin, a specific translational inhibitor. Under control conditions the protein was found to be fairly stable for at least 24 h after addition of lincomycin (Fig. 3B). However, in salt-acclimated cells the GgpS protein content decreased after the addition of lincomycin. In contrast to the stable GgpS under low-salt conditions, where the protein is biochemically inactive, a half-life of about 24 h was estimated for the active GgpS in salt-treated cells. Therefore, the stability of the protein is obviously related to its activity state, while the half-lives of ggpS mRNA are not significantly different in control and salt-acclimated cells.
Induction of ggpS transcription after salt shock is proportional to stress.
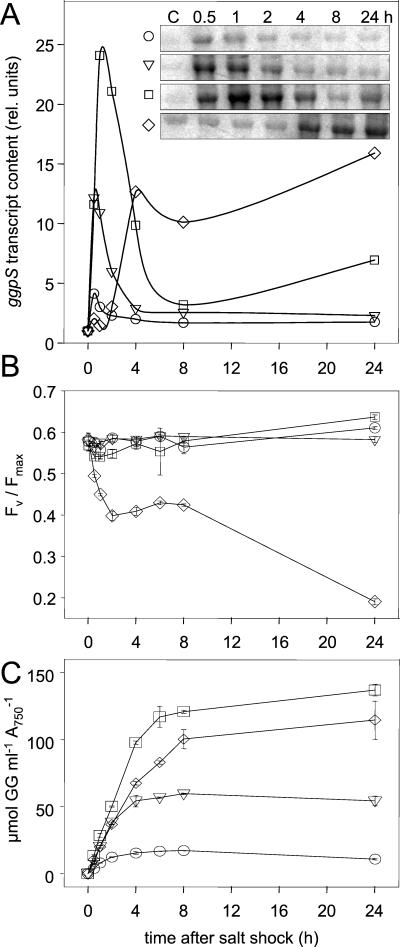
In order to analyze whether ggpS transcription depends on the strength of the salt stress, the ggpS mRNA contents were compared in Northern blot experiments after salt shocks consisting of 171, 342, 684, and 1,026 mM NaCl (1, 2, 4, and 6%) were applied (Fig. 4A). The shock treatments with NaCl concentrations up to 684 mM resulted in immediate increases in the ggpS mRNA and the same initial rate. The ratio of the observed induction maxima correlated with the salt concentration used for the shock treatment. However, after a 1,026 mM NaCl salt shock was applied, ggpS induction showed a lag phase and the induction maximum was not related to the strength of the shock in the first 24 h. The observed slower and lower induction of ggpS transcription with 1,026 mM NaCl could be explained by an overall strong disturbance of the metabolism of Synechocystis cells confronted by a salt concentration close to the maximal tolerance limit, 1.2 M NaCl (20). This interpretation is supported by the strong breakdown of photosynthesis as measured by photosystem II fluorescence in such cells. Compared to the other shock treatments, only cells treated with the highest salt concentration did not recover within 24 h from the initial drastic drop in the variable fluorescence/maximum fluorescence ratio (Fig. 4B). With external NaCl concentrations up to 684 mM, cells accumulated specific salt-related amounts of GG (Fig. 4C). Again, cells shocked with NaCl at a concentration of 1,026 mM exhibited a clearly reduced GG accumulation rate during the first few hours. In 24 h these cells were able to accumulate only about 30% of the amount of GG observed in completely acclimated cells (Fig. 2). In contrast, cells shocked with lower salt concentrations accumulated in 24 h nearly the same levels of GG that were seen in completely acclimated cells.
FIG. 4.
Comparison of the effects of salt shock strength on the accumulation of ggpS mRNA (A), photosynthetic efficiency (B), and GG accumulation (C). (A) Signals obtained by Northern blot analysis (insets) and results of the densitometric estimation to measure ggpS mRNA steady-state contents. rel. units, relative units. (B) Photosynthetic efficiency in cells shocked by different salt concentrations, as estimated by photosystem II fluorescence (excitation at 440 nm and emission at 680 nm). FV, variable fluorescence; Fmax, maximal fluorescence in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea ([FV − Fmax]/Fmax). (C) Accumulation of GG in cells shocked by different salt concentrations. Symbols: ○, 171 mM NaCl; ▿, 342 mM NaCl; □, 684 mM NaCl; ⋄, 1,026 mM NaCl.
Mapping of the ggpS promoter.
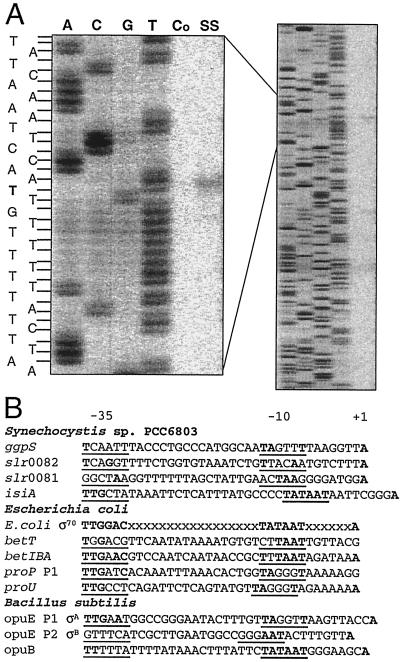
By using primer extension analysis the putative start point of transcription of the ggpS gene was determined. The 5′ end of the ggpS transcript was found to be 488 bp in front of the translational start codon (Fig. 5A). Like other cyanobacterial genes (25), the ggpS mRNA has a rather long nontranslated 5′ part. In agreement with the Northern blot analysis, the signal was detectable only with total RNA isolated from salt-shocked cells and not with RNA from control cells. Moreover, this transcriptional start point predicts an estimated size of the ggpS transcript of about 2,000 nucleotides (the coding sequence of the ggpS gene covers 1,500 bp), corresponding well to the size found in Northern blot experiments. In the upstream promoter region typical −35 and −10 boxes were proposed based on an alignment with the E. coli σ70 consensus promoter sequence. Furthermore, the promoter meets the criteria observed in more than 70% of the promoter sequences known for cyanobacteria (i. e., a conserved motif in the −10 region TANNNT and a purine as a transcriptional initiation site 7 ± 1 bp downstream from the −10 motif) (2). An alignment with the promoter sequences of genes with known salt-dependent expression in Synechocystis (25) or other organisms did not reveal well-conserved regions which are shared by all of the sequences and which might be related to the common salt induction of the sequences (Fig. 5B). A comparison of the ggpS promoter region with the proU and proP P1 promoters of E. coli (4, 18) showed the consensus motif TAGNNT in the −10 region, in which the first T is essential for salt-dependent induction of these genes in E. coli (18). Close similarity was also found with the σA-dependent P1 promoter consensus motif in the −10 region of TAGNTT region of the opuE gene from B. subtilis (24). However, a second promoter, dependent on an alternative sigma factor like the σB-dependent opuE P2 promoter in B. subtilis, could not be identified in front of the ggpS gene in Synechocystis.
FIG. 5.
Promoter mapping of the ggpS gene by primer extension analysis (A) and comparison of the ggpS promoter with promoter sequences of genes with known salt-dependent expression from Synechocystis, E. coli, and B. subtilis (B). (A) For the sequencing reaction (lanes A, C, G, and T) and cDNA synthesis (lanes Co and SS) the same primer was used. Total RNA was isolated from cells grown under control conditions (lane Co) or from cells shocked for 2 h by 684 mM NaCl (lane SS). (B) Promoter sequences are indicated by the names of the genes, as follows: ggpS (this study); slr0081, slr0082, and isiA, encoding proteins of unknown function and iron stress-induced protein A (25); E. coli σ70, the consensus sequence for σ70-dependent promoters; betT and betIBA, encoding choline uptake system BetT (15); proP P1 and proU, encoding proline uptake systems (18); opuE P1 σA and opuE P2 σB, encoding proline uptake systems (24); and opuB, encoding a choline uptake system (14).
σF is involved in regulation of ggpS expression.
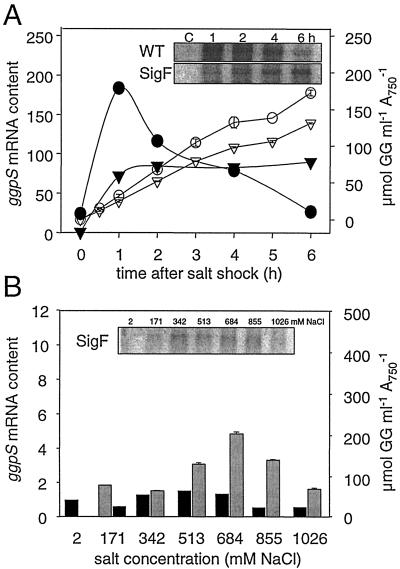
To identify regulatory components, we analyzed the ggpS transcription in a σF mutant known for diminished synthesis of many salt stress proteins (10). In cells of the σF mutant overall weaker ggpS induction lacking the transient maximum characteristic of wild-type cells was found (Fig. 6A). Despite the different ggpS expression, accumulation of GG was similar in mutant and wild-type cells after a salt shock consisting of 684 mM NaCl was applied. However, in completely salt-acclimated cells clear differences between the wild type and the σF mutant were found. In the σF mutant the ggpS transcript level showed no clear dependence on the external salt concentration, as observed in wild-type cells (compare Fig. 6B and 2). An almost constant weak transcription level was observed. The GG contents were found to be similar in wild-type and mutant cells only up to an external NaCl concentration of 684 mM. Cells of the σF mutant failed to acclimate to higher salt concentrations, since they were not able to accumulate the necessary amounts of GG at these high salinities like the wild-type cells. The diminished salt tolerance of σF cells was also documented by the decreasing growth rates and photosynthetic activities after longer exposure to NaCl concentrations higher than 684 mM (data not shown).
FIG. 6.
Comparison of salt-dependent ggpS expression and GG accumulation in cells of the Synechocystis wild type (WT) or a σF mutant (SigF) after a salt shock (A) or complete acclimation to different salt concentrations (B). (A) Changes in the ggpS mRNA steady-state contents after densitometric evaluation (solid symbols) and GG content (open symbols) after salt shock consisting of 684 mM NaCl for cells of the wild type (circles) or the σF mutant (triangles). Signals for ggpS mRNA obtained by Northern blot analysis are shown in the insets. (B) Changes in the ggpS mRNA steady-state content (solid bars) and the amount of accumulated GG (grey bars) in cells of the σF mutant completely acclimated to different salt concentrations. Signals for ggpS mRNA obtained by Northern blot analysis are shown in the inset (compare to Fig. 2).
DISCUSSION
The data presented here clearly show that in addition to the already known direct biochemical regulation of GgpS activity, salt-related alteration of the expression of the ggpS gene contributes to successful salt acclimation of Synechocystis. ggpS expression was altered at the level of transcript content and mRNA stability, as well as protein content and protein stability. In salt-shocked cells a transient increase in the ggpS mRNA level was observed, which was related to the shock strength. Hengge-Aronis et al. (9) observed a comparable situation for expression of the ots genes in E. coli. While the induction factor for ggpS after salt shock was 25-fold, expression of the transporters betT and proP of E. coli was stimulated about 5-fold, while expression of proU was stimulated about 100-fold (15, 18). In E. coli or B. subtilis salt-induced expression has usually been measured by using promoter fusions, in which the amount or the activity of the reporter protein was monitored and the mRNA content was not directly monitored as it was in our study. The initial increase in the ggpS mRNA was not translated directly into GgpS protein. Whether such short-term induction of genes occurs cannot be detected by promoter fusions. The inability of Synechocystis to convert the high ggpS mRNA level directly into GgpS protein indicates that the translation apparatus is more disturbed than the transcription apparatus after salt shock and the remaining translation rate allows only a linear increase in the protein content.
In salt-acclimated cells the ggpS transcript level depends on the salt concentration of the medium and is the basis for higher protein levels and finally higher GG contents. Expression of the opuE gene involved in proline uptake in B. subtilis is also linearly related to the external salt concentration (24), but no information is available for OpuE protein content and activity. Giæver et al. (3) demonstrated that expression of the ots genes in E. coli was linearly linked to the salt concentration of the medium up to an NaCl concentration of 400 mM.
The primary signal perceived and the sensors involved in regulating ggpS expression after salt shock are completely unknown. However, the signaling pathway obviously could involve the alternative σ factor σF. Induction of many salt stress proteins (10) was almost absent and ggpS induction was strongly reduced in the σF mutant. This did not affect the kinetics of GG accumulation, since the synthesizing enzymes were preformed and could be activated by a biochemical mechanism. However, in salt-acclimated cells of the σF mutant the ggpS expression level did not increase parallel to an increase in the external salt concentration, resulting in insufficient GG accumulation in cells confronted with NaCl concentrations higher than 684 mM, which explains the reported salt sensitivity of the σF mutant near the resistance limit (10). In E. coli expression of the otsBA operon depends on σS. Elimination of σS resulted in a complete failure of osmosis-stimulated ots transcription (9). Expression of the proP gene also depends on this σ factor (18). However, the σF of Synechocystis and the σS of E. coli are not closely related, since they do not cluster in one group. Rather, the σB of B. subtilis seems to be similar to σF of Synechocystis (27). In a σB mutant of B. subtilis opuE expression is not induced after shock treatments, while in salt-acclimated cells of this mutant opuE expression is still related to the salt concentration, with a σA-dependent promoter (24). Obviously, in Synechocystis both shock-induced ggpS induction and ggpS accumulation proportional to the salt content in acclimated cells are σF related, since in completely acclimated cells of the σF mutant ggpS transcription also remains at a constitutive low level. At present, the consensus sequence for SigF in Synechocystis is not known. Therefore, the possibility that the putative ggpS promoter is not directly recognized by SigF cannot be ruled out, and the reduced ggpS expression in the SigF mutant might be a rather indirect effect. All attempts to define further promoter elements responsible for the salt induction of ggpS failed when the putative ggpS promoter was compared with promoter sequences of genes with known salt-dependent expression from Synechocystis, E. coli, and B. subtilis.
In summary, as in other bacterial systems, the fine regulation of osmolyte biosynthesis in Synechocystis comprises two obvious regulatory levels, alteration of gene expression and biochemical modulation of enzyme activities to ensure GG synthesis proportional to stress. Our data on expression of the ggpS gene and salt regulation of GgpS activity, together with the current knowledge concerning cyanobacterial physiology, can be used to construct the following model of regulatory events in salt-stressed and salt-acclimated cyanobacteria (Table 1).
TABLE 1.
Key events involved in cooperation of transcriptional and biochemical control for salt-dependent regulation of GG synthesis in control, salt-shocked, and salt-acclimated cells of Synechocystis
| Cells | Transcriptional control
|
Biochemical control
|
GG synthesis | ||
|---|---|---|---|---|---|
| ggpS mRNA level | GgpS protein level | Ion concn | GgpS activity | ||
| Control | Weak, constitutive expression, σF independent | Constitutive level, very stable protein | Low | Completely inactive | Switched off |
| Salt shocked | Stress proportional, σF-dependent transient increase, transient mRNA stabilization | Linear protein accumulation, up to 10-fold increase | Transiently high | Completely active | Linear accumulation, maximal rate |
| Salt acclimated | Salt proportional, σF-dependent increase | Salt proportional, increased level | Salt proportional, weakly increased | Partially active | Salt proportional |
In cells grown under control conditions the ggpS gene is constitutively expressed at a low rate. Despite the very low level of transcription, a significant level of GgpS protein accumulates that is sufficient for an immediate high rate of GG synthesis after salt shock. The GgpS accumulation is promoted by the stability of the inactive protein. Nevertheless, control cells do not synthesize GG, since the internal ion concentrations are below the threshold for GgpS activation and the system is switched off at the biochemical level (6). The ggpS expression under control conditions is not dependent on σF, since almost identical GgpS contents were found in the σF mutant and the wild type, which allowed the same initial GG synthesis rates.
Application of salt shocks results in maximal activation of the GG-synthesizing system on the transcriptional level, as well as the biochemical regulatory level. Salt shocks consisting of more than 300 mM salt result in an immediate influx of inorganic ions and activation of GG synthesis (21). In vitro experiments showed that 200 mM NaCl was sufficient for complete activation of the GgpS pool on the biochemical level, which occurred even in the presence of chloramphenicol (6). From the GG accumulation in salt-shocked cells (data from this study and our previous studies) the initial GG synthesis rate was calculated. For cells shocked by NaCl concentrations greater than 300 mM, a rate of approximately 20 μmol of GG h−1 ml−1 A750−1 was calculated for the first 4 h (342 mM NaCl, 19.2 μmol of GG h−1 ml−1 A750−1; 684 mM NaCl, 21.3 μmol of GG h−1 ml−1 A750−1; 1,026 mM NaCl, 17.3 μmol of GG h−1 ml−1 A750−1). After treatment with lower salt concentrations (e.g., 171 mM NaCl) only a slight increase in the ion concentration in the cytoplasm is expected, which should not result in complete GgpS activation. Indeed, a lower GG synthesis rate (6.5 μmol of GG h−1 ml−1 A750−1) was calculated, reflecting incomplete activation. During the subsequent acclimation process, first Na+ is exchanged for K+, the ion concentration is reduced, and the ions are replaced by the accumulating GG (22). Therefore, the activity of the GgpS should decrease with time. However, parallel to the biochemical activation, dramatically increased ggpS transcription occurs in salt-stressed cells. The large amount of ggpS mRNA might be necessary to guarantee sufficiently high GgpS protein translation in spite of the general disturbed protein synthesis machinery. This results in an increased GgpS protein content, which compensates for losses in GgpS activity by protein turnover and decreased activity parallel to the decreased ion concentration. Thus, rapid GG accumulation is ensured.
Finally, in cells completely acclimated to enhanced NaCl concentrations there is a close correlation among ggpS mRNA, GgpS protein, and GG contents. Despite the approximately sevenfold-higher GgpS content in cells acclimated to 684 mM NaCl, an approximately threefold-lower GG synthesis rate was calculated for acclimated cells (6.6 μmol of GG h−1 ml−1 A750−1) compared to the rate in shocked cells. In completely salt-acclimated cells the concentration of ions is significantly lower than the concentration immediately after the shock event (22); therefore, the GG-forming system is only partially active. To allow a GG synthesis rate corresponding to the external salt concentration, the cells need to accumulate more protein.
The scenario of regulatory events involved in salt stress-related GG accumulation in Synechocystis described above indicates that biochemical regulation might be more important for the immediate reaction to the alarm situation after shock, which is characterized by strong disturbance of metabolism, including de novo protein synthesis. Transcriptional control seems to represent the basis for the fine-tuning of GG synthesis in salt-acclimated cells.
Acknowledgments
The technical assistance of B. Brzezinka and K. Sommerey is greatly appreciated.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft and a studentship to K.M. from the Hans Böckler Stiftung. Financial support provided by the Fond der Chemischen Industrie to M.H. is acknowledged.
Footnotes
This work is dedicated to Norbert Erdmann on his 65th birthday.
REFERENCES
- 1.Bremer, E., and R. Krämer. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria, p. 79-97. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 2.Curtis, S. E., and J. A. Martin. 1995. The transcription apparatus and the regulation of transcription initiation, p. 613-639. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Giæver, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strom. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gowrishankar, J. 1989. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J. Bacteriol. 171:1923-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagemann, M., and N. Erdmann. 1994. Activation and pathway of glucosylglycerol biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 140:1427-1431. [Google Scholar]
- 6.Hagemann, M., A. Schoor and N. Erdmann. 1996. NaCl acts as a direct modulator in the salt adaptive response: salt-dependent activation of glucosylglycerol synthesis in vivo and in vitro. J. Plant Physiol. 149:746-752. [Google Scholar]
- 7.Hagemann, M., A. Schoor, R. Jeanjean, E. Zuther and F. Joset. 1997. The stpA gene from Synechocystis sp. strain PCC 6803 encodes the glucosylglycerol-phosphate phosphatase involved in cyanobacterial osmotic response to salt shock. J. Bacteriol. 179:1727-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecker, M., W. Schumann, and U. Völker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 9.Hengge-Aronis, R., W. Klein, R. Lange, M. Rimmele, and W. Boos. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173:7918-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huckauf, J., C. Nomura, K. Forchhammer, and, M. Hagemann. 2000. Stress responses of Synechocystis sp. strain PCC 6803 mutants impaired in genes encoding putative alternative sigma factors. Microbiology. 146:2877-2889. [DOI] [PubMed] [Google Scholar]
- 11.Kaasen, I., P. Falkenberg, O. B. Styrvold, and A. R. Strom. 1992. Molecular cloning and physical mapping of the otsBA genes, which encode the osmoregulatory trehalose pathway of Escherichia coli: evidence that transcription is activated by katF (AppR). J. Bacteriol. 174:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaasen, I., J. McDougall, and A. R. Str/om. 1994. Analysis of the otsBA operon for osmoregulatory trehalose synthesis in Escherichia coli and homology of the OtsA and OtsB proteins to the yeast trehalose-6-phosphate synthase/phosphatase complex. Gene 145:9-15. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Nruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 14.Kappes, R. M., B. Kempf, S. Kneip, J. Boch, J. Gade, J. Meier-Wagner, and E. Bremer. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for the synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203-216. [DOI] [PubMed] [Google Scholar]
- 15.Lamark, T., T. P. Rokenes, J. McDougall, and A. R. Strøm. 1996. The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J. Bacteriol. 178:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 17.Marin, K., E. Zuther, T. Kerstan, A. Kunert, and M. Hagemann. 1998. The ggpS gene from Synechocystis sp. strain PCC 6803 encoding glucosyl-glycerol-phosphate synthase is involved in osmolyte synthesis. J. Bacteriol. 180:4843-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellies, J., A. Wise, and M. Villarejo. 1995. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J. Bacteriol. 177:144-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikkat, S., M. Hagemann, and A. Schoor. 1996. Active transport of glucosylglycerol is involved in salt adaptation of the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 142:1725-1732. [DOI] [PubMed] [Google Scholar]
- 20.Reed, R. H., and W. D. P. Stewart. 1985. Osmotic adjustment and organic solute accumulation in unicellular cyanobacteria from freshwater and marine habitats. Mar. Biol. 88:1-9. [Google Scholar]
- 21.Reed, R. H., D. L. Richardson, and W. D. P. Stewart. 1985. Na+ uptake and extrusion in the cyanobacterium Synechocystis PCC 6714 in response to hypersaline treatment. Evidence for transient changes in plasmalemma Na+ permeability. Biochim. Biophys. Acta 814:347-355. [Google Scholar]
- 22.Reed, R. H., S. R. C. Warr, D. L. Richardson, D. J. Moore, and W. D. P. Stewart. 1985. Multiphasic osmotic adjustment in a euryhaline cyanobacterium. FEMS Microbiol. Lett. 28:225-229. [Google Scholar]
- 23.Sakamoto, T., and D. A. Bryant. 1997. Temperature-regulated mRNA accumulation and stabilization for fatty acid desaturase genes in the cyanobacterium Synechococcus sp. strain PCC 7002. Mol. Microbiol. 23:1281-1292. [DOI] [PubMed] [Google Scholar]
- 24.Spiegelhalter, F., and E. Bremer. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the sigma A- and sigma B-dependent stress-responsive promoters. Mol. Microbiol. 29:285-296. [DOI] [PubMed] [Google Scholar]
- 25.Vinnemeier, J., and M. Hagemann. 1999. Identification of salt-regulated genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 by subtractive RNA hybridization. Arch. Microbiol. 172:377-386. [DOI] [PubMed] [Google Scholar]
- 26.Whatmore, A. M., J. A. Chudek, and R. H. Reed. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527-2535. [DOI] [PubMed] [Google Scholar]
- 27.Wösten, M. M. S. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]