Abstract
We screened actinomycete strains for dinitrogen (N2)-producing activity and discovered that Streptomyces antibioticus B-546 evolves N2 and some nitrous oxide (N2O) from nitrate (NO3−). Most of the N2 that evolved from the heavy isotope ([15N]NO3−) was 15N14N, indicating that this nitrogen species consists of two atoms, one arising from NO3− and the other from different sources. This phenomenon is similar to codenitrification in fungi. The strain also evolved less, but significant, amounts of 15N15N from [15N]NO3− in addition to 15N15NO with concomitant cell growth. Prior to the production of N2 and N2O, NO3− was rapidly reduced to nitrite (NO2−) accompanied by distinct cell growth, showing that the actinomycete strain is a facultative anaerobe that depends on denitrification and nitrate respiration for anoxic growth. The cell-free activities of denitrifying enzymes could be reconstituted, supporting the notion that the 15N15N and 15N15NO species are produced by denitrification from NO3− via NO2−. We therefore demonstrated a unique system in an actinomycete that produces gaseous nitrogen (N2 and N2O) through both denitrification and codenitrification. The predominance of codenitrification over denitrification along with oxygen tolerance is the key feature of nitrate metabolism in this actinomycete.
The biological process of dinitrogen (N2) gas formation from fixed nitrogen compounds such as nitrate (NO3−) plays an important role in maintaining homeostasis of the global environment. Bacterial denitrification was long considered the sole biological reaction responsible for this condition, and its systems have been characterized in detail (3, 8, 20). Denitrification physiologically functions as anaerobic respiration in which NO3− is used as the terminal electron acceptor when oxygen (O2) is unavailable. Known bacterial denitrifying systems consist of four steps that successively reduce NO3− to N2 and involve nitrite (NO2−), nitric oxide (NO), and nitrous oxide (N2O) as intermediates. Production of the enzymes catalyzing each step is induced by nitrogen oxides and suppressed by O2 (3, 20). During the past decade, denitrifiers have been discovered in a variety of taxa including filamentous fungi (9, 10), yeasts (15), and actinomycetes (1, 11). All of these novel denitrifiers produce N2O as the major denitrification product. Both nitrogen atoms in the N2O product are derived from nitrate (or nitrite) (1, 9-11, 15). The phenomenon is therefore defined as denitrification, since the process should include the formation of an N—N bond (20). This system in several fungi is localized at respiring mitochondria, where it acts as anaerobic respiration, as it does in bacterial systems (5, 13, 16). Another unique feature of the fungal system is the involvement of cytochrome P450 (P450nor) as NO reductase (Nor) (6).
Biological processes other than bacterial denitrification evolve N2 from fixed nitrogen compounds, but their molecular mechanisms and physiological significance remain to be elucidated. We identified simultaneous fungal codenitrification and denitrification, in which a hybrid N2 species is formed by combining two nitrogen atoms derived from NO2− and from other nitrogen compounds (10, 14). The denitrifying fungi Fusarium solani and Cylindrocarpon tonkinense evolve hybrid N2 species (10). The fact that the denitrifying fungus Fusarium oxysporum evolves N2O instead of N2 by codenitrification only when a nitrogen compound in addition to NO2− such as azide, salicylhydroxamic acid, or ammonium (NH4+) is available (14) suggests that the mechanisms of this process differ among fungal species. Anammox is a third N2-generating metabolic process that has been identified in the strictly anaerobic chemolithotrophic Planctomycetales (12), in which NH4+ is combined with NO2− to form N2.
The actinomycetes form a unique taxon among gram-positive bacteria. Although actinomycetes naturally proliferate in soil and in aqueous environments, little is known about how they accomplish denitrification. Denitrifiers also occur among actinomycetes (1, 11), and the system of Streptomyces thioluteus has been characterized previously (11). All of the actinomycete denitrifiers found in these studies evolve N2O from NO3− or NO2−, and thus, no known actinomycete strains contain a complete denitrifying system that can thoroughly reduce NO3− to N2.
The present study continues screening for denitrifying actinomycetes (11) by using a highly sensitive N2 detection system equipped with an isotope mass spectrometer and NO3− labeled with a stable isotope ([15N]NO3−). The results showed that Streptomyces antibioticus B-546 has N2-producing activity and that most of the N2 molecules are formed via intracellular codenitrification that is induced simultaneously with denitrification.
MATERIALS AND METHODS
Strains and media.
The strains used in this study originated from the JCM (Japan Collection of Microorganisms, RIKEN) or IFO (Institute for Fermentation, Osaka, Japan) type culture collections, except for strains A, B, C, and N. We isolated the latter four strains from soil, and S. antibioticus B-546 originated from a different culture collection in the Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo. All cells were seeded, precultured, or cultured in glycerol-peptone (GP) medium (3% glycerol, 0.2% peptone, 10 mM KH2PO4, 0.02% MgSO4·7H2O, and inorganic salts) (11) unless otherwise stated.
Batch cultures in flasks.
Actinomycetes were batch cultured in flasks as follows. Seed cultures (10 ml) in 50-ml tubes were inoculated into 300 ml of GP medium in 500-ml Erlenmeyer flasks and rotary shaken at 30°C for 4 days at 120 rpm (preculture). Finally, portions of the preculture (100 ml) were transferred to 500-ml flasks containing 100 ml of GP medium supplemented with 10 mM NaNO3 and incubated as described above (for preculture). Denitrifying actinomycetes were screened under initially aerobic conditions that were attained by sealing the flask after inoculation with a rubber stopper without replacing headspace air. NaNO3 was labeled with 10% heavy isotope nitrogen (15N). Time-dependent changes in nitrogen oxides were investigated under two aerating conditions. Initially aerobic cultures were incubated in the same manner as for screening. Anaerobic cultures were incubated in the same manner, but the headspace air was replaced with argon gas before the flask was sealed. Both cultures used [15N]NaNO3 instead of [14N]NaNO3. When necessary, acetylene gas was added to a ratio of 10% of the headspace volume.
Quantification of nitrogenous oxides, dinitrogen, and cell weight.
We determined NO3− and NO2− with an ion-pair, high-performance liquid chromatograph equipped with a TSK gel IC-Anion PW column (Tosoh, Tokyo, Japan) as described previously (9). Oxygen, N2O, and N2 concentrations in the gas phase were determined by gas chromatography (GC) as described previously (9). 15N-labeled N2 gas was analyzed by isotope mass spectrometry (MS) with a Finnigan DELTA Plus isotope mass spectrometer as described previously (15). Collected cells were dried at 80°C for 2 h and then weighed (cell dry weight).
Preparation of crude extract and subcellular fractionation.
Denitrifying cells of S. antibioticus B-546 were harvested by centrifugation, resuspended in buffer (100 mM potassium phosphate [pH 7.2], 0.1 mM EDTA, 0.1 mM dithiothreitol, 10% glycerol, 0.25 μM N-tosyl-l-phenylalanyl chloromethyl ketone, and 0.25 μM phenylmethanesulfonyl fluoride), and sonicated (Sonifier 250; Branson) for 60 min with occasional cooling. The sonicate was separated by centrifugation at 10,000 × g for 20 min to obtain the supernatant (crude extract), which was further sedimented by centrifugation at 100,000 × g for 60 min. The resulting supernatant (soluble fraction) and pellet (membranes) were analyzed as follows.
Enzyme assay.
Nitrate reductase (Nar) was assayed as described previously (5) with methyl viologen-dithionite as the electron donor. Nitrite reductase (Nir) was assayed with NADH-phenazine methosulfate as the electron donor by determining the amount of NO produced by the P450nor-trap method (5). Nor activity was determined by measuring NADH-dependent N2O formation by GC as described previously (6). Nitrous oxide reductase (N2Or) was assayed as described previously (4) with the following modification. The reaction mixture (0.85 ml of 10 mM potassium phosphate [pH 7.2] and 0.05 ml of 10 mM methyl viologen) in 1-ml Zumberg-type cuvettes was flushed with argon and an appropriate volume of 1 mM sodium dithionite to give an A600 of 1.0 ± 0.2. An aliquot of the enzyme was injected, and the A600 was monitored for 1 min to measure the background oxidation rate before starting the reaction by injecting 0.08 ml of N2O gas. The reaction was measured by monitoring the N2O-dependent oxidation of reduced methyl viologen at 600 nm. The protein concentration was determined with a protein assay reagent (Bio-Rad Laboratories, Inc., Richmond, Calif.).
RESULTS
Screening actinomycetes that produce N2.
We examined denitrification in the following strains: Saccharopolyspora gregorii IFO15045T, Microbispora grisealba IFO14840T, Nocardia asteroides IFO3384, Nocardia brasiliensis IFO14402T, Streptomyces prunicolor IFO13075T, Streptomyces roseolus IFO12816T, Streptomyces acrimycini IFO12736T, Streptomyces badius IFO12745T, Nocardia carnea IFO14403T, Streptomyces daghestanicus IFO12762T, Streptomyces umbrinus IFO13091T, Streptomyces variabilis IFO12825T, Micromonospora citrae IFO14025T, Streptomyces coelicolor JCM4375, Streptomyces lividans JCM4783, Nocardia salmonicida JCM4826, Streptomyces rubescens IAM0074, S. antibioticus B-546, strain B, strain C, strain E, and strain N. Each strain was initially cultured under aerobic conditions (see Materials and Methods) in medium containing NO3−, 10% of which was labeled with a heavy isotope ([15N]NO3−). The headspace gas of the culture flask was analyzed after the incubation by GC-MS for isotopically labeled N2 gas. Among the strains tested, only S. antibioticus B-546 produced a significant amount (9.5 × 101 μmol) of N2 species (14N15N) after a 4-day incubation, indicating N2 production from NO3−. We then screened four other S. antibioticus strains (JCM3117, JCM4690, IFO12652, and IFO13271) and found that strain JCM3117 evolved up to 1.9 × 101 μmol of N2 species (14N15N) after a 4-day incubation. These results showed that strains of this actinomycete species evolve more N2 than do the other species tested.
Codenitrification and denitrification by S. antibioticus.
We investigated N2 production by S. antibioticus B-546 in more detail. The strain was initially incubated under the aerobic conditions described above but with the stable isotope species of NO3−, in which 99% of N atoms are labeled with 15N. We changed the content of 15N with the expectation that the 14N15N-N2 species observed in the previous experiment would be replaced by 15N15N-N2 species. However, the major N2 species produced remained 14N15N (Fig. 1) with a very small but distinct amount of 15N15N. Furthermore, the N2O that was also formed was in fact a 15N15NO species (data not shown). Neither 15N15N nor 15N15NO was observed in the control experiment without cells (data not shown), indicating that 15N15N and 15N15NO were formed because of denitrification. Since almost all of the added NO3− was of the heavy isotope species, the 14N15N-N2 species should be generated by a combination of two nitrogen atoms, one derived from NO3− and the other from other nitrogen sources (codenitrification). The hybrid N2 species was not detected in the control experiment without the cells. The medium contained 10 mM phosphate buffer (pH 7.2), and the final pH of the medium after 5 days of incubation was above 6.0. These results rule out the possibility that the hybrid N2 species was formed by chemical means (2) but indicate that N2 arises from a physiological reaction like that in fungal denitrifiers (10, 14).
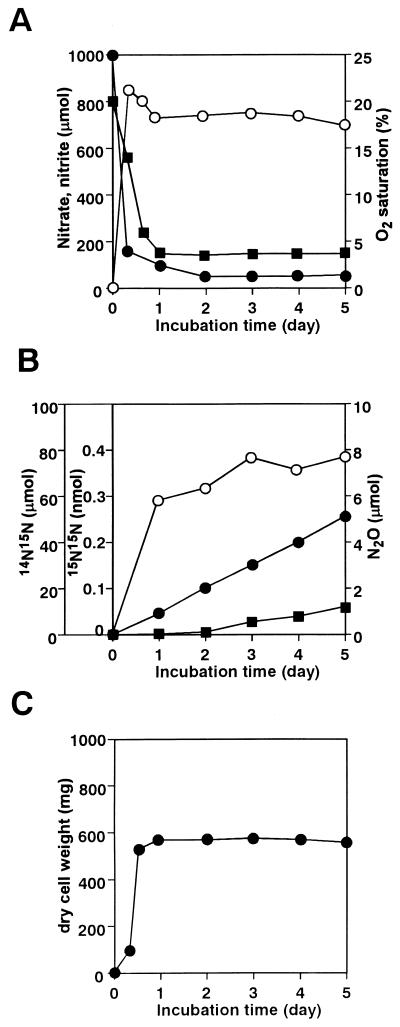
FIG. 1.
N2 production by S. antibioticus B-546 under aerobic conditions. Flask headspace was filled with air to initially maintain aerobic conditions. (A) Amounts of NO3− (solid circles) and NO2− (open circles) in medium and O2 in the gas phase (solid squares) were determined at indicated times. (B) Amounts of 14N15N (solid circles), 15N15N (open circles), and N2O (solid squares) in the gas phase were determined at indicated times. (C) Cell growth. Results are representative values from three experiments.
Time-dependent changes in each component during the initial aerobic culture show that the culture was divided into two phases (Fig. 1). In the first phase, NO3− and O2 were rapidly consumed with the concomitant production of NO2− (Fig. 1A) and 15N15N-N2 species (Fig. 1B), as well as cell growth (Fig. 1C). Most (70%) nitrogen atoms of the consumed NO3− seemed to be recovered into NO2−. However, these activities almost stopped or significantly declined within 24 h. During the second phase after the 1-day incubation, N2O began to be evolved whereas the emission of 15N15N-N2 species declined. By contrast, the hybrid 14N15N-N2 species was emitted during the first and the second phases throughout the incubation. Some portion of the nitrogen atoms of consumed NO3− were recovered into 14N15N-N2 (6%), 15N15N-N2 (0.0001%), and N2O (0.3%). In contrast, NO was not detected throughout the incubation. The consumption of O2 stopped in the second phase, although O2 still accounted for 2% of the headspace gas. These results showed that the strain can reduce 15NO3− to produce 15N14N, 15N15N, N2O, and NO2− species under aerobic conditions, which is in sharp contrast to other bacterial denitrifiers.
Nitrate metabolism by S. antibioticus under anoxic conditions.
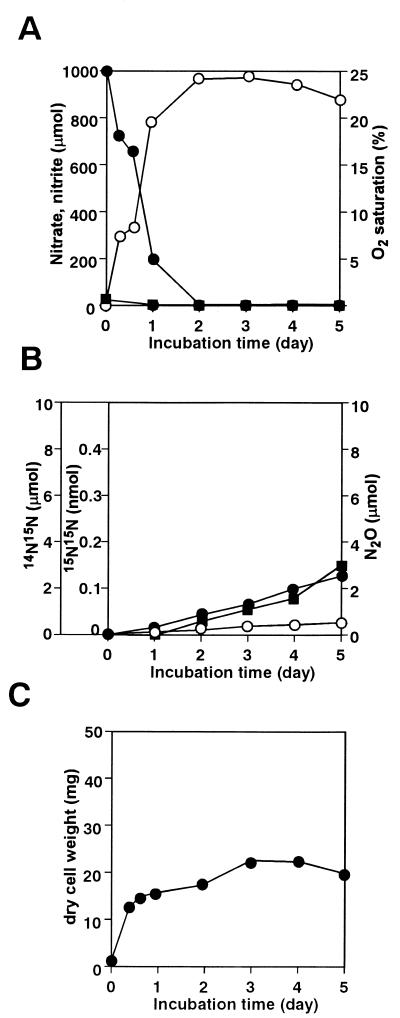
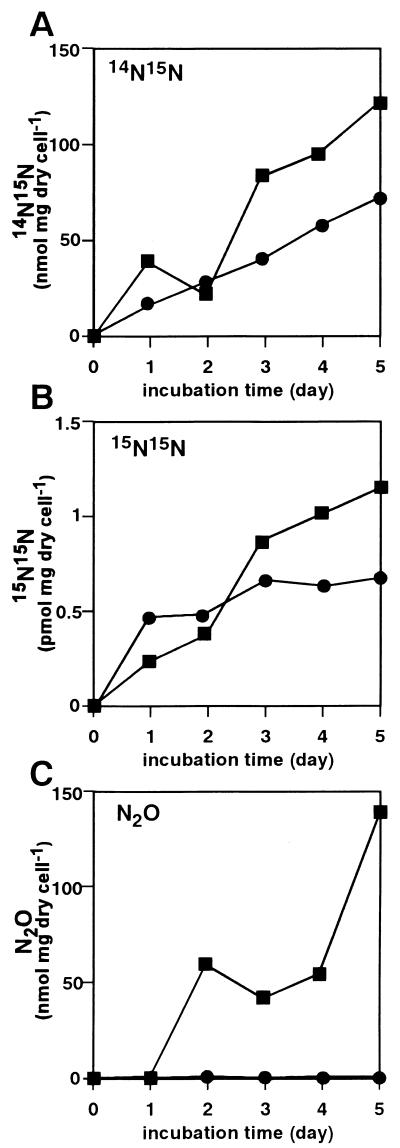
The aerating (initial aerobic) conditions above (Fig. 1) mimicked those required for fungal denitrification (18). Next we examined similar cultures under anoxic conditions. Figure 2 shows that NO3− was first converted to NO2− as in the aerobic culture (Fig. 1), that the conversion was almost stoichiometric, and that 15N14N, 15N15N, and N2O gas species evolved. Cell growth was distinct even under anoxic conditions (Fig. 2C) although cell yield was much lower than that under aerobic conditions (Fig. 1C). Figure 3 shows the time-dependent accumulation of each gas species per milligram of cells (dry weight) during aerobic (Fig. 1) and anoxic (Fig. 2) cultures. More of all of the gas species accumulated (per milligram of cells) under anoxic conditions. In particular, the amounts of N2O evolved were increased by 500- to 1,000-fold under anoxic, compared with aerobic, conditions.
FIG. 2.
N2 production by S. antibioticus B-546 under anaerobic conditions. Flask headspace was filled with argon to maintain anaerobic conditions. Symbols are as described for Fig. 1. Results are representative values from three experiments.
FIG. 3.
Denitrification by S. antibioticus B-546. Time-dependent accumulation of each gas species per milligram of dry cells was calculated and plotted. Circles, aerobic culture (Fig. 1); rectangles, anoxic culture (Fig. 2).
Denitrifying enzyme activities.
The formation of 15N15N, 15N15NO, and NO2− from 15NO3− suggests that S. antibioticus B-546 contains a conventional denitrifying system common to bacteria. We therefore examined the reconstitution of cell-free activities of enzymes that should be involved in the system. Table 1 shows that nitrate reductase (Nar), nitrite reductase (Nir), and nitric oxide reductase (Nor) activities were detected in crude extract or subcellular fractions prepared from the denitrifying cells (on day 3 in Fig. 1). Significant Nar, Nir, and Nor activities were recovered in the membranes as well as in the soluble fraction. In contrast, we could not reconstitute nitrous oxide reductase (N2Or) activity.
TABLE 1.
Denitrifying enzyme activities of S. antibioticus B-546a
| Denitrifying enzyme | Total activity (nmol min−1)
|
||
|---|---|---|---|
| Crude extract | Soluble fraction | Membranes | |
| NO−3 reductase | 12.5 | 6.7 | 2.7 |
| NO−2 reductase | 396 | 260 | 49 |
| NO reductase | 56 | 13 | 6.0 |
S. antibioticus B-546 was initially cultured under aerobic conditions for 3 days, homogenized, and then fractionated as described in Materials and Methods.
Effect of acetylene on denitrification of S. antibioticus.
Since cell-free N2Or activity could not be reconstituted, we further examined this step with respect to the in vivo activity of the actinomycete strain. N2Or activities both in vivo and in vitro are generally inhibited by acetylene; thus, in vivo incubation of the denitrifying system usually results in N2O accumulation (17). When S. antibioticus B-546 was initially cultured under aerobic conditions, acetylene did not affect the in vivo production of 14N15N, 15N15N, and N2O from 15NO3− (data not shown), showing that the production of N2 by strain B-546 is insensitive to acetylene.
Dissimilar nitrate reduction by S. antibioticus.
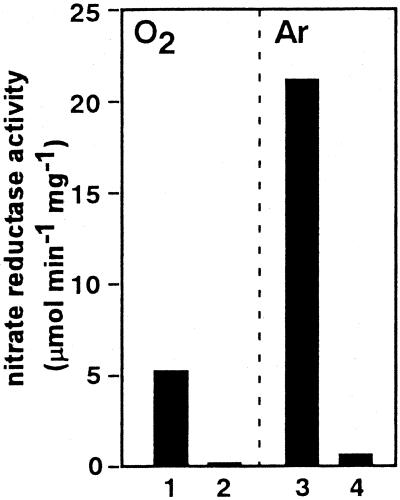
The above results indicate that nitrate metabolism by S. antibioticus supports anoxic cell growth (Fig. 2C). This notion was further supported by the following findings. The cell-free specific activity of Nar was higher in the anoxic cells than in those grown under aerobic conditions (Fig. 4), which is consistent with the complete conversion of NO3− to NO2− (Fig. 2A). The addition of tungstate, an inhibitor of molybdenum enzymes, to the culture medium considerably inhibited the cell-free enzyme activity in both the aerobic and anoxic cells, indicating that Nar activity is dependent on a molybdenum enzyme (Fig. 4). These properties are similar to those of bacterial Nar, which is involved in nitrate respiration (20).
FIG. 4.
Nitrate reductase activity of S. antibioticus B-546 under anaerobic conditions. Flask headspace was filled with air (bars 1 and 2) or argon (bars 3 and 4) and incubated for 24 h (the first phase) in medium with (bars 2 and 4) or without (bars 1 and 3) 10 mM sodium tungstate. Results are typical of over three experiments.
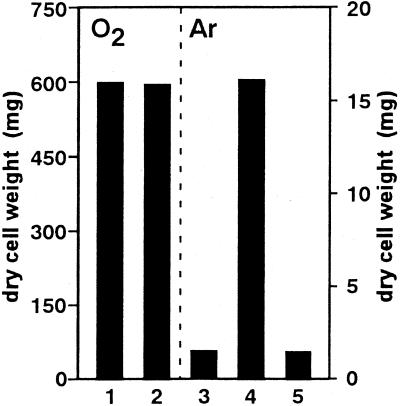
NO3− reduction during the first phase accompanied cell growth even in the absence of O2 (Fig. 2). When NO3− was omitted from medium that still contained another nitrogen source (peptone), little growth was attained by the anoxic culture in sharp contrast to the normal growth attained under (initial) aerobic conditions in the absence of NO3− (Fig. 5). The addition of tungstate to the culture inhibited the NO3−-dependent cell growth under anoxic conditions (Fig. 5, bar 5), again contrasted with the absence of a tungstate effect on the aerobic growth (data not shown). These results unequivocally demonstrated that anoxic cell growth depended on the Nar-dependent, dissimilatory reduction of NO3−, whereas the high cell yield by the aerobic culture was mostly supported by O2 respiration.
FIG. 5.
Nitrate-dependent cell growth of S. antibioticus B-546 under anaerobic conditions. Flask headspace was filled with air (bars 1 and 2) or argon (bars 3 to 5) and incubated for 24 h (the first phase in Fig. 1 and 2). GP medium was supplemented with (bars 2, 4, and 5) or without (bars 1 and 3) 10 mM NaNO3. Sodium tungstate (10 mM) was added (bar 5). Typical results of more than three experiments are shown.
The anoxic cell growth (Fig. 2C) consisted of two phases, consistent with the initial reduction of NO3− to NO2− and the following initiation of N2O evolution (Fig. 2A and B). These results support the notion that this actinomycete strain prefers, as the anoxic energy-yielding process, the reduction step from NO3− to NO2− by Nar to the subsequent reduction steps. When NO3− became unavailable due to consumption, the strain began to utilize the accumulated NO2− for further reduction, which resulted in denitrification. The strain was selective about the denitrification product, preferring N2O to N2 (detected above as 15N15N). The Nir and Nor activity detected in the cell extracts (Table 1) should correspond to the further reduction of NO2− to N2O.
DISCUSSION
We are the first to demonstrate that denitrifiers produce N2 as the denitrification product in actinomycetes. We also showed that the denitrifying system of S. antibioticus B-546 is distinct from other known systems of bacteria in that most N2 production depends on codenitrification, where N2 is formed by combining two nitrogen atoms, one from NO3− and one from other nitrogen sources. The hybrid N2 species appears to be formed from NO2− (or NO) as it is in fungal codenitrification (10) but not directly from NO3−, since the formation continued long after NO3− was consumed (Fig. 1 and 2). Another N2 species (15N15N from 15NO3−) should be formed by reducing N2O since both nitrogen atoms of N2O were derived from NO3− (15N15NO when 15NO3− was applied), showing that both products (N2O and homo-N2 species) are formed by normal denitrification. Thus, N2 is formed in S. antibioticus B-546 via dual pathways of nitrate metabolism, namely, denitrification and codenitrification. Less hybrid N2 species than denitrification product (N2O) is produced by known fungal codenitrification processes (10). Therefore, the unique feature of nitrate metabolism by the actinomycete strain is that codenitrification predominates over, or at least is comparable to, denitrification.
These results are also the first to show that an actinomycete strain can attain growth even when the O2 supply is completely lost (Fig. 2) and that anoxic growth depends on denitrification (nitrate respiration) (Fig. 2 and 4). Another unique feature of this type of nitrate metabolism is its aerobic or O2-resistant nature, which is in sharp contrast to the nitrate metabolic processes of other bacteria that are suppressed by atmospheric O2 (3). NO3− was converted to NO2− and all the gas species were formed under both aerobic and anoxic conditions (Fig. 1 and 2), although the formation rate for each metabolite differed between the conditions. The formation rate per milligram of cell (Fig. 3) for the denitrification products (N2O and 15N15N) was much higher under anoxic than under aerobic conditions, supporting the importance of denitrification for anoxic growth. By contrast, the rates for conversion of NO3− to NO2− or the formation of the hybrid N2 gas species (14N15N) were comparable between the conditions. The aerobic nitrate metabolism does not apparently contribute to cell growth (Fig. 1C and 5). However, this does not rule out the possibility that nitrate metabolism is an energy-yielding process, since NO3− was converted to NO2− (possibly supported by Nar) simultaneously with O2 uptake, and O2 respiration should be much more effective than nitrate respiration for energy production. This means that, under conditions such as the first stage shown in Fig. 1, suppressing nitrate respiration would be energetically even more favorable because nitrate should compete for electrons with O2. In the second stage of the aerobic culture, the cell mass remained almost constant (Fig. 1C). The number of cells grown under aerobic conditions should have been too large for the weak denitrifying activity to support more growth (compare Fig. 1C and 2C).
Codenitrification was expressed in S. antibioticus irrespective of the extent of aeration. Thus, codenitrification appears to be associated with the formation of NO2− from NO3−. However, the present results cannot confirm whether codenitrification contributes to cell growth. Another nitrogen source for codenitrification could be the peptone that was added to the medium as well as nitrate. The O2-resistant nature of Nar along with codenitrification in the actinomycete strain has provided new insight into the dissimilar metabolism of nitrate by microorganisms, whereas their physiological significance or mechanism(s) remains to be elucidated.
We recently showed that many soil fungi are not obligatory aerobes but facultative anaerobes and that facultative anaerobiosis is supported not only by denitrification but also by ammonia fermentation, which is a novel anoxic metabolism of nitrate by fungi (19). Most known actinomycetes reside in soils, and like fungi, they have been considered obligatory aerobes. Therefore, the present study has revealed the possibility that many soil actinomycetes are in fact facultative anaerobes.
Our characterization indicates that the denitrifying system of the actinomycetes is more similar to that of fungal systems than bacterial systems. Codenitrification (10, 14), aerobic qualities (9, 18), formation of N2O as the main product of denitrification (9, 10, 15), and rapid excretion of NO2− into the medium after conversion from NO3− (18) are all features of fungal denitrification. On the other hand, the P450nor involved in the fungal systems (Nor) seems not to occur among actinomycetes, although P450nor is classified into the same phylogenetic branch as other actinomycete P450s within the P450 superfamily (7). The phylogenetic relationship among the denitrifying systems of fungi, actinomycetes, and other bacteria requires further clarification. The contributions of the novel denitrifying systems of actinomycetes and of fungal systems to the nitrogen cycle in nature remain to be assessed.
Acknowledgments
This study was supported by PROBRAIN (Program for Promotion of Basic Research Activities for Innovative Biosciences), RITE (Research Institute of Innovative Technology for the Earth), and the Structural Biology Sakabe Project at KEK, Tsukuba, Japan.
REFERENCES
- 1.Albrecht, A., J. G. G. Ottow, and G. Benckiser. 1997. Incomplete denitrification (NO and N2O) from nitrate by Streptomyces violaceoruber and S. nitrosporeus revealed by acetylene inhibition and 15N gas chromatography-quadruple mass spectrometry analyses. Naturwissenschaften 84:145-147. [Google Scholar]
- 2.Chalk, P. M., and C. J. Smith. 1983. Chemodenitrification, p. 65-89. In J. R. Freney and J. R. Simpson (ed.), Gaseous loss of nitrogen from plant-soil systems. Nijhoff/Junk, The Hague, The Netherlands.
- 3.Ferguson, S. J. 1994. Denitrification and its control. Antonie Leeuwenhoek 66:89-110. [DOI] [PubMed] [Google Scholar]
- 4.Ferretti, S., J. G. Grossman, S. S. Hasnain, R. R. Eady, and B. E. Smith. 1999. Biochemical characterization and solution structure of nitrous oxide reductase from Alcaligenes xylosoxidans (NCIMB 11015). Eur. J. Biochem. 259:651-659. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi, M., Y. Matsuo, T. Tanimoto, S. Suzuki, F. Maruo, and H. Shoun. 1996. Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 271:16263-16267. [DOI] [PubMed] [Google Scholar]
- 6.Nakahara, K., T. Tanimoto, K. Hatano, K. Usuda, and H. Shoun. 1993. Cytochrome P-450 55A1 (P-450dNIR) acts as nitric oxide reductase employing NADH as the direct electron donor. J. Biol. Chem. 268:8350-8355. [PubMed] [Google Scholar]
- 7.Nelson, D. R., L. Koymans, T. Kamataki, J. J. Stegeman, R. Feyereisen, D. J. Waxman, M. R. Waterman, O. Gotoh, M. J. Coon, R. W. Estabrook, I. C. Gunsalus, and D. W. Nebert. 1996. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6:1-42. [DOI] [PubMed] [Google Scholar]
- 8.Payne, W. J. 1973. Reduction of nitrogenous oxides by microorganisms. Bacteriol. Rev. 37:409-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoun, H., and T. Tanimoto. 1991. Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J. Biol. Chem. 266:11078-11082. [PubMed] [Google Scholar]
- 10.Shoun, H., D.-H. Kim, H. Uchiyama, and J. Sugiyama. 1992. Denitrification by fungi. FEMS Microbiol. Lett. 94:277-281. [DOI] [PubMed] [Google Scholar]
- 11.Shoun, H., M. Kano, I. Baba, N. Takaya, and M. Matsuo. 1998. Denitrification by actinomycetes and purification of dissimilatory nitrite reductase and azurin from Streptomyces thioluteus. J. Bacteriol. 180:4413-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strous, M., J. A. Fuerst, E. H. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 13.Takaya, N., S. Suzuki, S. Kuwazaki, H. Shoun, F. Maruo, M. Yamaguchi, and K. Takeo. 1999. Cytochrome P450nor, a novel class of mitochondrial cytochrome P450 involved in nitrate respiration in the fungus Fusarium oxysporum. Arch. Biochem. Biophys. 372:340-346. [DOI] [PubMed] [Google Scholar]
- 14.Tanimoto, T., K. Hatano, D.-H. Kim, H. Uchiyama, and H. Shoun. 1992. Co-denitrification by the denitrifying system of the fungus Fusarium oxysporum. FEMS Microbiol. Lett. 93:177-180. [Google Scholar]
- 15.Tsuruta, S., N. Takaya, L. Zhang, H. Shoun, K. Kimura, M. Hamamoto, and T. Nakase. 1998. Denitrification by yeasts and occurrence of cytochrome P450nor in Trichosporon cutaneum. FEMS Microbiol. Lett. 168:105-110. [DOI] [PubMed] [Google Scholar]
- 16.Usuda, K., N. Toritsuka, Y. Matsuo, D.-H. Kim, and H. Shoun. 1995. Denitrification by the fungus Cylindrocarpon tonkinense: anaerobic cell growth and two isozyme forms of cytochrome P-450nor. Appl. Environ. Microbiol. 61:883-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshinari, T., and R. Knowles. 1976. Acetylene inhibition of nitrous oxide reduction by denitrifying bacteria. Biochem. Biophys. Res. Commun. 69:705-710. [DOI] [PubMed] [Google Scholar]
- 18.Zhou, Z., N. Takaya, M. A. Sakairi, and H. Shoun. 2001. Oxygen requirement for the denitrification by the filamentous fungus Fusarium oxysporum. Arch. Microbiol. 175:19-25. [DOI] [PubMed] [Google Scholar]
- 19.Zhou, Z., N. Takaya, A. Nakamura, M. Yamaguchi, K. Takeo, and H. Shoun. 2002. Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi. J. Biol. Chem. 277:1892-1896. [DOI] [PubMed] [Google Scholar]
- 20.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]