Abstract
The Mesorhizobium loti strain R7A symbiosis island is a 502-kb chromosomally integrated element which transfers to nonsymbiotic mesorhizobia in the environment, converting them to Lotus symbionts. It integrates into a phenylalanine tRNA gene in a process mediated by a P4-type integrase encoded at the left end of the element. We have determined the nucleotide sequence of the island and compared its deduced genetic complement with that reported for the 611-kb putative symbiosis island of M. loti strain MAFF303099. The two islands share 248 kb of DNA, with multiple deletions and insertions of up to 168 kb interrupting highly conserved colinear DNA regions in the two strains. The shared DNA regions contain all the genes likely to be required for Nod factor synthesis, nitrogen fixation, and island transfer. Transfer genes include a trb operon and a cluster of potential tra genes which are also present on the strain MAFF303099 plasmid pMLb. The island lacks plasmid replication genes, suggesting that it is a site-specific conjugative transposon. The R7A island encodes a type IV secretion system with strong similarity to the vir pilus from Agrobacterium tumefaciens that is deleted from MAFF303099, which in turn encodes a type III secretion system not found on the R7A island. The 414 genes on the R7A island also include putative regulatory genes, transport genes, and an array of metabolic genes. Most of the unique hypothetical genes on the R7A island are strain-specific and clustered, suggesting that they may represent other acquired genetic elements rather than symbiotically relevant DNA.
The symbiosis between legumes and the root nodule bacteria collectively known as rhizobia is of critical agronomic and environmental importance, accounting for the majority of the nitrogen fixed through biological processes. Rhizobia are phylogenetically diverse, falling into five genera of α-proteobacteria (Rhizobium, Bradyrhizobium, Sinorhizobium, Azorhizobium, and Mesorhizobium) (64, 70) and at least two genera of β-proteobacteria (Burkholderia and Ralstonia) (35). It is thought that the rhizobial lineages diverged well before the evolution of legumes and that the genes required for the formation of the symbiosis were subsequently acquired by lateral transfer from undefined sources (8, 33). Reflecting the accessory nature of the traits, several species of rhizobia contain the genes required for nodulation and nitrogen fixation on large plasmids that can be cured under laboratory conditions without affecting the survival of the bacteria (31). Exceptions in which the symbiosis genes are encoded on the chromosome include Bradyrhizobium species, in which the symbiosis genes are clustered but not known to be mobile (18), and at least one strain of Mesorhizobium loti, strain ICMP3153, in which the genes are located on a mobile symbiosis island (57).
M. loti is the microsymbiont of several Lotus species, including Lotus corniculatus and L. japonicus. The symbiosis island of M. loti strain ICMP3153 was discovered through its ability to transfer to nonsymbiotic mesorhizobia in the environment and convert them to symbionts (57). Four genomic species of nonsymbiotic mesorhizobia were subsequently isolated from rhizosphere samples of field-grown L. corniculatus (56), and the transfer of the island to three of these species was demonstrated in the laboratory (58). The ability of the symbiosis island to convert a soil saprophyte to a symbiont suggests that it contains a large proportion of the microsymbiont genes required for the symbiosis. Characterization of the island showed that it is an ∼500-kb genetic element that integrates into a phenylalanine tRNA gene, reconstructing the gene at one end (arbitrarily defined as the left end) and producing a 17-bp direct repeat of the 3′ end of the tRNA gene at the right end. Within the left end of the island, a gene, intS, that encodes a product with similarity to members of the phage P4 integrase subfamily is located 198 bp downstream of the tRNA gene (58). The integrase is required for integration and excision of the island (J. T. Sullivan and C. W. Ronson, unpublished data).
The element was termed a symbiosis island on the basis of its similarities to pathogenicity islands of gram-negative bacteria that have played a key role in the evolution of these pathogens. Pathogenicity islands are defined regions of chromosomal DNA containing clusters of genes required for virulence which are absent from benign isolates of the same or related species (21). Most are situated adjacent to tRNA genes, and many contain an integrase of the P4 family. The available evidence suggests that pathogenicity islands were acquired by lateral transfer, but it has proved difficult to demonstrate their transfer under laboratory conditions; in contrast, the symbiosis island is readily transferable.
Several considerations led us to determine the nucleotide sequence of the symbiosis island of M. loti strain R7A, a derivative of strain ICMP3153. The sequence would give insights into the symbiotic complement of M. loti and also into the signals and mechanisms underlying horizontal transfer of the symbiosis island and perhaps of other genomic islands in the environment. Moreover, M. loti is the natural microsymbiont of L. japonicus, which is one of two model legumes presently subject to intensive studies designed to elucidate the plant contribution to the symbiosis (53). DNA sequence analysis would also rapidly provide information on the bacterial input to the symbiosis that is crucial to complement genetic studies of the plant. Comparison of the island sequence with those of the symbiotic regions of other rhizobia would provide insights into the evolution of both the island and the symbiosis. Sequences available for comparison include those of the 536-kb symbiotic plasmid pNGR234a of Sinorhizobium sp. strain NGR234 (14), a 400-kb symbiotic region of Bradyrhizobium japonicum USDA110 (18), and the entire genomes of Sinorhizobium meliloti strain 1021 (16) and M. loti strain MAFF303099 (25).
The 7.6-Mb strain MAFF303099 genome consists of a chromosome and two plasmids, pMLa and pMLb. A comparison with strain R7A indicates that the MAFF303099 chromosome contains a putative symbiosis island integrated adjacent to the phenylalanine tRNA gene, although the island differs at both ends from the R7A island and it is not known whether it is transmissible (25). Here we report a sequence analysis of the R7A symbiosis island and a comparative analysis of the R7A island with the putative strain MAFF303099 symbiosis island and with the other rhizobial sequences. This analysis has revealed potentially novel symbiotic genes and has given significant insight into the evolution of the two M. loti symbiosis islands.
MATERIALS AND METHODS
Bacterial strain.
The symbiosis island of M. loti strain R7A, a field reisolate of strain ICMP3153 (57), was sequenced. Strain ICMP3153 is also known as NZP2238, and culture collection isolates of these strains may differ by the presence or absence of a plasmid (57). Strain R7A lacks plasmids (57).
BAC cloning.
Three SpeI fragments of approximately 80, 120, and 140 kb contained within the island had previously been linked together and with the ends of the island by pLAFR1- or pIJ3200-based cosmid clones (58). The nucleotide sequence of the island was obtained from the three SpeI fragments cloned into a bacterial artificial chromosome (BAC) vector and from eight of the cosmid clones that, together with the SpeI fragments, spanned the island. To clone the SpeI fragments, the BAC vector pBeloBacII (69) was modified by digestion with NotI to remove the lacZα fragment, blunt ended, and ligated to a blunt-ended BspLU11I/HpaI lacZα fragment from the vector Litmus 28 (New England Biolabs, Beverley, Mass.) to produce pLitBac. This vector contains a unique SpeI-compatible AvrII site in the polylinker. The SpeI fragments were isolated by pulsed-field gel electrophoresis (PFGE) using a Bio-Rad DRIII electrophoresis cell and 1% SeaPlaque GTG low-melting-point agarose (FMC Bioproducts) gels in 1× Tris-acetate-EDTA buffer. Electrophoresis of a SpeI digest of R7A DNA was carried out at 6 V/cm at an included angle of 120° by using a linearly ramped pulse of 1 to 13 s for 18 h at 14°C. DNA fractions of between 80 and 200 kb were isolated by electroelution as described previously (54). Fractions were ligated into pLitBac which had been digested with AvrII and dephosphorylated. Ligations were transformed into Escherichia coli DH10B by electroporation as described previously (52). Sequences obtained from plasmid subclones of cosmids which overlapped the SpeI fragments were used as described previously (11) to design PCR primers to screen BAC clones directly from cultures of pooled clones. Selected BAC clones were sized on PFGE gels and used to probe Southern blots of PFGE gels containing genomic SpeI digests of diverse strains of mesorhizobia containing the island, to confirm that they hybridized to island SpeI fragments of the expected size.
Random library construction and sequencing.
To obtain random subclones for sequencing, CsCl gradient-purified cosmid and BAC clones were sheared by nebulization, end repaired using T4 DNA polymerase and Klenow polymerase, and size fractionated by agarose gel electrophoresis. Fractions of approximately 2.5 kb were extracted from gels, ligated into pUC8 which had been digested with SmaI and dephosphorylated, and transferred by electroporation into DH10B. Colony hybridizations were carried out by hybridizing membranes successively with the BAC or cosmid vector and the relevant BAC or cosmid clone to eliminate random clones containing vector DNA or nonisland sequences.
All sequencing was carried out using plasmid DNA or PCR products as templates and dye-terminator chemistry on ABI377 sequencers. Random pUC8 clones obtained from cosmids and BACs were sequenced using Universal forward and, in selected cases, reverse primers. Sequencing of individual BACs and cosmids was continued until an approximately 4 times coverage level was achieved. To complete the project, directed sequencing with custom primers was carried out on selected random clones and PCR products amplified from genomic DNA. PCR products were also used to examine possible frameshifts, and all frameshifts in pseudogenes were verified. Approximately 4,500 sequences were used for the final assembly.
Assembly and annotation.
Sequences were manually edited and then assembled using Seqman (DNAStar Inc., Madison, Wis.) and Sequencher (GeneCodes Corp., Ann Arbor, Mich.). Open reading frames (ORFs) were identified using BlastX (1) and GeneMark.hmm programs trained on a heuristic model (7) and also a FrameD (50) program trained on a set of 50 previously identified symbiosis island ORFs. The minimum size for ORFs without database homologues was 150 bp. Predicted ORFs and intergenic regions were used to interrogate nonredundant protein databases by using the Blast programs (1) via the website http://www.ncbi.nlm.nih.gov/blast. Sequence annotation and analysis were performed utilizing the Web-based microbial genome annotation system iANT (61). Proteins were categorized using a modified Riley classification (46). In general, the minimal level of similarity leading to assignment of putative gene products as homologues was defined as 30% amino-acid identity over an alignment spanning 80% or more of both the putative gene product and its database homologue; gene products that shared >40% amino acid identity were considered likely orthologues. Comparisons to the MAFF303099 sequence were carried out using the website http://www.kazusa.or.jp/rhizobase.
Nucleotide sequence accession number
The sequence reported in this paper has been deposited in the EMBL database under accession number AL672111. The complete annotated genetic map and classification of the R7A island are available on the Web at http://sequence.toulouse.inra.fr/msi.
RESULTS AND DISCUSSION
General features of the island sequence.
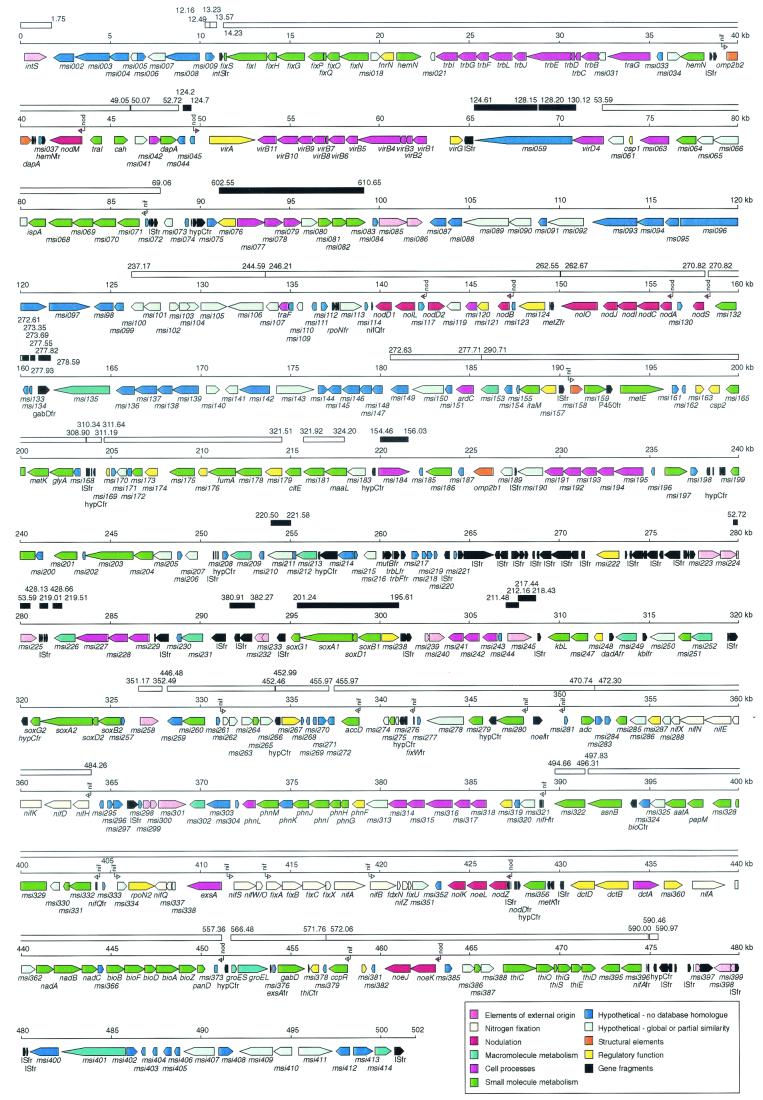
The island sequence was determined to be 501,801 bp in length, with an average G+C content of 59.3%. Variation in G+C content ranged from 47.3% for the nodulation gene nolL (msi116) to ∼69% for the cytochrome P450 gene cluster (msi071 to msi064). The genetic organization of the island is shown in Fig. 1. A total of 414 genes were predicted, giving a gene density of 1 per 1.21 kb. The genes were grouped into 11 categories based on functional class (46), with the addition of those of nodulation and nitrogen fixation, and compared with genes on the putative MAFF303099 island, pNGR234a, the symbiotic region of B. japonicum, and the complete S. meliloti genome (Table 1). pNGR234a contained more genes conserved with the M. loti islands than did the symbiotic plasmids of S. meliloti or the symbiotic region of B. japonicum. Apart from seven nod and 16 nif genes, only five genes encoded likely orthologues conserved across all the rhizobial symbiotic regions. These were the C4-dicarboxylate transport gene dctA, which is required for nitrogen fixation (28), msi337 and msi338, which may be involved in nitrogen fixation (see below), and msi300 and msi301, which are part of an insertion sequence (IS). Interestingly, 96 of the 109 hypothetical genes detected in the R7A island that have no database matches to other bacteria are not present in the M. loti MAFF303099 genome, indicating that they are strain rather than species specific.
FIG. 1.
Detailed map of the strain R7A symbiosis island. Coordinates are given in kilobases, starting from the first base pair downstream of the phenylalanine tRNA gene. Putative genes are colored according to functional class, while gene fragments are colored black (see key). The locations of potential nod-box and NifA-regulated promoters are shown by arrows with “nod” or “nif” above them. Regions conserved with the putative MAFF303099 symbiosis island are indicated by bars above the scale bars. Open bars are colinear regions, and closed bars represent potentially translocated regions. The coordinates (in kilobases) of the MAFF303099 island, also starting from the first base pair downstream of the phenylalanine tRNA gene, are indicated next to the bars.
TABLE 1.
Functional classes and likely orthologues in other rhizobia of symbiosis island genes
| Functional class | No. of genes in R7A island | No. of genesa also in:
|
|||
|---|---|---|---|---|---|
| MAFF303099 symbiotic island | S. meliloti 1021 genomeb | pNGR234a | B. japonicum symbiotic cluster | ||
| Nodulation | 16 | 16 | 8a, 1b | 15 | 10 |
| N fixation | 20 | 20 | 16a | 20 | 20 |
| Regulatory function | 27 | 21 | 2a, 6b, 6c | 8 | 2 |
| Small molecule metabolism | 95 | 78 | 13a, 14b, 14c | 20 | 9 |
| Macromolecule metabolism | 13 | 8 | 2a, 1b | 1 | 2 |
| Cell processes | 51 | 31 | 5a, 5b, 9c | 14 | 1 |
| Elements of external origin | 18 | 10 | 5a, 2b | 6 | 8 |
| Structural elements | 3 | 2 | 1c | 1 | 1 |
| Hypothetical | |||||
| Globalc | 37 | 31 | 3a, 4b, 2c | 14 | 4 |
| Partial | 25 | 7 | |||
| No matches | 109 | 13 | |||
| Total | 414 | 237 | 119 (54a, 33b, 32c) | 99 | 57 |
Likely orthologues were defined as sharing at least 40% amino acid identity over at least 80% of both subject and query sequences.
a, b, and c represent S. meliloti pSyma, pSymb, and chromosome respectively.
Defined as sharing similarity over at least 80% of both subject and query sequences and an “Expect” value of less than 1e−6.
In addition to intact genes, 36 pseudogenes of IS origin and 30 pseudogenes of non-IS origin were identified. These pseudogenes often contained stop codons and had undergone deletions and frameshifts compared to their intact database orthologues. Some pseudogenes have intact orthologues on the island, whereas others do not (e.g., metZfr). Several pseudogenes were also found on pNGR234a (14), and some of these have intact orthologues on the R7A island (e.g., accD, hemN, and msi064).
A feature of the R7A island was the relatively low number of intact ISs in comparison to those of the sequenced symbiotic regions of other rhizobia, with most IS-related sequences being present as pseudogenes. The IS-related sequences were concentrated in two areas, a region of low coding density from 263 to 308 kb and a region from 476 to 480 kb that seems to comprise three copies of an IS, two of which inserted sequentially into the original copy. Similar clustering of ISs into noninformational regions was observed for pNGR234a and may reflect a mechanism to preserve the integrity of coding regions (14).
Comparison between the R7A and MAFF303099 symbiosis islands.
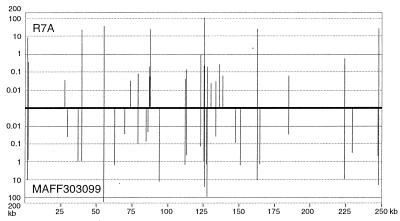
The putative MAFF303099 island was defined by comparison with the R7A island and, at 610,975 bp, is about 109 kb larger than the R7A island (25). The two islands have similar G+C contents (59.3% and 59.7%), which are lower than that of the remainder of the MAFF303099 chromosome (63.0%). The integrase gene downstream of the phenylalanine tRNA gene is the first gene on both islands, but at the other end the two islands share only 24 bp, including the 17-bp direct repeat of the 3′ end of the phenylalanine tRNA gene. However, the two islands share conserved backbone sequences of 248 kb with about 98% DNA sequence identity (Fig. 1), indicating that they evolved from a common ancestor. The backbone has a G+C content of 60.3% and contains all the genes identified below as likely to be required for Nod factor synthesis, nitrogen fixation, and island transfer. The backbone is interrupted by numerous strain-specific “islets” that represent DNA either lost or gained by each strain and range in size from <50 bp up to 168 kb (Fig. 2). The few noncolinear regions that encode similar proteins, such as the region containing msi076 to msi083 on the R7A island (Fig. 1), are less conserved (<90% nucleotide identity). This suggests that these regions were acquired from different sources by each island subsequent to their divergence from the common ancestor, rather than arising through translocation of regions present in the ancestral island.
FIG. 2.
Distribution of strain-specific islets more than 50 bp in size on the two symbiosis islands. The horizontal axis (linear scale in kilobases) represents the conserved backbone, and the vertical axis (logarithmic scale in kilobases) represents the R7A (above horizontal axis) or MAFF303099 (below horizontal axis) strain-specific islets.
Differential decay of some of the pseudogenes present on both islands accounts for some of the islets. For example, both strains contain fragments of a P4 integrase-like gene adjacent to fixS (msi010 on the R7A island) (Fig. 1) that show about 70% identity at the amino acid level to regions of the symbiosis island integrase IntS. However, the portions of the gene remaining differ in each strain and do not overlap, giving rise to islets in both strains. Similarly, both strains have fragments of a nifA gene adjacent to msi396, but 460 bp more of the gene remains in MAFF303099, giving rise to a MAFF303099-specific islet. Four other MAFF303099-specific islets, but no R7A-specific islets, consist solely of ISs.
A substantial portion of the DNA not in common between the two islands comprises hypothetical genes and IS-related sequences. About 8% (42 kb) of the R7A island consists of transposase/integrase/resolvase genes or fragments thereof compared to 19% (114 kb) for MAFF303099 (25), which accounts for a significant portion of the size difference between the two islands. Apart from intS and a few IS fragments, none of the genes or fragments of external origin are in regions colinear between the two islands. Indeed, the products of many of these genes are more similar to those from other bacteria than to those of the other island or, in the case of R7A, those of the MAFF303099 genome. These observations suggest that each island was independently invaded by ISs and other mobile elements subsequent to their evolution from a common pristine ancestor. It seems likely that these mobile elements were also involved in the acquisition of the larger strain-specific islets.
A common theme of microbe-host interactions is the secretion by the microbe of effector proteins directly into the cytoplasm of host cells via a type III or IV secretion system (9, 22). For example, pNGR234a encodes a type III system that is regulated by a nod-box promoter and is required for host-specific nodulation (30, 67). The R7A island has a gene cluster (msi046 to msi058 and msi060) with strong similarity to those vir genes from Agrobacterium tumefaciens that encode the type IV pilus through which T-DNA and proteins are transferred to the plant (9, 24). This cluster is not present in MAFF303099, but MAFF303099 has a gene cluster on a 168-kb strain-specific islet with strong similarity to the clusters encoding type III secretion systems on pNGR234a (14) and in B. japonicum (18). The R7A vir cluster includes virB1 to B11, which form the mating pore virD4, which couples the transferred moiety to the pore, and homologues of virA and virG, the two-component regulatory system required for expression of the vir system. No T-DNA processing genes are present on the R7A island, suggesting that the island vir system may function to secrete proteins, as observed for several type IV systems (9, 66), rather than a DNA-protein complex into plant cells. The virA gene is preceded by a nod box, the promoter sequence responsible for nodulation gene induction, suggesting that the system may have a symbiotic role. Comparison of the two islands indicates that the vir system has been lost from MAFF303099. Downstream of dapA (msi043 in R7A), MAFF303099 has an ORF comprising an integrase-recombinase gene fused to a 173-bp region identical to the 5′ end of the R7A virD4 (msi060) gene in antisense orientation. The intervening DNA in R7A, containing the entire vir cluster and msi059, is deleted from the corresponding location in MAFF303099, resulting in a 23.6-kb R7A-specific vir islet. However, the MAFF303099 island has a DNA region nearly identical to the R7A region, comprising msi059 and the adjacent 3′ 120 nucleotides of virD4, located adjacent to its type III gene cluster. Hence, it seems likely that this region was translocated to its new location in MAFF303099, perhaps as part of the recombination event that led to the deletion of the vir system from MAFF303099. Msi059 encodes a large protein with an internal repetitive structure that, because of its location in the strains, is a candidate for transfer to plant cells via the type III and IV systems in both strains.
The plasticity of the genetic complement of the two symbiosis islands is illustrated by the findings that a number of gene clusters found on the R7A island are also present on plasmid pMLa in MAFF303099 and that some of these are absent from the MAFF303099 island. For example, the cluster msi362 to msi374 (nad and bio operons) is syntenous on both islands and pMLa, as are msi325 to msi327, which encode metabolic enzymes. Clusters present on R7A and pMLa but absent from the putative MAFF303099 island include msi203 to msi205 and msi246 to msi247, which encode enzymes involved in amino-acid metabolism, and msi305 to msi319, which encode gene products involved in phosphonate transport and metabolism, as well as an ABC oligopeptide transporter.
Nodulation genes.
The Nod factor produced by M. loti strain R7A is an N-acetylglucosamine pentasaccharide in which the nonreducing residue carries a carbamoyl residue and is N-methylated and N-acylated and the reducing residue is substituted at the C-6 position with O-acetylfucose (29). The island carries the 12 genes required for synthesis of this structure plus the nodIJ genes required for Nod-factor transport (41). The genes are arranged in seven operons, each of which is preceded by a nod box. Four of the operons are in one cluster, while nodM and the two operons (noeKJ and nodZnoeLnolK) required for the synthesis of the O-acetylfucose moiety are in different regions of the island, suggesting independent acquisition. Two copies of NodD, the regulator that binds nod boxes and activates gene expression, are encoded on the island.
Nitrogen fixation.
Most of the known structural nif and fix genes on the island fall into three clusters. The organization of the nifH operon is identical to that of the nifH1 operon of pNGR234a, including the gene downstream of nifX, msi288 (y4vQ in pNGR234a) (14). This gene is also found in the same position in B. japonicum (18), indicating that it is likely to have a function in nitrogen fixation, despite its not being present in S. meliloti (3). The organization of the fixA-fixU cluster is identical to those of pNGR234a and S. meliloti, except that nifZ is not present in S. meliloti. The symbiosis island also contains orthologues of most other genes located in the pNGR234a nif-fix cluster. The island cluster msi332 to msi328 is similar to y4vGHIJ with the addition of a ferredoxin gene and, as all genes in the cluster have homologues implicated in oxidation-reduction reactions, may have a role in electron transfer. ORFs msi276 to msi274 are similar to y4xE, fdxB, and y4xQ, respectively; msi262 is similar to y4vC; msi334 is similar to y4vD, and msi338 to msi336 are similar to y4xE, y4vS, and y4vE (nifQ), respectively. Given their respective locations and likely regulation by NifA in both organisms (see reference 14 and below), these genes may all have a role, as yet unknown, in nitrogen fixation.
The third island fix cluster is absent from pNGR234a and includes fixNOPQ, which encodes a cbb (3)-type heme-copper oxidase complex, and fixGHIS, which may participate in the uptake and metabolism of copper required for the oxidase complex (13, 42, 43). These genes are associated with a conserved hypothetical gene, msi018, the oxygen-responsive regulatory gene fnrN, and a gene required for anaerobic heme synthesis, hemN. The entire cluster is duplicated on the chromosomes of M. loti MAFF303099 (25) and R7A (J. T. Sullivan and C. W. Ronson, unpublished data). An island fixI::Tn5 mutant forms Fix+ nodules (S. H. Miller and C. W. Ronson, unpublished data), suggesting that the chromosomal copy can substitute for the island copy in symbiosis. The duplication of the cluster is consistent with its being required for microaerobic respiration in nonsymbiotic bacteria rather than being symbiosis-specific and with the island having acquired most genes with a specific symbiotic role. The fixNOQP operon is also reiterated in S. meliloti (3), Rhizobium leguminosarum (51), and R. etli (17), perhaps reflecting a similar evolutionary history.
While the regulation of symbiotic nitrogen fixation has not been studied in mesorhizobia, the location of fnrN indicates it is likely to be involved in activating fixNOQP expression in the absence of oxygen, as shown in R. leguminosarum and B. japonicum (13, 19, 51). The island also contains two copies of the nitrogen fixation transcriptional regulator nifA, nifA1, which is located between fixX and nifB (as also found on pNGR234a) and in S. meliloti and R. leguminosarum (3, 14, 23), and nifA2, which is not located in a nif-fix cluster. The products of the two nifA genes are more similar to NifA proteins from other rhizobia (i.e., NifA1 to that of R. etli and NifA2 to that of B. japonicum) than to each other, suggesting that the genes were separately acquired by the island rather than arising by duplication of a single gene. The genes are not functionally redundant, as a nifA2 mutant forms ineffective nodules (55). No potential activators of nifA gene expression were identified on the island. However, copies of fixLJ and fixK, key oxygen-responsive regulators of nitrogen fixation in S. meliloti and B. japonicum (4, 13), are located on the chromosome of M. loti MAFF303099 (25), as are copies of the two-component regulatory system regSR/actSR, which regulates nifA expression in B. japonicum (12). Hence nif-fix gene regulation in M. loti is likely to be complex and a variation of that characterized in other rhizobia.
NifA activates nif gene expression in association with the product of the rpoN gene, σ54 or RpoN. A copy of rpoN is present on the island in a nif gene region. RpoN-dependent promoters have a consensus 5′-TGGCAC-N5-TTGC-3′ sequence motif, with the GG and GC doublets at −24/−12 strongly conserved. NifA binds to an upstream activator sequence (UAS) (TGT-N10-ACA) 80 to 150 nucleotides upstream of the −24/−12 sequence (13). An initial search of the island for potential NifA-dependent promoter sequences revealed that the −12 doublet could be either GC or GA, as also found for R. etli (63). A search for promoters with a UAS for NifA together with the modified RpoN consensus revealed 16 likely candidates, including eight that together encompass all the potential nif gene operons that are in common with those of pNGR234a (Fig. 1).
Other genes potentially regulated by NifA.
Other genes associated with NifA-regulated promoters include two, msi071 and msi158, that are conserved with pNGR234a and B. japonicum. msi071 is the first gene in a cytochrome P450 cluster that may be involved in synthesis of a terpenoid, possibly the phytohormone gibberellin (62). Compared to M. loti, the pNGR234a and B. japonicum clusters are truncated by ISs at or within the msi064 homologue, which probably encodes isopentenyl pyrophosphate isomerase, and hence are likely to be nonfunctional although they are expressed in nodules. The B. japonicum, strain NGR234, and M. loti clusters all have a distinctive high G+C content of 69% (including msi064 in M. loti), and the gene products share >98% amino acid identity, suggesting that the three rhizobia acquired the cluster relatively recently from a common source.
msi158 encodes an outer membrane protein similar to Omp21 from Comamonas acidovorans that is expressed under microaerobic conditions and may be a porin or involved in adhesion (2). Its pNGR234a homologue is strongly expressed in nodules (40), which, taken together with the fact that no other homologues of the protein are present in the M. loti MAFF303099 genome, indicates that the protein may have a symbiosis-specific role.
Predicted functions of some of the other genes potentially regulated by NifA also indicate a role in symbiosis. msi036 encodes a protein with a sequence with strong similarity over its full length to those of the Omp2 porins of Brucella species (10) and is one of a five-member family of these genes in the M. loti MAFF303099 genome. msi188 is an additional member of this family not present in MAFF303099. It is interesting that two of the three genes encoding outer membrane proteins on the symbiosis island are likely to be NifA regulated, as these proteins are at the interface of the plant-microbe interaction.
The msi273 product is 1-aminocyclopropane-1-carboxylate (AAC) deaminase, which catalyses the conversion of AAC, the metabolic precursor of the phytohormone ethylene, to ammonia and α-ketobutyrate. The plant-growth-promoting bacteria Pseudomonas fluorescens and Enterobacter cloacae promote root elongation through expression of ACC deaminase (26, 68). A role for ACC deaminase in the symbiosis is indicated by the finding that inhibition of ethylene biosynthesis enhances nodulation of L. japonicus, especially when the plant roots are exposed to light (37, 65).
Msi380 shows strong similarity to cytochrome C peroxidase from Pseudomonas aeruginosa. In P. aeruginosa, the enzyme is located in the bacterial periplasm, where its likely function is to provide protection against toxic peroxides (45). A similar function could be envisaged for the island gene, as an oxidative burst is an early response in the alfalfa-S. meliloti symbiotic interaction (49) and in addition, hydrogen peroxide will be formed as a by-product of respiration.
Island transfer.
The island is unlikely to replicate as a plasmid, as it lacks the highly conserved repABC genes. The phage P4-like integrase encoded by intS (msi001) is required for the integration of the island and its excision as a circle (J. T. Sullivan and C. W. Ronson, unpublished data). The island also contains a trbBCDEJLFGI operon and a traG gene, which show similarity to those required for conjugative transfer of the Agrobacterium Ti plasmid, pNGR234a and RP4 (27), except that two genes, trbK and trbH, found on these plasmids are missing. Interestingly, the island trb operon is most similar to the trb operon on M. loti MAFF303099 pMLb, including the hypothetical gene msi021 immediately downstream of trbI. The island trbGImsi021gene products are also very similar to those from the partially sequenced trb operon of Tn4371, a transposon from Ralstonia eutrophus containing a bph gene cluster required for degradation of biphenyl (34), and to a partial trb operon from Caulobacter crescentus that is disrupted by an IS (36). The strong similarities between, absence of trbH from, and presence of msi021 in these four operons suggest that they form a new subfamily of mating pore systems.
The island also contains an orthologue of traF (msi108) that encodes a protease which acts on TrbC, the precursor of the major pilin subunit (20). The island traF is part of a cluster of five genes (msi110 to msi106) that are also conserved as part of a cluster (mll9738 to mll9748) on pMLb. Msi107 shows strong similarity to lytic murein transglycosylases. The pMLb cluster also includes homologues of traG (msi032) and msi031 that are adjacent to the trb operon on the R7A island and a repA gene and adjacent hypothetical gene not found on the island. Thus, it seems likely that msi110 to msi106 are involved in the transfer of the island. The cluster on the putative MAFF303099 island is interrupted by insertion of a transposase gene, raising the possibility that the MAFF303099 island may not be self transmissible.
Msi152 is a homologue of the incW plasmid pSa antirestriction protein ArdC (6). Homologues are also encoded on several Agrobacterium plasmids: pNGR234a, pNL1 from Sphingomonas aromaticivorans, and M. loti pMLa and pMLb (14, 25, 47, 59). These proteins exhibit high similarity to the N-terminal 300 amino acids of the TraC1 primase of RP4, which is transferred to recipients during conjugation (44). A complete copy of the RP4-type traC gene is not located on the island, Ti plasmids, or pNGR234a. ArdC and TraC1 are able to bind single-stranded DNA and escort plasmid DNA during conjugation (6), and so ArdC may play a role in the conjugative transfer of the island.
Island transfer may be regulated by Msi173 and Msi174, as these show similarity to TraI (homoserine lactone synthase) and TraR, respectively, members of the LuxI/LuxR family of cell density-dependent regulators, which together regulate Agrobacterium Ti plasmid transfer (15). A second traI homologue (msi039) is also present on the island, and traI and traR homologues are present on pMLb.
Transport.
The R7A and MAFF303099 islands encode six and seven ABC transport systems, respectively, and apart from ExsA (Msi339), all are encoded on strain-specific islets. ExsA is an MsbA-like exporter that has been implicated in the biosynthesis of exopolysaccharide I in S. meliloti (5). msi077 to msi079 encode a sugar importer most similar to ribose transporters. The operon is also present on pNGR234a and as part of a noncolinear conserved cluster on the putative MAFF303099 island, and in each of these cases it is adjacent to a metabolic operon that includes genes encoding the two subunits of transketolase. The gene products of msi195 to msi191 share about 80% identity to products of y4OPQRS from pNGR234a and so are likely to transport the same oligopeptide substrates. msi318 to msi314 also encode an oligopeptide transporter. The other two R7A ABC transporters, Msi229 to Msi227 and Msi243 to Msi241, are similar to polyamine and amino acid transporters, respectively. ABC transporters with similarity to oligopeptide and polyamine types are also encoded on the putative MAF303099 island but are not strongly conserved with the R7A operons. The MAFF303099 island does not encode a transporter similar to the Msi243 to Msi241 amino acid transporter. Thus, the exact complement of ABC transporters does not seem to be important for symbiosis.
In addition to ExsA, the products of four island genes are likely to be involved in small-molecule efflux. Two, Msi042 and Msi120, are members of the Rht family of amino acid efflux proteins, while Msi063 and Msi184 are members of the MFS family of transport proteins. All five efflux proteins are conserved between the two islands.
Metabolic genes.
The symbiosis island contains a wide variety of metabolic genes, including several with products that show strong similarity to enzymes involved in carbon metabolism, the synthesis and breakdown of amino acids, phosphonate degradation, and cofactor biosynthesis (Table 2). Of the seven nonsymbiont mesorhizobial strains isolated from the environment, all were auxotrophic for biotin and thiamine and all but one were auxotrophic for nicotinic acid (56). Transfer of the symbiosis island to these strains rendered them prototrophic, confirming the functionality of the bio, nad, and thi operons on the island (Table 2). The role, if any, of these cofactor biosynthetic genes in the symbiosis process remains unknown, as they are not required for nodulation or nitrogen fixation (reference 55 and N. G. McCallum and C. W. Ronson, unpublished data). The bio and nad operons are duplicated on pMLa in strain MAFF303099, providing further evidence of the accessory nature of the operons in these soil bacteria.
TABLE 2.
Metabolic genes of defined function on the R7A symbiosis island
| Msi number functional category and gene no. | Gene/operon mnemonic | Gene product(s) or predicted function |
|---|---|---|
| Amino acid metabolism related to 1-carbon metabolism | ||
| msi156 | ltaM | Low specificity l-threonine aldolase |
| msi160 | metE | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase |
| msi166 | metK | SAM synthetase |
| msi167 | glyA | Serine hydroxymethyltransferase |
| msi204 | cysM | Cysteine synthase |
| msi237-msi234 | soxBDAG1 | Sarcosine oxidase |
| msi246 | kbl | 2-amino-3-ketobutyrate coenzyme A ligase |
| msi256-msi253 | soxBDAG2 | Sarcosine oxidase |
| Other amino acid metabolism | ||
| msi043 | dapA | Dihydrodipicolinate synthase |
| msi182 | maaL | Methylaspartate ammonia-lyase |
| msi203 | Fusion protein containing putative ligase and arginosuccinate lyase | |
| msi323 | asnB | Asparagine synthetase |
| msi326 | aatA | Aspartate aminotransferase |
| Carbon metabolism | ||
| msi040 | cah | A-type carbonic anhydrase |
| msi177 | fumA | Fumarate hydratase Class I |
| msi282 | adc | Acetoacetate decarboxylase |
| msi327 | pepM | Phosphoenolpyruvate phosphomutase |
| Phosphate metabolism | ||
| msi311-msi305 | phnGHIJKML | Phosphonate degradation |
| Cofactor synthesis | ||
| msi363-msi365 | nadABC | Nicotinate biosynthesis |
| msi367-msi371 | bioBFDAZ | Biotin biosynthesis |
| msi372 | panD | Aspartate 1-decarboxylase |
| msi389-msi394 | thiCOSGED | Thiamine biosynthesis |
The island contains several genes that encode products involved in one-carbon metabolism, including the synthesis of glycine, methionine, S-adenosylmethionine (SAM), and the cofactor tetrahydrofolate and its methylene and methyl derivatives (Table 2), suggesting a demand for these compounds in symbiosis. Previously only glyA and a gene involved in methionine synthesis, metZ, have been implicated in symbiosis (48, 60). Tetrahydrofolate or its derivatives are required for glycine, methionine, and purine synthesis, and purine auxotrophs also form symbiotically defective nodules (28). In addition, tetrahydrofolate is required for the formylation of Met-tRNAfMet to initiate translation. It is possible that the presence of some of the C1 genes is an adaptation to the high levels of nif-specific protein synthesis in nodules. SAM is required for several functions, including DNA methylation, biotin biosynthesis, and in M. loti, the NodS-mediated methylation of Nod factors. Most of the genes implicated in C1 metabolism have homologues elsewhere in the MAFF303099 genome; however, the metE gene, which encodes a cobalamin-independent methionine synthase, is only present on the island. A metE gene is also present in the symbiotic region of B. japonicum (18), and its presence may therefore represent a symbiosis-specific adaptation.
Two other metabolic genes that are unique to the islands are maaL, which encodes a methyl-aspartate ammonia lyase involved in glutamate degradation, and fumA, which encodes a Class I fumarase that may be involved in anaerobic respiration. maaL has only been found in a few bacteria, including E. coli strain O157:H7, in which it is part of an O-specific island together with a fumA homologue (39). msi040 encodes an α-type carbonic anhydrase and is also unique to the islands. The function, if any, of these and the other metabolic genes on the islands remains to be determined, but it seems likely that they contribute to the symbiosis by maintaining and fine-tuning metabolic fluxes through the various phases of the rhizobium-plant interaction.
Conclusions and perspectives.
Our sequence analysis has shown that the strain R7A symbiosis island is a large, site-specific conjugative transposon that is likely to transfer via a type IV mating pore in a mechanism analogous to plasmid-mediated conjugation. Comparison of the R7A island with the putative symbiosis island of M. loti MAFF303099 revealed that the two islands share 248 kb of DNA, showing near-perfect colinearity, and about 98% nucleotide identity, indicating that this DNA was present in a common ancestral island. Nevertheless, the two islands show a surprising degree of divergence, mainly due to the deletion and acquisition of multiple DNA segments that interrupt the colinear regions. The dynamic nature of the islands is also emphasized by the presence of several pseudogenes at various stages of decay in the two islands and the presence of a number of gene clusters on the R7A island that are plasmid borne in MAFF303099. Most of the unique hypothetical genes on the R7A island are clustered and are on strain-specific segments, suggesting that they represent genetic elements of unknown function rather than symbiotically relevant DNAs. This hypothesis is supported by the finding that most of the ISs are also strain specific. The overall pattern of colinear segments interrupted by strain-specific segments is similar to that observed between the genomes of related bacterial pathogens (32, 38, 39) and presumably reflects common evolutionary mechanisms.
The DNAs common to both islands include the putative transfer genes, most if not all genes known to be required for nodulation and nitrogen fixation, and several genes with deduced functions that indicate they may have a symbiotic role. These include genes also present on pNGR234a that may be involved in nitrogen fixation. The symbiosis island also confers upon the host bacterium a wide variety of metabolic and transport functions, at least some of which are likely to augment the symbiotic interaction, as they are also found on other symbiotic replicons. Some functions, such as the ability to synthesize cofactors and the ability to catabolize sarcosine and phosphonates, may contribute to survival in the rhizosphere environment, although they are not required for the survival of the nonsymbiotic saprophytic mesorhizobia. The comparative analysis described here provides the foundation for future analysis that will provide insight into the function of these newly revealed genes. Questions of particular interest include the significance of the type IV secretion system in R7A versus the type III secretion system in MAFF303099, the signals and mechanisms underlying horizontal transfer of the symbiosis island to nonsymbionts in the environment, and more generally, the evolution and transmission of genomic islands.
Acknowledgments
This work was supported by grants from the Marsden Fund, administered by the Royal Society of New Zealand, the U.S. Department of Education (DE FG02-91ER200021), the University of Otago, and Michigan State University.
We thank the Australian Genome Research Facility, Brisbane, Australia, and the Michigan State University Plant Research Laboratory Sequencing Unit for assistance with the sequencing and P. DeHaan and S. Strzalkowski for technical assistance.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldermann, C., A. Lupas, J. Lubieniecki, and H. Engelhardt. 1998. The regulated outer membrane protein Omp21 from Comamonas acidovorans is identified as a member of a new family of eight-stranded β-sheet proteins by its sequence and properties. J. Bacteriol. 180:3741-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batut, J., and P. Boistard. 1994. Oxygen control in Rhizobium. Antonie Leeuwenhoek 66:129-150. [DOI] [PubMed] [Google Scholar]
- 5.Becker, A., H. Kuster, K. Niehaus, and A. Puhler. 1995. Extension of the Rhizobium meliloti succinoglycan biosynthesis gene cluster: identification of the exsA gene encoding an ABC transporter protein, and the exsB gene which probably codes for a regulator of succinoglycan biosynthesis. Mol. Gen. Genet. 249:487-497. [DOI] [PubMed] [Google Scholar]
- 6.Belogurov, A. A., E. P. Delver, O. V. Agafonova, N. G. Belogurova, L. Y. Lee, and C. I. Kado. 2000. Antirestriction protein Ard (Type C) encoded by IncW plasmid pSa has a high similarity to the “protein transport” domain of TraC1 primase of promiscuous plasmid RP4. J. Mol. Biol. 296:969-977. [DOI] [PubMed] [Google Scholar]
- 7.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broughton, W. J., and X. Perret. 1999. Genealogy of legume-Rhizobium symbioses. Curr. Opin. Plant Biol. 2:305-311. [DOI] [PubMed] [Google Scholar]
- 9.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloeckaert, A., J. M. Verger, M. Grayon, and N. Vizcaino. 1996. Molecular and immunological characterization of the major outer membrane proteins of Brucella. FEMS Microbiol. Lett. 145:1-8. [DOI] [PubMed] [Google Scholar]
- 11.Dewar, K., L. Sabbagh, G. Cardinal, F. Veilleux, F. Sanschagrin, B. Birren, and R. C. Levesque. 1998. Pseudomonas aeruginosa PAO1 bacterial artificial chromosomes: strategies for mapping, screening, and sequencing 100 kb loci of the 5.9 Mb genome. Microb. Comp. Genomics 3:105-117. [DOI] [PubMed] [Google Scholar]
- 12.Emmerich, R., H. Hennecke, and H. M. Fischer. 2000. Evidence for a functional similarity between the two-component regulatory systems RegSR, ActSR, and RegBA (PrrBA) in alpha-Proteobacteria. Arch. Microbiol. 174:307-313. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, H. M. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 15.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lalaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 17.Girard, L., S. Brom, A. Davalos, O. Lopez, M. Soberon, and D. Romero. 2000. Differential regulation of fixN-reiterated genes in Rhizobium etli by a novel fixL-fixK cascade. Mol. Plant-Microbe Interact. 13:1283-1292. [DOI] [PubMed] [Google Scholar]
- 18.Gottfert, M., S. Rothlisberger, C. Kundig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutiérrez, D., Y. Hernando, J.-M. Palacios, J. Imperial, and T. Ruiz-Argüeso. 1997. FnrN controls symbiotic nitrogen fixation and hydrogenase activities in Rhizobium leguminosarum biovar viciae UPM791. J. Bacteriol. 179:5264-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haase, J., and E. Lanka. 1997. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J. Bacteriol. 179:5728-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 22.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iismaa, S. E., P. M. Ealing, K. F. Scott, and J. M. Watson. 1989. Molecular linkage of the nif/fix and nod gene regions in Rhizobium leguminosarum biovar trifolii. Mol. Microbiol. 3:1753-1764. [DOI] [PubMed] [Google Scholar]
- 24.Kado, C. I. 2000. The role of the T-pilus in horizontal gene transfer and tumorigenesis. Curr. Opin. Microbiol. 3:643-648. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 26.Li, J., D. H. Ovakim, T. C. Charles, and B. R. Glick. 2000. An ACC deaminase minus mutant of Enterobacter cloacae UW4 no longer promotes root elongation. Curr. Microbiol. 41:101-105. [DOI] [PubMed] [Google Scholar]
- 27.Li, P. L., D. M. Everhart, and S. K. Farrand. 1998. Genetic and sequence analysis of the pTiC58 trb locus, encoding a mating-pair formation system related to members of the type IV secretion family. J. Bacteriol. 180:6164-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long, S. R. 1989. Rhizobium genetics. Annu. Rev. Genet. 23:483-506. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Lara, I. M., J. D. van den Berg, J. E. Thomas-Oates, J. Glushka, B. J. Lugtenberg, and H. P. Spaink. 1995. Structural identification of the lipo-chitin oligosaccharide nodulation signals of Rhizobium loti. Mol. Microbiol. 15:627-638. [DOI] [PubMed] [Google Scholar]
- 30.Marie, C., W. J. Broughton, and W. J. Deakin. 2001. Rhizobium type III secretion systems: legume charmers or alarmers? Curr. Opin. Plant Biol. 4:336-342. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Romero, E., and J. Caballero-Mellado. 1996. Rhizobium phylogenies and bacterial genetic diversity. Crit. Rev. Plant Sci. 15:113-140. [Google Scholar]
- 32.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 33.Mergaert, P., M. Van Montagu, and M. Holsters. 1997. Molecular mechanisms of Nod factor diversity. Mol. Microbiol. 25:811-817. [DOI] [PubMed] [Google Scholar]
- 34.Merlin, C., D. Springael, and A. Toussaint. 1999. Tn4371: a modular structure encoding a phage-like integrase, a Pseudomonas-like catabolic pathway, and RP4/Ti-like transfer functions. Plasmid 41:40-54. [DOI] [PubMed] [Google Scholar]
- 35.Moulin, L., A. Munive, B. Dreyfus, and C. Boivin-Masson. 2001. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 411:948-950. [DOI] [PubMed] [Google Scholar]
- 36.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nukui, N., H. Ezura, K. Yuhashi, T. Yasuta, and K. Minamisawa. 2000. Effects of ethylene precursor and inhibitors for ethylene biosynthesis and perception on nodulation in Lotus japonicus and Macroptilium atropurpureum. Plant Cell Physiol. 41:893-897. [DOI] [PubMed] [Google Scholar]
- 38.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 39.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Y. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157: H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 40.Perret, X., C. Freiberg, A. Rosenthal, W. J. Broughton, and R. Fellay. 1999. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 32:415-425. [DOI] [PubMed] [Google Scholar]
- 41.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preisig, O., R. Zufferey, and H. Hennecke. 1996. The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Arch. Microbiol. 165:297-305. [DOI] [PubMed] [Google Scholar]
- 43.Preisig, O., R. Zufferey, L. Thöny-Meyer, C. A. Appleby, and H. Hennecke. 1996. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178:1532-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rees, C. E., and B. M. Wilkins. 1990. Protein transfer into the recipient cell during bacterial conjugation: studies with F and RP4. Mol. Microbiol. 4:1199-1205. [DOI] [PubMed] [Google Scholar]
- 45.Ridout, C. J., R. James, and C. Greenwood. 1995. Nucleotide sequence encoding the di-haem cytochrome c551 peroxidase from Pseudomonas aeruginosa. FEBS Lett. 365:152-154. [DOI] [PubMed] [Google Scholar]
- 46.Riley, M. 1993. Functions of the gene products of Escherichia coli. Microbiol. Rev. 57:862-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossbach, S., and H. Hennecke. 1991. Identification of glyA as a symbiotically essential gene in Bradyrhizobium japonicum. Mol. Microbiol. 5:39-47. [DOI] [PubMed] [Google Scholar]
- 49.Santos, R., D. Herouart, S. Sigaud, D. Touati, and A. Puppo. 2001. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact. 14:86-89. [DOI] [PubMed] [Google Scholar]
- 50.Schiex, T., P. Thébault, and D. Kahn. 2000. Recherche des gènes et des erreurs de séquençage dans les génomes bactériens GC-riches (et autres…), p. 321-328. In O. Gascuel and M.-F. Sagot (ed.), JOBIM Conference Proceedings. Springer-Verlag, Montpellier, France.
- 51.Schluter, A., T. Patschkowski, J. Quandt, L. B. Selinger, S. Weidner, M. Kramer, L. Zhou, M. F. Hynes, and U. B. Priefer. 1997. Functional and regulatory analysis of the two copies of the fixNOQP operon of Rhizobium leguminosarum strain VF39. Mol. Plant-Microbe Interact. 10:605-616. [DOI] [PubMed] [Google Scholar]
- 52.Sheng, Y., V. Mancino, and B. Birren. 1995. Transformation of Escherichia coli with large DNA molecules by electroporation. Nucleic Acids Res. 23:1990-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stougaard, J. 2001. Genetics and genomics of root symbiosis. Curr. Opin. Plant Biol. 4:328-335. [DOI] [PubMed] [Google Scholar]
- 54.Strong, S. J., Y. Ohta, G. W. Litman, and C. T. Amemiya. 1997. Marked improvement of PAC and BAC cloning is achieved using electroelution of pulsed-field gel-separated partial digests of genomic DNA. Nucleic Acids Res. 25:3959-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan, J. T., S. D. Brown, R. R. Yocum, and C. W. Ronson. 2001. The bio operon on the acquired symbiosis island of Mesorhizobium sp. strain R7A includes a novel gene involved in pimeloyl-CoA synthesis. Microbiology 147:1315-1322. [DOI] [PubMed] [Google Scholar]
- 56.Sullivan, J. T., B. D. Eardly, P. van Berkum, and C. W. Ronson. 1996. Four unnamed species of nonsymbiotic rhizobia isolated from the rhizosphere of Lotus corniculatus. Appl. Environ. Microbiol. 62:2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan, J. T., H. N. Patrick, W. L. Lowther, D. B. Scott, and C. W. Ronson. 1995. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. USA 92:8985-8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki, K., Y. Hattori, M. Uraji, N. Ohta, K. Iwata, K. Murata, A. Kato, and K. Yoshida. 2000. Complete nucleotide sequence of a plant tumor-inducing Ti plasmid. Gene 242:331-336. [DOI] [PubMed] [Google Scholar]
- 60.Tate, R., A. Riccio, E. Caputo, M. Iaccarinoi, and E. J. Patriarca. 1999. The Rhizobium etli metZ gene is essential for methionine biosynthesis and nodulation of Phaseolus vulgaris. Mol. Plant-Microbe Interact. 12:24-34. [DOI] [PubMed] [Google Scholar]
- 61.Thébault, P., F. Servant, T. Schiex, D. Kahn, and J. Gouzy. 2000. L'environnement iANT: integrated annotation tool, p. 361-365. In O. Gascuel and M.-F. Sagot (ed.), JOBIM Conference Proceedings. Springer-Verlag, Montpellier, France.
- 62.Tully, R. E., P. van Berkum, K. W. Lovins, and D. L. Keister. 1998. Identification and sequencing of a cytochrome P450 gene cluster from Bradyrhizobium japonicum. Biochim. Biophys. Acta 1398:243-255. [DOI] [PubMed] [Google Scholar]
- 63.Valderrama, B., A. Dávalos, L. Girard, E. Morett, and J. Mora. 1996. Regulatory proteins and cis-acting elements involved in the transcriptional control of Rhizobium etli reiterated nifH genes. J. Bacteriol. 178:3119-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Berkum, P., and B. D. Eardly. 1998. Molecular evolutionary systematics of the Rhizobiaceae, p. 1-24. In H. P. Spaink, A. Kondorosi, and P. J. J. Hooykaas (ed.), The Rhizobiaceae. Molecular biology of plant-associated bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 65.van Spronsen, P. C., M. Gronlund, C. P. Bras, H. P. Spaink, and J. W. Kijne. 2001. Cell biological changes of outer cortical root cells in early determinate nodulation. Mol. Plant-Microbe Interact. 14:839-847. [DOI] [PubMed] [Google Scholar]
- 66.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. de Vlaam, T. J. Regensburg-Tuink, and P. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 67.Viprey, V., A. Del Greco, W. Golinowski, W. J. Broughton, and X. Perret. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381-1389. [DOI] [PubMed] [Google Scholar]
- 68.Wang, C., E. Knill, B. R. Glick, and G. Defago. 2000. Effect of transferring 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase genes into Pseudomonas fluorescens strain CHA0 and its gacA derivative CHA96 on their growth-promoting and disease-suppressive capacities. Can. J. Microbiol. 46:898-907. [DOI] [PubMed] [Google Scholar]
- 69.Wang, K., C. Boysen, H. Shizuya, M. I. Simon, and L. Hood. 1997. Complete nucleotide sequence of two generations of a bacterial artificial chromosome cloning vector. BioTechniques 23:992-994. [DOI] [PubMed] [Google Scholar]
- 70.Young, J. M., L. D. Kuykendall, E. Martinez-Romero, A. Kerr, and H. Sawada. 2001. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola, and R. vitis. Int. J. Syst. Evol. Microbiol. 51:89-103. [DOI] [PubMed] [Google Scholar]