Abstract
We have identified a cluster of six genes involved in trehalose transport and utilization (thu) in Sinorhizobium meliloti. Four of these genes, thuE, -F, -G, and -K, were found to encode components of a binding protein-dependent trehalose/maltose/sucrose ABC transporter. Their deduced gene products comprise a trehalose/maltose-binding protein (ThuE), two integral membrane proteins (ThuF and ThuG), and an ATP-binding protein (ThuK). In addition, a putative regulatory protein (ThuR) was found divergently transcribed from the thuEFGK operon. When the thuE locus was inactivated by gene replacement, the resulting S. meliloti strain was impaired in its ability to grow on trehalose, and a significant retardation in growth was seen on maltose as well. The wild type and the thuE mutant were indistinguishable for growth on glucose and sucrose. This suggested a possible overlap in function of the thuEFGK operon with the aglEFGAK operon, which was identified as a binding protein-dependent ATP-binding transport system for sucrose, maltose, and trehalose. The Kms for trehalose transport were 8 ± 1 nM and 55 ± 5 nM in the uninduced and induced cultures, respectively. Transport and growth experiments using mutants impaired in either or both of these transport systems show that these systems form the major transport systems for trehalose, maltose, and sucrose. By using a thuE′-lacZ fusion, we show that thuE is induced only by trehalose and not by cellobiose, glucose, maltopentaose, maltose, mannitol, or sucrose, suggesting that the thuEFGK system is primarily targeted toward trehalose. The aglEFGAK operon, on the other hand, is induced primarily by sucrose and to a lesser extent by trehalose. Tests for root colonization, nodulation, and nitrogen fixation suggest that uptake of disaccharides can be critical for colonization of alfalfa roots but is not important for nodulation and nitrogen fixation per se.
The gram-negative soil bacterium Sinorhizobium meliloti is able to interact with the roots of Medicago sativa (lucerne or alfalfa) to form nitrogen-fixing nodules and survive as a free-living saprophytic bacterium in the soil. To successfully survive as a saprophyte, it must have the ability to compete with other soil microorganisms for the limited nutrient resources and to cope with different abiotic stresses, such as osmotic stress. As a soil bacterium, S. meliloti can be exposed to many different molecules originating from other bacteria, insects, plants, and fungi. For example, more than 400 different molecules have been identified in alfalfa that may become available to S. meliloti through exudation or decomposition (36). One of these compounds is the disaccharide trehalose [α-d-glucopyranosyl-(1,1)-α-d-glucopyranoside] (13, 36), a well-known osmoprotectant found in many different organisms (11). Trehalose occurs in plant tissues colonized by microorganisms, such as mycorrhizal roots (41), and in other symbiotic structures, including nitrogen-fixing nodules formed by legumes in response to rhizobial infection (13, 34). Under nonstress conditions, S. meliloti can take up trehalose and use it as its sole carbon and energy source (17). Under various stress conditions, S. meliloti synthesizes and accumulates trehalose, which can serve as a compatible solute (7, 22). In cultures grown at high osmolarity, exogenous trehalose stimulates the accumulation of intracellular trehalose (43); this has been found to decrease the generation time (18).
A variety of mechanisms for trehalose transport have been documented for prokaryotes. In Salmonella enterica serovar Typhimurium, for example, trehalose is transported via two pathways: the mannose-specific IIMan enzyme of the phosphoenolpyruvate:carbohydrate phosphoenoltransferase system (PTS) and the galactose permease pathway (37). Escherichia coli has two systems for trehalose uptake which are differentially used depending upon the osmolarity of the medium. Under conditions of high osmolarity, trehalose is hydrolyzed by a periplasmic trehalase to glucose, which is taken up by enzyme II of the PTS specific for glucose (5). At low osmolarity, trehalose is taken up by the use of the PTS-dependent enzyme IITre (27). Evidence for a similar PTS has also been reported for Vibrio parahaemolyticus (28). In the hyperthermophilic archaeon Thermococcus litoralis, a gene cluster has been found that encodes a maltose/trehalose transport system (24) which has homology to the malEGFK cluster of E. coli. This cluster belongs to the family of binding protein-dependent ATP-binding cassette transport systems (BPD ABC transport systems) that normally consist of a high-affinity substrate-binding protein located in the periplasm, two hydrophobic membrane proteins, and two cytoplasmic ATP-hydrolyzing subunits (6). Willis and Walker (47) identified the operon aglEFGAK from S. meliloti which encodes a periplasmic binding protein-dependent sugar transport system involved in the uptake of the α-glucoside disaccharides sucrose, maltose, and trehalose. However, since the agl mutants were able to utilize these disaccharides, the existence of at least one additional transport system for α-glucosides in S. meliloti was suggested (47).
In the genome of S. meliloti there are a predicted 430 ABC transport systems and over half are found on the megaplasmid pSymB (12). In this report, we demonstrate that an operon of four genes located in pSymB, called thuEFGK, encoding proteins belonging to the superfamily of BPD ABC transporters, is the second system suggested by Willis and Walker (47). By mutational analysis and transport and growth studies, we show that the thuEFGK operon is also involved in transport of trehalose, maltose, and sucrose. Despite their substrate overlap for these disaccharides, thuEFGK is induced only by trehalose, whereas algEFGAK is induced by sucrose and presumably also by trehalose. Furthermore, we show here that an S. meliloti mutant of these two transport systems is severely impaired in its ability to colonize alfalfa roots, whereas nodulation and nitrogen fixation are not affected.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains and plasmids used in this study are presented in Table 1. E. coli strains DH5α, S17-1/λpir, and S327/λpir were grown in Luria broth (39). S. meliloti strain Rm1021 and S. meliloti strain Rm9628 were grown in tryptone yeast (TY) medium or M9 minimal medium with an appropriate carbon source (39). Unless otherwise stated, antibiotics were added, when appropriate, at the following final concentrations: kanamycin, 50 μg/ml; streptomycin, 500 μg/ml; tetracyline, 5 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, plasmid, or transposon | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. meliloti | ||
| Rm1021 | SU47 str-21 Strr | 31 |
| Sm7019 | Rm1021 thuE::pThuE629, Φ(thuE′-lacZ) transcriptional fusion, Strr Kmr | This work |
| Sm7023 | Rm1021 thuA::Tn5-1062, Φ(thuA′-luxAB) transcriptional fusion, Strr Kmr | This work |
| Sm7025 | Rm9628 thuE::pThuE690 Strr Kmr Tetr | This work |
| Rm9628 | Rm1021 aglE192::Tn5 Strr Kmr | 47 |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | |
| S17-1/λpir | RK2 tra regulon, λpir, host for pir-dependent plasmids | 42 |
| S327/λpir | λpir, host for pir-dependent plasmids | 33 |
| Plasmids | ||
| pRL1062 | Bearing Tn5-1062, oriT(RK2) Kmr | 48 |
| pRK2013 | ColE1 Tra+ Kmr | 9 |
| pApa7023 | ≈20-kb ApaI fragment from S. meliloti Sm7023 containing a thuA::Tn5-1062, Kmr | This work |
| pVIK112 | lacZY for transcriptional fusions, Kmr | 25 |
| pThuE629 | pVIK112, 629-bp thuE internal fragment, Φ(thuE′-lacZ) transcriptional fusion, Kmr | This work |
| pThuE690 | pSUP205, 690-bp thuE internal fragment, Tetr | This work |
| pSUP205 | mob λcos Cmr Tetr | 42 |
| Transposon | ||
| Tn5-1062 | Tn5 derivative, promoterless luxAB, oriV Kmr | 48 |
Transposon mutagenesis.
Mutagenesis of S. meliloti Rm1021 was performed using pRL1062. pRK2013 was used as helper plasmid and triparental matings were performed according to the method of de Bruijn and Rossbach (8).
DNA sequencing.
To recover genomic sequences contiguous with the inserted transposon, DNA extracted from Sm7023 was digested with ApaI restriction enzyme (which does not cut within the transposon), ligated, and transformed into DH5α by electroporation (48). Selection of transformants on TY-kanamycin plates resulted in the isolation of plasmid pApa7023, which carried a 6.1-kb genomic S. meliloti DNA and was sequenced by primer walking from the Tn5 insertion using the primer 5′-CTAAGCTGCCTCCATCCATG-3′ for the first sequencing reaction. The entire nucleotide sequence was determined on both strands by using Taq FS DNA polymerase and fluorescent dideoxy terminators in a cycle sequencing method, and the resultant DNA fragments were separated by electrophoresis and analyzed using an automated Applied Biosystems 377 DNA Sequencer.
Sequence data analysis.
Putative open reading frames (ORFs) and translation products were determined using the programs MacVector 5.0.1 (Eastman Kodak) and CodonUse 3.1 (Conrad Halling, Department of Molecular Genetics and Cell Biology, University of Chicago, Chicago, Ill.). Protein searches were run with the program BLASTP (1) on the National Center for Biotechnology Information server (Bethesda, Md.). Sequence alignments were done using the program ClustalX (44).
Insertional inactivation of thuE.
For insertional mutagenesis of thuE in S. meliloti Rm1021, an internal thuE fragment of 629 bp (from bp +301 to +929 of the thuE ORF) was amplified by PCR using plasmid pApa7023 as a template. Primers used in the PCR were 5′-TGGAATTCGTCGACCTGACC-3′ (EcoRI; forward) and 5′-GATCTAGACCGCGAGGTTCC-3′ (XbaI; reverse), introducing the indicated (underlined) restriction endonuclease recognition sites. The PCR product was digested with EcoRI and XbaI restriction enzymes and cloned into vector pVIK112 (25), resulting in plasmid pThuE629, which was electroporated into E. coli SY327/λpir, a readily transformable E. coli strain. pThuE629 was reisolated, electroporated into E. coli S17-1/λpir, which has on the chromosome the tra region of RP4, and finally introduced by conjugation into S. meliloti strain Rm1021 (8). For insertional mutagenesis of thuE in S. meliloti strain Rm9628 , an internal thuE fragment of 690 bp (from bp +301 to +990 of the thuE ORF) was amplified by PCR using plasmid pApa7023 as a template. Primers used in the PCR were 5′-TGGAATTCGTCGACCTGACC-3′ (EcoRI; forward) and 5′-ATGAATTCGGCCGAGCCCAG −3′ (EcoRI; reverse), introducing the indicated restriction endonuclease recognition sites. The PCR product was digested with EcoRI restriction enzyme and cloned into vector pSUP205 (42), resulting in plasmid pThuE690, which was electroporated into E. coli DH5α and finally introduced by conjugation into S. meliloti strain Rm9628. Transconjugants were screened by replica plating for their ability to grow on TY or M9 minimal medium with 0.4% (wt/vol) trehalose and containing appropriate antibiotics. Disruption of the thuE locus was confirmed by Southern blot hybridization using the internal fragment of thuE as a probe; the resulting strains were called Sm7019 and Sm7025, respectively.
For plasmid DNA isolation, restriction enzyme digestion, and ligation, standard methods were used as previously described (39). All synthetic oligonucleotides used in this study were purchased from MedProbe (Oslo, Norway). Electroporation was performed with a Bio-Rad Gene Pulser according to the manufacturer's specifications.
Disaccharide uptake assay.
Cells were grown to exponential phase in M9 minimal medium with 0.2% (wt/vol) mannitol in the presence of 0.2% (wt/vol) trehalose, 0.2% (wt/vol) maltose, or 0.2% (wt/vol) sucrose. The cells were harvested by centrifugation (3,000 × g for 10 min) at room temperature and washed three times with M9 salts without any carbon source and adjusted with the same medium to a turbidity at 600 nm of 0.2. [14C]trehalose (1.1 GBq/mmol), [14C]maltose (2.3 GBq/mmol), or [14C]sucrose (2.1 GBq/mmol) was added to a final concentration of 1 μM and the suspension was incubated at 25°C. At various time points after addition, 110-μl aliquot samples were removed and filtered through 0.45-μm-pore-size filters (Millipore) under vacuum. The filters were washed with 5 ml of M9 salts and transferred to scintillation vials containing 4 ml of Ultima Gold scintillation liquid (Packard). The radioactivity trapped in the cells was measured with a scintillation counter (2300TR; Packard Instruments Co., Meriden, Conn.). [14C]maltose and [14C]sucrose were purchased from Amersham-Pharmacia (Little Chalfont, Buckinghamshire, England), whereas [14C]trehalose was prepared according to the method of Horlacher et al. (23). The protein content of the cultures was estimated using a Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.) with bovine serum albumin as the standard. Uptake rates were calculated for the first 4 min, during which time uptake was found to be linear.
Competition experiment.
Mid-log-phase cells were harvested as described above, washed three times with M9 salts without any carbon source, and resuspended in the same medium to an optical density at 600 nm (OD600) of approximately 0.2. Aliquots of the washed cells (650 μl) were transferred to 1.5-ml microcentrifuge tubes containing M9 medium with various carbon sources (see Table 2) at a final concentration of 100 μM along with [14C]trehalose (1.1 GBq/mmol) or [14C]sucrose (2.1 GBq/mmol) at a final concentration of 1 μM. At 2 min after addition, 110-μl aliquot samples were removed and treated as described above.
TABLE 2.
Effects of various competitors on the uptake of [14C]trehalose and [14C]sucrose in S. meliloti strains Rm1021, Sm7019, and Rm9628
| Competitor | Inhibition (%) of [14C]trehalose uptakea
|
Inhibition (%) of [14C]sucrose uptakea
|
||||||
|---|---|---|---|---|---|---|---|---|
| Sucrose-induced
|
Trehalose-induced
|
Sucrose-induced
|
Trehalose-induced
|
|||||
| Sm7019 | RM1021 | Rm9628 | Rm1021 | Sm7019 | Rm1021 | Rm9628 | Rm1021 | |
| None | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cellobiose | 89 | 70 | 0 | 17 | 84 | 91 | 5 | 65 |
| Glucose | 13 | 18 | 55 | 33 | 39 | 54 | 79 | 68 |
| Lactose | 4 | 0 | 10 | 4 | 9 | 18 | 0 | 23 |
| Maltose | 91 | 90 | 88 | 89 | 84 | 93 | 82 | 94 |
| Mannitol | 0 | 2 | 11 | 9 | 15 | 0 | 14 | 6 |
| Sucrose | 81 | 72 | 12 | 26 | 0 | 0 | 0 | 0 |
| Trehalose | 0 | 0 | 0 | 0 | 81 | 95 | 82 | 95 |
The results are expressed as mean percent inhibition of trehalose or sucrose uptake, and each value represents the mean for three different experiments. The transport rates with 1 μM concentrations of radiolabeled sugars served as control values. For competition studies, S. meliloti cultures were mixed with a 100-fold excess of the unlabeled sugar (100 μM) plus [14C]trehalose (1 μM) or [14C]sucrose (1 μM).
Induction assay.
Cells were grown in 100 ml of M9 minimal medium with 0.4% (wt/vol) mannitol at 28°C to an OD600 of between 0.2 and 0.3. The culture was divided into 10-ml aliquots in 50-ml flasks and incubated at 28°C on a rotary shaker (150 rpm), and various carbon sources were added to a final concentration of 0.4% (wt/vol). At different time points following the addition of carbon sources, samples were removed and assayed for β-galactosidase activity by the method of Miller (32).
Plant nodulation tests.
Nodulation assays were performed in Leonard jar assemblies containing sterile vermiculite and nitrogen-free Hoagland's medium (45). Five M. sativa (Moapa 69) seedlings were planted in each jar. Each treatment had three replicates, with three jars per replicate. Plants were grown with photosynthetically active photon (400 to 700 nm) flux density of 250 mol m−2 s−1 using a 12-h photoperiod at 25 and 20°C and 30 to 40% relative humidity. Nodulation was assessed 5 weeks after planting. A random sampling of nodules from each jar was checked for S. meliloti identity on TY extract plates with appropriate antibiotic selections. Symbiotic nitrogen fixation was assessed by dry weight determination. Roots and shoots were harvested separately from each jar and dried to a constant weight in an oven set at 75°C.
Root colonization test.
Root colonization assays were performed in Mejanta jars (Sigma Chemical Company) containing washed sterile vermiculite saturated with nitrogen-free Hoagland's medium (45). Vermiculite was washed two times with tap water and soaked overnight in nitrogen-free Hoagland's medium and used to fill the jars after the excess liquid was decanted. Five sterile alfalfa seedlings (48 h old) were planted in each jar and grown as described above. Each treatment had three replicates with three jars per replicate. The plants were inoculated at the time of planting with 100 to 200 bacterial cells per plant. The inoculum was prepared from early-stationary-phase cultures of S. meliloti grown in Vincent's medium (45) with glucose as the carbon source. Cells were harvested, washed once with sterile water, and suspended in sterile water diluted to contain approximately 103 cells/ml, and 100 μl was used to inoculate each plant. Roots were harvested 1 week after inoculation under sterile conditions and sonicated for 10 min in ice-cold buffer (0.05 M phosphate [pH 7.2]) containing 0.01% (wt/vol) Tween and vortexed for 30 s at maximum speed. Appropriate dilutions of these suspensions were plated onto selective media. All colonization experiments were repeated at least twice.
Statistical analysis.
Statistical analysis of the root colonization data was done using statistical software from Minitab Inc. Chi-square tests were performed to assess the significance between the observed and expected ratios of wild type to mutant cells in the harvested suspensions (46).
Nucleotide sequence accession number.
Sequence data for thuR, thuEFGK, and thuA have been deposited in the GenBank (National Center for Biotechnology Information) database under accession no. AF175299.
RESULTS
Identification of trehalose transport genes.
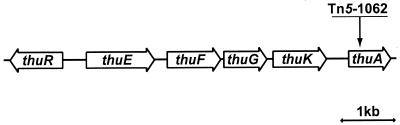
To isolate trehalose transport genes, transposon mutagenesis was performed using a derivative of Tn5 containing promoterless luxAB genes (48). Approximately 5,000 S. meliloti Rm1021 mutants were tested for their ability to grow on trehalose as sole carbon and energy source. One Tn5 insertion mutant, Sm7023, was isolated which was severely reduced in its ability to utilize trehalose as its sole carbon and energy source. Sm7023 was tested for its ability to utilize other carbon sources and was found to grow as well as the wild type in sucrose, maltose, glucose, succinate, and mannitol (data not presented). Computer-aided analysis of the DNA sequences surrounding this Tn5 insertion site revealed that the insertion disrupted an ORF. Comparison of the deduced amino acid sequence encoded by this ORF revealed high sequence identity (70%) and similarity (88%) to a protein with unknown function encoded by the mll3015 gene of Mesorhizobium loti (26). Since the phenotype of the mutant Sm7023 suggested that this gene is involved in trehalose utilization, we named the mutated ORF thuA. Upstream of the Tn5 insertion site, four ORFs that have strong homology to BDP ABC transport systems and a putative regulatory protein were detected (Fig. 1). Because of the similarity to trehalose transport genes of T. litoralis and the proximity to the gene thuA required for trehalose utilization, we have given this operon the provisional name thu for trehalose utilization. According to the published sequence data of the S. meliloti genome, the thu locus was found to be located on the pSymB megaplasmid (12).
FIG. 1.
Gene map of the trehalose utilization region of S. meliloti Rm1021. The insertion site of transposon Tn5-1062 in mutant Sm7023 is indicated.
The predicted amino acid sequence of another ORF exhibits similarity to various solute-binding protein components of BPD ABC transport systems. In particular, the putative protein has 37% identity and 71% similarity with the MalE trehalose/maltose-binding protein of T. litoralis (24). After the convention established for E. coli and T. litoralis maltose/trehalose uptake systems, we designate this ORF as thuE.
Downstream from thuE are two ORFs, thuF and thuG, whose protein products have a high degree of similarity to hydrophobic transmembrane components in the BPD ABC transport systems. Hydropathic profiles, determined according to the method of Kyte and Doolittle (29), and analysis for predicted transmembrane helices (3) suggest that both ThuF and ThuG contain six hydrophobic regions, corresponding to six membrane-spanning α-helical segments found in the inner membrane proteins of BPD ABC transport systems (6, 20). In addition to these six hydrophobic regions, the highly conserved sequence motif termed the EAA loop (EAA-X3-G-X9-I-X-LP), present in all inner membrane proteins from BPD ABC transport systems (6), was identified in ThuF and ThuG with minor variations. Downstream from thuG is thuK, which encodes a polypeptide whose predicted amino acid sequence reveals similarity to various ATP-binding proteins known to be components that energize the ABC transport systems (2, 21). Similarities between these proteins include conserved regions known as the Walker motif A (consensus G-X2-G-X-G-K-S/T), Walker motif B (consensus hhhh-D-E-P, where h stands for hydrophobic), and the linker peptide (consensus L-S-G-G-Q-Q/R/K-Q-R) that is unique to the ABC transport family (6, 40). An ORF upstream of thuE, designated thuR, extends in the opposite orientation from a GTG initiation codon (Fig. 1). The putative translation product of thuR is a polypeptide with an N-terminal sequence (KLKEFAKOLGLSPTTVS) that contains a helix-turn-helix motif characteristic of many DNA-binding proteins. Alignment of ThuR with other protein sequences present in GenBank revealed high homology to many of the known and putative transcriptional regulators involved in carbohydrate utilization which belong to the LacI-GalR family of regulatory proteins (35). The highest homology was found with the S. meliloti AlgR protein (49% identity and 78% similarity), a putative repressor involved in the regulation of the aglEFGAK operon encoding an α-glucosidase and a periplasmic BPD ABC transport system for α-glucosides (47).
thuEFGK is involved in trehalose, maltose, and sucrose uptake.
Due to the similarity between ThuE and the high-affinity trehalose/maltose transport system found in T. litoralis, we analyzed the parameters of trehalose transport in the wild-type strain, Rm1021. It was grown in M9 medium containing either mannitol or mannitol and trehalose as carbon sources. [14C]trehalose uptake was measured at concentrations ranging from 0.01 to 0.5 μM. Rm1021 had Km values of 8 ± 1 and 55 ± 5 nM in the uninduced and trehalose-induced cultures, respectively. The Vmaxs for trehalose transport were 1.0 ± 0.1 and 15 ± 0.1 nmol/min/mg of protein in these cultures. Km values in the submicromolar range confirmed that Rm1021 has a high-affinity transport system for trehalose.
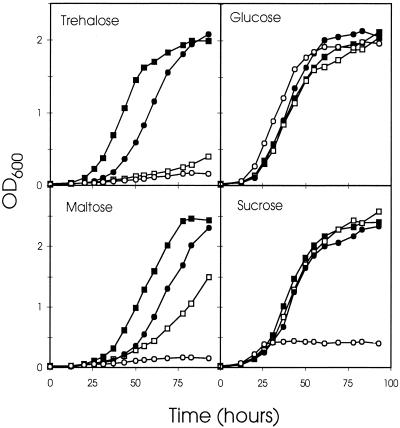
To test if thuE is involved in the uptake of trehalose and/or maltose in S. meliloti, we disrupted the thuE locus by creating a transcriptional fusion to the E. coli lacZ. The resultant strain, Sm7019, and the wild-type strain Rm1021 were tested for their ability to utilize trehalose as sole carbon and energy source. They were grown in M9 liquid medium containing 0.4% (wt/vol) glucose, sucrose, maltose, or trehalose. As shown in Fig. 2, the wild type and Sm7019 had indistinguishable growth curves on glucose. However, the mutant was clearly impaired in its ability to grow with trehalose as carbon source. Growth on maltose was also impaired, albeit to a lesser extent. No significant growth retardation was observed in sucrose. These results indicate that thuE encodes a protein that is involved in utilization of trehalose and maltose.
FIG. 2.
Growth of S. meliloti wild type and transport mutants on various disaccharides. Shown is the growth of Sm7019 (□), Sm7025 (○), Rm9628 (•), and Rm1021 (▪) in M9 medium containing various carbon sources at 0.4% (wt/vol), as indicated in the individual panels.
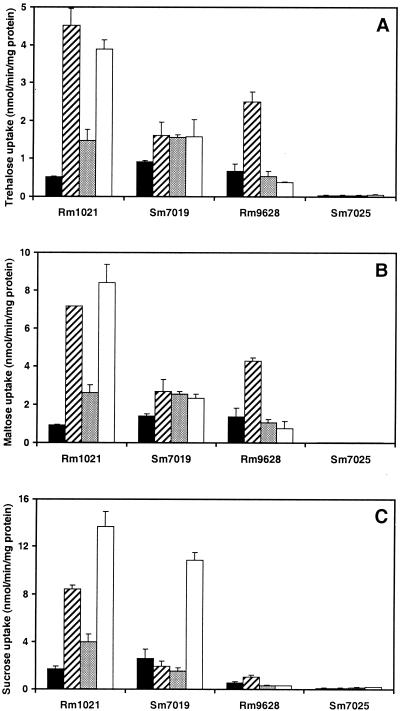
Uptake experiments were carried out with [14C]trehalose or [14C]maltose with S. meliloti Rm1021 and Sm7019 that had been grown in M9 minimal medium containing mannitol and various carbon sources. Because Sm7019 does not grow well on trehalose, Sm7019 and wild-type strains were cultured on mannitol in order to measure basal transport rates. Basal transport rates of trehalose and maltose for the wild type and Sm7019 were found to be the same when these strains were grown in minimal medium (Fig. 3A and B). When grown in minimal medium plus maltose, both the wild type and Sm7019 mutant showed a slightly enhanced rate of transport for both trehalose and maltose compared to cells grown on mannitol alone (Fig. 3A and B). However, the most significant change in transport rate was observed in the wild-type strain when it was grown in minimal medium containing mannitol plus trehalose. Uptake of both trehalose and maltose was enhanced eightfold in these treatments (Fig. 3A and B). The Sm7019 cultures grown in mannitol plus trehalose did not show this eightfold enhancement, suggesting that the thuEFGK operon is indeed responsible for this enhanced transport of both trehalose and maltose in Rm1021.
FIG. 3.
Uptake of [14C]trehalose, [14C]maltose, or [14C]sucrose by Rm1021, Sm7019, Rm9628, and Sm7025. Cultures were grown on mannitol plus trehalose (hatched), mannitol plus maltose (stippled), mannitol plus sucrose (white), or mannitol only (black). Each uptake value represents the mean for three different uptake experiments ± standard error.
Due to earlier findings suggesting that sucrose, maltose, and trehalose share a common uptake system in S. meliloti strains (17, 47), we wished to test the involvement of thuE in the uptake of sucrose. Uptake experiments were carried out with [14C]sucrose with S. meliloti Rm1021 and Sm7019 cells that had been grown in M9 media containing mannitol alone or with maltose, trehalose, or sucrose in addition to mannitol. Both the wild type and Sm7019 took up sucrose at a basal rate when grown on medium containing mannitol alone. Maltose enhanced the rate of sucrose transport twofold over the uninduced basal level (Fig. 3C). The presence of sucrose in the medium, however, resulted in a sixfold induction of sucrose uptake in the wild type and fourfold induction in the thuE mutant (Fig. 3C). Significant differences were also apparent when the thuE mutant and wild-type strains were grown in medium containing mannitol and trehalose (Fig. 3C). In Sm7019 cultures, the sucrose transport rate was comparable to the uninduced basal rate, while in the wild-type strain the presence of trehalose induced sucrose transport to up to 3.5 times over the basal rate (Fig. 3C). These results suggest that the thuEFGK operon may be involved in sucrose uptake as well as in the uptake of trehalose and maltose.
Sucrose-inducible trehalose uptake system.
The eightfold increase in maltose and trehalose uptake in cells cultured in the presence of sucrose indicates the presence of a sucrose-inducible uptake system for trehalose, maltose, and sucrose. Genes encoding such a sucrose-inducible uptake system could be the aglEFGAK operon from S. meliloti, shown by Willis and Walker (47) to be involved in uptake of the α-glucoside disaccharides sucrose, maltose, and trehalose. To test this hypothesis, uptake experiments with [14C]trehalose or [14C]maltose were carried out with S. meliloti Rm9628, a strain containing a mutation in the aglE gene that encodes the binding protein of the AglEFGAK transport system (47). Uptake experiments showed that Rm9628 cells grown in a trehalose-containing medium had increased rates of uptake of trehalose and maltose (four- and threefold, respectively) compared to the basal uninduced level (Fig. 3A and B). In contrast, only a twofold increase in the uptake of [14C]sucrose was seen in Rm9628 cells that had been grown on trehalose. No increase was apparent in maltose- or sucrose-containing media (Fig. 3C). These results indicate that even though the thuEFGK operon is involved in sucrose uptake, aglEFGAK is the operon which codes for the major transport system for sucrose. The results of the uptake experiments from Rm9628 and Sm7019 also indicate that sucrose and trehalose are the primary inducers of the aglEFGAK and thuEFGK operons, respectively. The fivefold increase in sucrose uptake by Rm1021 grown in trehalose-containing media suggests that trehalose may also induce the agl operon, albeit rather less efficiently (compare sucrose uptake between Rm1021 and Rm9628 grown in trehalose medium in Fig. 3C).
In order to test if there were additional transport systems for trehalose, maltose, and sucrose uptake in S. meliloti, we constructed an S. meliloti Rm9628 strain disrupted in the thuE locus as well. As expected, the resultant double mutant, Sm7025, was more strongly impaired in its ability to import these three disaccharides than was its parent strain, Rm9628, or the thuE mutant Sm7019 (Fig. 3). Regardless of the carbon sources supplied, the quantities of trehalose and maltose imported by Sm7025 amounted to less than 10% of the amounts imported by the wild type, its parent strain Rm9628, or the thuE mutant Sm7019 cultured under similar conditions. Sucrose import by Sm7025 was reduced to 25, 12, 55, and 60% of the amount imported by Rm9628 cells grown in mannitol or mannitol and trehalose, maltose, and sucrose, respectively (Fig. 3C). These results confirm that thuEFGK and aglEFGAK together code for the two major transport systems in S. meliloti involved in the uptake of the disaccharides trehalose, maltose, and sucrose.
Inhibition of trehalose and sucrose uptake by other disaccharides.
The fact that trehalose induces both its own uptake, the uptake of maltose, and to a lesser extent uptake of sucrose suggests that these two uptake systems have an overlapping specificity for uptake and induction. Glenn et al. (16) had shown in S. meliloti strain WU60 that sucrose, maltose, and trehalose shared a common uptake system, but we wished to confirm this finding in S. meliloti Rm1021. To determine if trehalose, maltose, and maybe other structurally related disaccharides use a common uptake system, inhibition experiments were carried out with cultures of Rm1021, Rm9628, and Sm7019 grown in M9 minimal medium containing mannitol and sucrose or trehalose. Mid-log-phase cells were assayed for [14C]trehalose or [14C]sucrose uptake in the presence of a 100-fold excess of unlabeled competitor. As shown in Table 2, the disaccharides cellobiose, sucrose, and maltose acted as strong inhibitors of [14C]trehalose uptake, and cellobiose, trehalose, and maltose inhibited [14C]sucrose uptake when Sm7019 and wild-type Rm1021 were grown in medium containing sucrose to induce the aglEFGAK operon. Lactose and the monosaccharides mannitol and glucose did not exhibit such strong inhibitory effects. These results suggest that AglEFGAK transports cellobiose, sucrose, maltose, and trehalose. When Rm1021 and Rm9628 were grown in media containing trehalose to induce the thuEFGK operon, [14C]trehalose uptake was strongly inhibited by the disaccharide maltose but not by the disaccharides cellobiose and sucrose. The monosaccharide glucose also inhibited trehalose uptake to a lesser extent (Table 2). These results indicate that the ThuEFGK system is involved primarily in the uptake of trehalose and maltose but not of sucrose or cellobiose.
The uptake of sucrose in cultures in which the thuEFGK operon was induced, on the other hand, presented a more complex picture. [14C]sucrose uptake by wild-type strain Rm1021 cultured in medium containing trehalose was inhibited >90% by maltose and trehalose and 65% by cellobiose (Table 2). This inhibition pattern suggests that both the agl and thu uptake systems are induced in these cultures. Uptake of [14C]sucrose by Rm9628 cultured in the presence of trehalose was inhibited only by maltose and trehalose, and not by cellobiose. These observations confirm that the AglEFGAK system is involved in the uptake of cellobiose. That strain Rm9628 had a much longer lag phase (>24 h) than Sm7019 and Rm1021 (<13 h) when cultured in the presence of cellobiose as the only source of carbon further supports this conclusion.
Growth tests.
When testing the Sm7025 strain for its ability to utilize trehalose, maltose, or sucrose as sole carbon and energy source, it was found that the mutant showed almost no growth on trehalose and maltose and sucrose (Fig. 2). These results confirm that, even though the ThuE system is not primarily involved in sucrose transport, ThuEFGK may be the alternative transport system suggested by Willis and Walker (47) for sucrose uptake.
Induction of thuE operon.
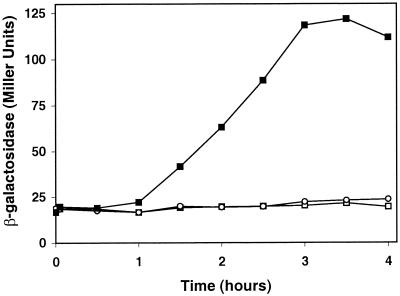
Since expression of a binding protein in S. meliloti may be regulated by its substrate (4, 30), we tested whether thuE expression is induced by trehalose and/or other sugars. Induction experiments were carried out with strain Sm7019 grown in M9 liquid medium containing 0.4% (wt/vol) mannitol to which trehalose, sucrose, or maltose was added to a final concentration of 0.4% (wt/vol). We assumed that residual uptake of the disaccharides in strain Sm7019 (Fig. 3) would allow for induction of the thuEFGK operon. β-Galactosidase activity was measured over time. In Fig. 4, it is shown that 1 h after transfer to trehalose-containing medium, thuE′-lacZ gene expression increased, whereas bacteria in the maltose- or sucrose-containing medium did not show any increased expression of β-galactosidase activity even after 4 h of incubation. To study the specificity of induction of this operon, similar experiments were carried out with Sm7019 cells exposed to compounds having structural similarities to trehalose. Among the nine different sugar compounds tested, only trehalose and lactose induced lacZ expression after the 4-h incubation period (by a factor of eight and two, respectively). Cellobiose, sucrose, glucose, lactose, maltopentaose, maltose, and mannitol failed to induce this operon (results not shown).
FIG. 4.
Induction of thuE′-lacZ gene expression by trehalose (▪), maltose (□), and sucrose (○) in S. meliloti Sm7019.
Effects of transport mutations on plant nodulation.
The symbiotic phenotypes of the mutant Sm7019 and the thuE algE double mutant were tested on alfalfa plants. The interaction resulted in the formation of pink normal-looking nodules similar to nodules seen on plants inoculated with the wild-type Rm1021 strain. The numbers of nodules formed per plant and plant dry weight were also not significantly different between the mutants and the wild type (data not presented). These nodulation tests indicate that the thuEFGK and aglEFGAK uptake systems are not critical for nodule development or symbiotic nitrogen fixation in alfalfa. However, these results do not rule out a role for sucrose and trehalose transport in competitive nodulation.
Effects of transport mutations on root colonization.
Since disaccharides have been shown to be a component of alfalfa root exudate (36), we compared the abilities of mutants Sm7019 and Rm9628 and double mutant Sm7025 to compete with the wild-type Rm1021 for growth on alfalfa root. In spite of considerable scatter in the total number of bacteria recovered from the roots between experiments, the results show (Table 3) that Sm7019, Rm9628, and Sm7025 were able to colonize alfalfa root as well as Rm1021 was when they were not competed against the wild type. In treatments where they were competed with Rm1021, Sm7019 and Rm9628 were significantly impaired in their ability to compete with the wild type, and Sm7025 was even more impaired than Sm7019 and Rm9628 in its ability to compete with the wild type (Table 3). These results suggest that disaccharides may be an important source of energy for S. meliloti growth on alfalfa roots.
TABLE 3.
Colonization of alfalfa root by S. meliloti strainsa
| Strain mixture | Mixture ratio (WT/MU) | Total cells per root (103)b | Rm1021 cells per root (103) | Mutant cells per root (103) | WT/MU (% ratio)c
|
|
|---|---|---|---|---|---|---|
| Expected | Observed | |||||
| Rm1021 + Sm7019 | 1:0 | 44.8 ± 8.0 B | NA | NA | ||
| 0:1 | 35.6 ± 8.3 B | NA | NA | |||
| 1:1 | 29.8 ± 5.1 B | 20.9 ± 4.5 | 7.8 ± 1.6 | 50:50 | 68:32* | |
| Rm1021 + Rm9628 | 1:0 | 38.6 ± 2.2 B | NA | NA | ||
| 0:1 | 64.4 ± 5.6 C | NA | NA | |||
| 1:1 | 70.0 ± 9.8 C | 43.8 ± 7.5 | 26.2 ± 3.3 | 50:50 | 60:40* | |
| Rm1021 + Sm7025 | 1:0 | 23.6 ± 2.0 A | NA | NA | ||
| 0:1 | 22.3 ± 3.5 A | NA | NA | |||
| 1:1 | 36.1 ± 6.2 B | 28.1 ± 4.7 | 7.7 ± 2.0 | 50:50 | 84:16* | |
WT, wild-type Rm1021; MU, mutant strain; NA, not applicable.
Treatments followed by the same letter were not statistically significant at the 95% confidence level. Data are means ± standard errors.
Chi-square test performed with Minitab statistical software;
P = <0.001.
DISCUSSION
In this paper, we report the identification of an operon in S. meliloti strain Rm1021 that is composed of a group of four genes, thuEFGK, which encodes a BDP ABC transport system for the disaccharides trehalose, sucrose, and maltose and is inducible by trehalose. To our knowledge this is the first report of a trehalose-inducible transport system for trehalose in S. meliloti. Willis and Walker (47) reported the presence of another operon, aglEFGAK, for uptake of the disaccharides trehalose, sucrose, and maltose. In spite of the fact that there are two transport systems for trehalose transport, our observations indicate that the thuE operon must be functional for optimal growth in trehalose. When the aglEFGAK operon alone is active, growth is significantly delayed and reduced, and no growth was observed when both operons were disrupted (Fig. 2). Our results show that the thuEFGK operon is involved primarily in the uptake of trehalose and maltose and only to a much lesser extent in sucrose uptake. For example, more than 95% of the total sucrose uptake in S. meliloti Rm1021 can be accounted for by the AglEFGAK system and only less than 5% of the total sucrose uptake by Rm1021 cultures can be attributed to the ThuEFGK system (Fig. 3C). The aglEFGAK operon thus has a broader specificity for disaccharide transport. In addition to sucrose, the aglEFGAK operon is also involved in the transport of trehalose, cellobiose, and maltose, as evidenced by results of the competitive inhibition studies on trehalose and sucrose uptake (Table 2).
The thuEFGK operon is induced only by trehalose and not by sucrose (Fig. 4). From the results of transport and competition experiments, we can deduce that the aglEFGAK operon is induced not only by sucrose but may also be induced by high concentrations of trehalose in the cell. Uptake of sucrose by Rm1021 cultures grown in medium containing trehalose is inhibited by cellobiose (65%), maltose (94%), and trehalose (95%). This pattern of inhibition is consistent with the aglEFGAK operon as well as the thuEFGK operon being induced by trehalose in these cultures. Uptake of sucrose by Rm9628 cultures grown in the presence of trehalose (where the agl operon is not active) is not inhibited by cellobiose. Our contention is that the induction of the aglEFGAK operon in Rm1021 grown in the presence of trehalose is effected through the trehalose transported by the functional ThuEFGK transport system in these cultures. The following observations are also consistent with this conclusion. (i) Sucrose transport is induced about fivefold in Rm1021 cultures grown with trehalose as the inducer (Fig. 3C). (ii) This induced uptake of sucrose in cultures where trehalose is the inducer requires that both the thuEFGK and aglEFGAK operons are functional (Fig. 3C); it is not observed in either trehalose-grown Rm9628 or Sm7019.
Our observations that more than 98% of the sucrose uptake in S. meliloti is due to the functional AglEFGAK system (Fig. 3C) confirm Willis and Walker's observation that this system is the major transporter for sucrose uptake (47). Nevertheless, the growth of Rm9628 in sucrose is not impaired (Fig. 2). This implies that the residual transport of sucrose in these cultures is sufficient for the growth observed and that sucrose must be a very efficient source of carbon for growth of S. meliloti. Our results that Sm7025 is unable to grow with sucrose as the sole source of carbon confirms that these two transport systems may be the only transport systems involved in the uptake of sucrose in S. meliloti. Willis and Walker (47) reported that the aglEFGAK operon encodes a transport system for the α-glucosides sucrose, trehalose, and maltose. Our results, however, suggest that this system also imports cellobiose, which is a β-glucoside (Table 2). Structural characteristics of the four disaccharides that strongly compete with the uptake of sucrose in Rm1021 are that they all contain a 1-linked glucosyl residue (trehalose, maltose, and cellobiose) or a glucosyl residue linked to a fructosyl residue (sucrose). Lactose, in contrast, which contains a galactosyl residue linked 1-4 to glucose, does not compete (Table 2). Whether the four strong competitors of sucrose uptake are reducing or nonreducing sugars seems to be unimportant, since both reducing (cellobiose and maltose) and nonreducing (trehalose and sucrose) disaccharides are equally competitive. Linkage of the monosaccharide moiety does not seem to play an important role, since the β-glucoside cellobiose is as strong a competitor of maltose and trehalose uptake as the α-glucosides maltose, sucrose, and trehalose. From the results given above, one can deduce that the aglEFGAK operon codes for a transport system that takes up a variety of disaccharides containing a 1-linked glucosyl residue. The presence of transporters with broad substrate specificities has been reported in different organisms, including an α-glucoside transporter capable of transporting several α-glucosides that has been documented in Saccharomyces (19).
Plant nodulation tests showed that the uptake of trehalose and sucrose does not play a crucial role in nodule development. However, its role in competitiveness for nodule occupancy cannot be ruled out. This finding is similar to the finding that other rhizobial mutants unable to utilize different hexoses formed effective nitrogen-fixing nodules (10, 16, 30, 38), but still, hexose import can be critical for competition for nodule occupancy (15). Lack of a nodulation phenotype for the mutants in disaccharide transport suggests that the functions may be important for the free-living rhizobium. We can only speculate on the possible advantages for S. meliloti to have two BPD ABC transport systems for disaccharides and particularly for trehalose sucrose and maltose. S. meliloti normally lives in a habitat where both the biotic and abiotic environments are in a state of flux. The availability of carbon sources can suddenly become limited, and therefore the presence of a high-affinity transport system may be necessary for S. meliloti to utilize these scarce carbon sources efficiently. It is thus not surprising that disaccharide uptake and utilization appears to be important in rhizosphere competition. Our observations show that Sm7019, Rm9628, and Sm7025 were impaired not in their ability to colonize the root per se (Table 3) but only in their ability to compete with the wild type. Sm7025, which is unable to import trehalose, maltose, and sucrose, was more impaired in competitive colonization than Sm7019 and Rm9628, suggesting that these disaccharides are important sources of carbon in alfalfa root exudate. We have observed induction of the thuE′-lacZ fusion when Sm7019 is growing on the root surface (data not presented). This observation and earlier reports (36) show that trehalose is available as a carbon source in the root exudate. The role of maltose in rhizobium-root interactions, however, is less obvious because the presence of maltose in alfalfa root exudates is not documented. To our knowledge this is the first report of disaccharide transport being critical for competitive root colonization by any rhizobium. Trehalose- utilizing Pseudomonas fluorescens strains were preferentially found in the mycorrhizosphere of Douglas fir (14). The ability to utilize trehalose likewise endows a competitive edge for root colonization in S. meliloti. These observations support the hypothesis of Finan et al. (12) that genes located on the pSymB megaplasmid are important, especially in enhancement of the competitive abilities of S. meliloti in the natural habitat.
Acknowledgments
This work was supported by a grant from the Norwegian Research Council.
The sequencing was performed in the W. M. Keck Facility at Yale University, New Haven, Conn. We also thank Leslie Wanner, U.S. Department of Agriculture, Beltsville, Md., for critically reading the manuscript. Part of this work was done at the Department of Agronomy and Range Sciences at the University of California—Davis. T.V.B. thanks D. A. Phillips and his colleagues for support during her stay at the University of California—Davis. The technical help of Eli Robertsen is gratefully acknowledged.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Mayers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ames, G. F.-L. 1986. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu. Rev. Biochem. 55:397-425. [DOI] [PubMed] [Google Scholar]
- 3.Bairoch, A., P. Bucher, and K. Hofmann. 1997. The PROSITE database, its status in 1997. Nucleic Acids Res. 25:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boncompagni, E., L. Dupont, T. Mignot, M. Østerås, A. Lambert, M.-C. Poggi, and D. Le Rudulier. 2000. Characterization of a Sinorhizobium meliloti ATP-binding cassette histidine transporter also involved in betaine and proline uptake. J. Bacteriol. 182:3717-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boos, W., U. Ehmann, H. Forkl, W. Klein, M. Rimmele, and P. Postma. 1990. Trehalose transport and metabolism in Escherichia coli. J. Bacteriol. 172:3450-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boos, W., and J. M. Lucht. 1996. Periplasmic binding protein-dependent ABC transporters, p. 1175-1209. In R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 7.Breedveld, M. W., C. Dijkema, L. P. T. M. Zevenhuizen, and A. J. B. Zehnder. 1993. Response of intracellular carbohydrates to a NaCl shock in Rhizobium leguminosarum biovar trifolii TA-1 and Rhizobium meliloti SU-47. J. Gen. Microbiol. 139:3157-3163. [Google Scholar]
- 8.de Bruijn, F. J., and S. Rossbach. 1994. Transposon mutagenesis, p. 387-417. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington D.C.
- 9.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan, M. J. 1981. Properties of Tn5-induced carbohydrate mutants in Rhizobium meliloti. J. Gen. Microbiol. 122:61-67. [Google Scholar]
- 11.Elbein, A. D. 1974. The metabolism of trehalose. Adv. Carbohydr. Chem. Biochem. 30:227-256. [DOI] [PubMed] [Google Scholar]
- 12.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fougére, F., D. Le Rudulier, and J. G. Streeter. 1991. Effects of salt stress on amino acid, organic acid, and carbohydrate composition of roots, bacteroids, and cytosol of alfalfa (Medicago sativa L.). Plant Physiol. 96:1228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frey, P., P. Frey-Klett, J. Garbaya, O. Berge, and T. Heulin. 1997. Metabolic and genotypic fingerprinting of fluorescent pseudomonads associated with the Douglas fir-Laccaria bicolor mycorrhizophere. Appl. Environ. Microbiol. 63:1852-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fry, J., M. Wood, and P. S. Poole. 2001. Investigation of myo-inositol catabolism in Rhizobium leguminosarum bv. viciae and its effect on nodulation competitiveness. Mol. Plant-Microbe Interact. 14:1016-1025. [DOI] [PubMed] [Google Scholar]
- 16.Glenn, A. R., R. Arwas, I. A. McKay, and M. J. Dilworth. 1984. Fructose metabolism in wildtype, fructokinase-negative and revertant strains of Rhizobium leguminosarum. J. Gen. Microbiol. 130:231-237. [Google Scholar]
- 17.Glenn, A. R., and M. J. Dilworth. 1981. The uptake and hydrolysis of disaccharides by fast- and slow-growing species of Rhizobium. Arch. Microbiol. 129:233-239. [Google Scholar]
- 18.Gouffi, K., N. Pica, V. Pichereau, and C. Blanco. 1999. Disaccharides as a new class of nonaccumulated osmoprotectants for Sinorhizobium meliloti. Appl. Environ. Microbiol. 65:1491-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, E.-K., F. Cotty, C. Sottas, H. Jiang, and C. A. Michels. 1995. Characterization of AGT1 encoding a general α-glucoside transporter from Saccharomyces. Mol. Microbiol. 17:1093-1107. [DOI] [PubMed] [Google Scholar]
- 20.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 21.Higgins, C. F., I. D. Hiles, K. Whalley, and D. J. Jamieson. 1985. Nucleotide binding by membrane components of bacterial periplasmic binding protein-dependent transport systems. EMBO J. 4:1033-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoelzle, I., and J. G. Streeter. 1990. Increased accumulation of trehalose in rhizobia cultured under 1% oxygen. Appl. Environ. Microbiol. 56:3213-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horlacher, R., R. Peist, and W. Boos. 1996. Improved method for the preparative synthesis of labeled trehalose of high specific activity by Escherichia coli. Appl. Environ. Microbiol. 62:3861-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horlacher, R., K. B. Xavier, H. Santos, J. DiRuggiero, M. Kossmann, and W. Boos. 1998. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 180:680-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalogeraki, V. S., and S. C. Winans. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188:69-75. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 27.Klein, W., R. Horlacher, and W. Boos. 1995. Molecular analysis of treB encoding the Escherichia coli enzyme II specific for trehalose. J. Bacteriol. 177:4043-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubota, Y., S. Iuchi, and S. Tanaka. 1982. Evidence for the existence of a trehalose-specific enzyme II of the phosphoenolpyruvate:sugar phosphotransferase system in Vibrio parahaemolyticus. FEMS Microbiol. Lett. 13:5-7. [Google Scholar]
- 29.Kyte, J., and R. F. Doolittle. 1985. A simple method for displaying the hydropathy character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 30.Lambert, A., M. Østerås, K. Mandon, M.-C. Poggi, and D. Le Rudulier. 2001. Fructose uptake in Sinorhizobium meliloti is mediated by a high-affinity ATP-binding cassette transport system. J. Bacteriol. 183:4709-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller, J., Z.-P. Xie, C. Staehelin, R. B. Mellor, T. Boller, and A. Wiemken. 1994. Trehalose and trehalase in root nodules from various legumes. Physiol. Plant. 90:86-92. [Google Scholar]
- 35.Nguyen, C. C., and M. H. Saier, Jr. 1995. Phylogenetic, structural and functional analyses of the LacI-GalR family of bacterial transcription factors. FEBS Lett. 377:98-102. [DOI] [PubMed] [Google Scholar]
- 36.Phillips, D. A., and W. R. Streit. 1997. Applying plant-microbe signalling concepts to alfalfa: roles for secondary metabolites, p. 319-342. In B. D. McKersie and D. C. W. Brown (ed.), Bio/technology and the improvement of forage legumes. CAB International, Wallingford, England.
- 37.Postma, P. W., H. G. Keizer, and P. Koolwijk. 1986. Transport of trehalose in Salmonella typhimurium. J. Bacteriol. 168:1107-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronson, C. W., and S. B. Primrose. 1979. Carbohydrate metabolism in Rhizobium trifolii: identification and symbiotic properties of mutants. J. Gen. Microbiol. 112:77-88. [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Saurin, W., and E. Dassa. 1994. Sequence relationships between integral inner membrane proteins of binding protein-dependent transport systems: evolution by recurrent gene duplications. Protein Sci. 3:325-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubert, A., P. Wyss, and A. Weimken. 1992. Occurrence of trehalose in vasicular-arbuscular mycorrhizal fungi and in mycorrhizal roots. J. Plant Physiol. 140:41-45. [Google Scholar]
- 42.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 43.Smith, L. T., G. M. Smith, M. R. D'Souza, J.-A. Pocard, D. Le Rdulier, and M. A. Madkour. 1994. Osmoregulation in Rhizobium meliloti: mechanism and control by other environmental signals. J. Exp. Zool. 268:162-165. [Google Scholar]
- 44.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vincent, J. M. 1970. A manual for the practical study of root nodule bacteria. IBP handbook 15. Blackwell, Oxford, United Kingdom.
- 46.Wardlaw, A. C. 2000. Practical statistics for experimental biologists, 2nd ed. John Wiley & Sons, Ltd., New York, N.Y.
- 47.Willis, L. B., and G. C. Walker. 1999. A novel Sinorhizobium meliloti operon encodes an α-glucosidase and a periplasmic-binding-protein-dependent transport system for α-glucosides. J. Bacteriol. 181:4176-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolk, C. P., Y. Cai, and J.-M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 88:5355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]