Abstract
Utilization of tightly linked ovo-A vs. ovo-B germline promoters results in the expression of OVO-A and OVO-B, C2H2 transcription factors with different N –termini, and different effects on target gene transcription and on female germline development. We show that two sex-determination signals, the X chromosome number within the germ cells and a female soma, differentially regulate ovo-B and ovo-A. We have previously shown that OVO regulates ovarian tumor transcription by binding the transcription start site. We have explored the regulation of the ovo-B promoter using an extensive series of transgenic reporter gene constructs to delimit cis-regulatory sequences as assayed in wild-type and sex-transformed flies and flies with altered ovo dose. Minimum regulated expression of ovo-B requires a short region flanking the transcription start site, suggesting that the ovo-B core promoter bears regulatory information in addition to a “basal” activity. In support of this idea, the core promoter region binds distinct factors in ovary and testis extracts, but not in soma extracts, suggesting that regulatory complexes form at the start site. This idea is further supported by the evolutionarily conserved organization of OVO binding sites at or near the start sites of ovo loci in other flies.
GERMLINE sex determination in Drosophila requires ovo+ (Oliver 2002). There are two primary germline sex-determination signals, an autonomous X chromosome karyotype signal (2X; the Y chromosome is not sex determining in Drosophila) and a nonautonomous inductive signal from the surrounding soma. The ovo gene acts downstream of these primary sex-determination signals to control 2X germ cell differentiation functions via ovarian tumor (otu) and ultimately Sex-lethal (Sxl) and to provide for 2X germ cell viability through an undefined pathway.
The ovo genes of Drosophila and mice encode C2H2 zinc-finger transcription factors (Mevel-Ninio et al. 1991; Garfinkel et al. 1992) required for germ cell and epidermal development (Oliver et al. 1987; Dai et al. 1998; Payre et al. 1999). Understanding the regulatory circuits upstream and downstream of ovo is complicated by the alternative mRNA the locus produces. Transcription from two closely linked start sites, ovo-A and ovo-B, gives rise to mRNA encoding two major C2H2 transcription factor isoforms and multiple variants due to alternative splicing (Mevel-Ninio et al. 1996; Andrews et al. 1998, 2000; Salles et al. 2002). The choice of promoters used is critical, as OVO-A is a negatively acting and OVO-B is a positively acting transcription factor (Andrews et al. 2000). Either the absence of OVO-B encoding transcripts (Andrews et al. 2000) or excessive and/or precocious expression of OVO-A encoding transcripts (Mevel-Ninio et al. 1996; Andrews et al. 1998, 2000) results in female sterility due to various degrees of defective oogenesis, including the complete absence of germ cells. Production of positive and negative transcription factors from ovo loci may also be conserved, as dual ovo promoters and ORFs are found in the distantly related olive fruit fly, Bactrocera oleae (Khila et al. 2003).
Ovo-A and ovo-B promoters are active in the female germline, but show differences in overall expression levels and perhaps pattern—the ovo-A promoter being considerably weaker and perhaps active later in oogenesis. RT-PCR and reporter gene expression show that both promoters are also active in the male germline (Mevel-Ninio et al. 1996; Andrews et al. 2000; Andrews and Oliver 2002). However, male germline development requires neither OVO-A nor OVO-B encoding transcripts (Andrews et al. 2000). The direct downstream target locus, otu, is strongly upregulated by OVO-B expression and strongly downregulated by OVO-A expression (Lu and Oliver 2001). The otu locus is, in turn, required for the regulation of germline sexual identity and oogenesis by Sxl (Oliver 2002).
While we know that the ovo locus (the sum of ovo-A and ovo-B expression) responds to sex-determination signals and regulates a downstream sex-determination pathway, we are just beginning to explore the specific roles of the ovo-A and ovo-B isoforms in germline sex determination. For example, the ovo locus (Oliver et al. 1994; Hinson and Nagoshi 1999; Waterbury et al. 2000), and the ovo-B promoter in particular (Andrews and Oliver 2002), is regulated by both a 2X germline karyotype (Oliver et al. 1994; Hinson and Nagoshi 1999; Andrews and Oliver 2002) and a female soma (Waterbury et al. 2000; Andrews and Oliver 2002). The regulation of the ovo-A promoter by sex-determination signals is unexplored. In this article we present evidence that the two alternative ovo promoters are differentially controlled by primary sex-determination signals.
Regulated transcription of genes typically depends on a basal core promoter and cis-regulatory modules that enhance or silence transcription from that core promoter. To better understand the cis-regulation of the ovo-B promoter, we compare the transcriptional activity of a series of deletion constructs stably introduced into flies by P-element-mediated transformation. In wild-type flies, a compact region surrounding the ovo-B core promoter is sufficient for the correct pattern of ovo-B expression, although flanking regions clearly augment ovo-B expression. Thus, the required sequences for ovo expression are very close to, or indeed at, the transcription start site, as is the case with the similarly organized otu locus (Lu et al. 1998; Lee and Garfinkel 2000; Lu and Oliver 2001). There are OVO DNA-binding sites at +1 of ovo-B and OVO footprints completely occlude the transcription start sites of both ovo-B and otu (Lu et al. 1998; Lee and Garfinkel 2000). This is an unusual location for a cis-regulatory element, as the polymerase complex must occupy this same region. We provide reporter, biochemical, and comparative genomic evidence suggesting that this binding site is functional.
MATERIALS AND METHODS
Drosophila culture and histology:
We used standard Drosophila techniques throughout (Ashburner 1989). Flies were grown at 25° ± 0.5° on PB or Gif media (KD Medical, Columbia, MD). Reporter gene expression was monitored in fixed gonads of flies heterozygous for the reporter that were stained with X-Gal as described by Pauli et al. (1993), except that the gonads where preincubated in staining buffer at ∼22° overnight. Alleles (with FlyBase identifications) used in this study were: ovoD1rv23 (FBal0012400), ovoD1 (FBal0013375), ovoΔap (FBal0104461), Df(2R)Trix (Fbab5072), tra-2B (FBal0017022), trahs.PB (FBal0035817), and traHsp83.PS (FBal0044393). See FlyBase (2003) for details and additional references. All DNA positions in the reporter constructs follow convention (Mevel-Ninio et al. 1991). The ovo-A start site is at +361 and the ovo-B start site is at +853. We also refer to positions relative to the specific transcription start sites in the text.
Transgenes:
Standard molecular biology techniques were use throughout (Sambrook et al. 1989). We used a series of transgenic flies bearing deletions of the ∼1-kb region, which replicates the wild-type pattern of ovo-B and ovo-A expression in wild-type adult gonads (Table 1). New reporter genes designed to map cis-regulatory regions were derived from previously described reporters (Andrews et al. 2000). They are: ovo::lacZ1.1 (FBal0104821), ovo::lacZΔa (FBal0104823), ovo-B::lacZ (FBal0123190), and ovo::lacZΔbp (FBal0104822). Deletions in the pCaSper-βgal-based plasmids were generated by removing the ovo BamHI fragments. Site-directed deletions were then introduced by PCR. Amplicons with deletions were directionally recloned at unique BglII and AgeI sites. We verified deletion boundaries by sequencing with fluorescent dye terminators (ABI-PRISM, dRhodamine Terminator cycle sequencing, and an ABI-377, Perkin-Elmer, Norwalk, CT). Completed constructs were stably introduced into flies using P-element-mediated transformation.
TABLE 1.
ovo reporter genes
| Reporter | Lines examined |
Deletiona (bp) |
|---|---|---|
| lacZΔap | 9 | 345–493 |
| lacZΔapΔ1 | 1 | 1–100, 345–493 |
| lacZΔapΔ2 | 3 | 1–228, 345–493 |
| lacZΔapΔ3 | 5 | 1–327, 345–493 |
| lacZΔapΔ4 | 2 | 1–589 |
| lacZΔapΔ5 | 3 | 1–728 |
| lacZΔapΔ6 | 6 | 1–814 |
| lacZΔapΔ7 | 3 | 1–100, 345–493, 971–1082 |
| lacZΔapΔ8 | 4 | 1–100, 345–493, 887–1082 |
| ovoB::lacZ | 3 | 1–781, 911–1082 |
| lacZΔbp | 3 | 831–978 |
| lacZΔbpΔ1 | 2 | 1–100, 831–978 |
| lacZΔbpΔ3 | 3 | 1–228, 831–978 |
| lacZΔbpΔ5 | 3 | 831–978, 987–1082 |
| lacZΔbpΔ6 | 2 | 631–1082 |
| lacZΔbpΔ8 | 3 | 416–1082 |
| lacZΔbpΔ9 | 1 | 536–730, 831–978 |
Relative to lacZ1.1.
Gel shifts:
Gel mobility shift assays of mutated OVO binding sites with bacterially expressed OVO DNA-binding domain polypeptides were performed according to Lu et al. (1998). Mutated binding sites were embedded in a 23-bp double-stranded oligonucleotide from the otu core promoter. For gel shifts using protein from fly tissues, the wild-type ovo-B transcription start site oligo was TCCTTTTTACAGTTACATAGCAA and the competing oligo was CTTAATTTAACGTTTAACAAATC (the nonamer corresponding to the putative site is underlined). The ovaries, testes, and carcasses of male and female adult flies were dissected and frozen in liquid nitrogen and stored at −70°. The tissues were homogenized in high-salt buffer (one volume tissue in three volumes buffer; 20 mm HEPES, pH 7.9, 25% glycerol, 800 mm KCl, 0.2 mm EDTA, 0.2 mm PMSF, 0.5 mm DTT) on ice. Following centrifugation (15,000 rpm in a microfuge for 15 min at 4°), the supernatants were collected and diluted with KCl-free high-salt buffer to a final KCl concentration of 100 mm. Diluted protein extract was centrifuged again, the supernatants were collected, and aliquots were stored at −70°. Protein (4 μg, as determined by Bradford assays) was used in the gel-shift assays with 1 ng of labeled oligo in the reaction buffer [10 mm Tris, pH 7.5, 2 μg of poly(dI-dC), 50 mm NaCl, 50 μm ZnCl2, 1 mm DTT, 2.5% Ficoll 400, 0.1% NP-40, and 50 μg BSA].
Informatics:
All sequences were obtained from GenBank (Benson et al. 2004). The Drosophila melanogaster sequence (Celniker et al. 2002) was AE003433 gi:22831713. Although the ovo gene sequence of several other Drosophila species is available, noncoding regulatory region sequence is not included. To obtain these sequences from other species, the putative regulatory region of ovo (−1 kb from the start of exon 2, which is common to both ovo-A and ovo-B transcripts) was compared with blastn (Altschul et al. 1990) against contigs from preliminary assemblies for D. pseudoobscura, D. virilis, D. yakuba, and D. simulans. While we were able to locate good hits to D. virilis ovo (tblastn, e = 1e-056) in the Agencourt provisional assembly (http://rana.lbl.gov/drosophila/assemblies/dvir_agencourt_arachne_12jul04.tar.gz), we failed to find significant matches within the noncoding region upstream of the ovo ORF and could not unambiguously identify the putative transcription start site for alignment with the ovo-B promoter of other ovo genes. We therefore excluded D. virilis from this study. Sequence 1 kb upstream of the start of exon 2 of B. oleae ovo (Khila et al. 2003) was extracted from BOL535757, accession AJ535757, gi:27656719. VISTA sequence alignments were used to compare each species to D. melanogaster (Mayor et al. 2000).
We compiled a list of OVO binding sites on the basis of previous DNAse protection “footprint” assays and SELEX (Lu et al. 1998; Lee and Garfinkel 2000), as well as derivative binding sites analyzed by DNA mobility shifts in this report. The resulting sites (36 of which are unique) were aligned and a position-specific scoring matrix was calculated. A pseudocount of 0.01 was added to each cell (King and Roth 2003). Additionally, we generated a position weight matrix (PWM) with background base frequency corrections of 0.3 (T, A) and 0.2 (C, G; Lenhard et al. 2003; Sandelin and Wasserman 2004). We wrote a perl script to calculate the PWM score of each possible nonamer in the promoter regions of ovo loci or non-ovo control sequences. To examine the significance of putative binding site enrichment, we compared results from the 5′-untranslated region (UTR), D. melanogaster core promoters, and random sequence. Raw and UTR sequences were downloaded from FlyBase (release 3.2.1). Random 100-bp sequences were extracted from the raw sequence via a perl script that uses the function “rand” to select random positions along the sequence. Core promoter sequences (Ohler et al. 2002) were downloaded from the Berkeley Drosophila Genome Project (http://www.fruitfly.org/seq_tools/datasets/Drosophila/promoter/prom/promoter_all_1941.fa). P-values are the frequency of control 100-mers showing equal or greater numbers of high-scoring nonamers when compared to the ovo sequences.
RESULTS
Expression of ovo-B and ovo-A promoters in wild-type and sex-transformed gonads:
We know little about promoter selection at the two closely linked promoters at the ovo locus. To test the hypothesis that primary sex-determination signals control the use of alternative transcription start sites, we compared the expression of the ovo-A and ovo-B promoters in response to a 2X karyotype and the sexual identity of the soma (Figure 1).
Figure 1.—
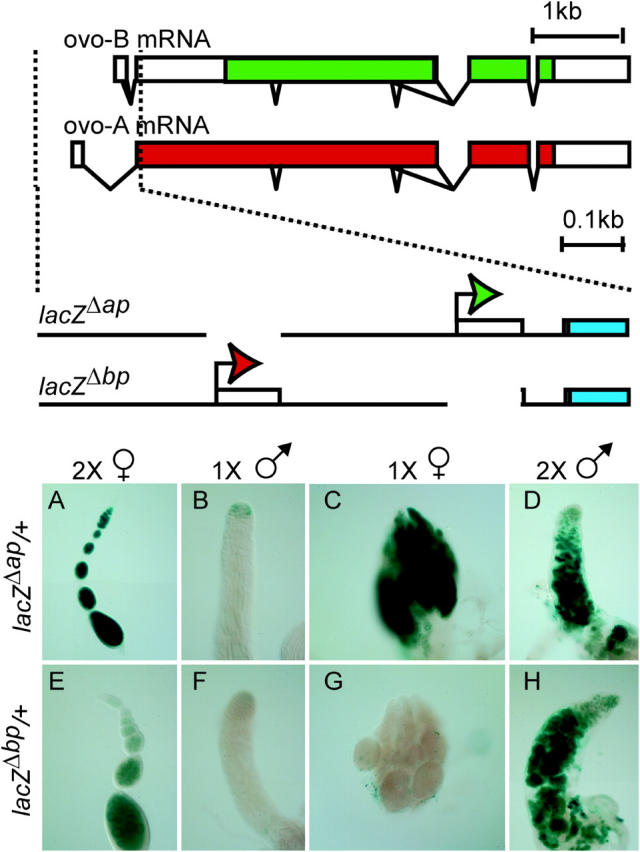
Expression of ovo-B- and ovo-A-specific reporters. The organization of the ovo locus and germline transcripts (top) and an expanded view outlining the structure of the base ovo-A- and ovo-B-specific reporters (bottom) are shown above the gonad photos. Shown are: ovo promoters (solid bent arrows), exons (bars), introns (bent lines), open reading frames (solid bars) encoding repressive (red) and activating (green) OVO isoforms, and lacZ encoding fragments (blue bars). The alternative exons defining the ovo-A and ovo-B mRNA are spliced to a common exon 2. Photomicrographs show expression of ovo-B and ovo-A reporters in wild-type and sex-transformed gonads. The sex chromosome karyotype and somatic differentiation phenotype are shown at the top and the reporter genotype is shown at the left. (A and E) Wild-type females. (B and F) Wild-type males. (C and G) Males transformed into females with traHsp83.PS. (D and H) Females transformed into males due to absence of tra-2 [tra-2B/Df(2R)Trix].
The functional unit of the ovary is the ovariole. From anterior to posterior, the ovarioles contain stem cells, dividing cystocytes, and young 16-cell egg chambers in the germarium, followed by progressively more advanced egg chambers and ultimately eggs, all arranged along the length of the ovariole (Spradling 1993). Wild-type females bearing the ovo-B reporter ovoΔap::lacZ (lacZΔap) showed robust expression in all the germline cells from the germarium to differentiated egg chambers, with later stages showing the strongest staining (Figure 1). The expression of the ovo-A reporter ovoΔbp::lacZ (lacZΔbp) was much weaker than that of lacZΔap with no overt staining in the anterior third of the germarium, light staining in middle regions of the germarium, and decreased staining in early egg chambers, followed by increased staining in later stages (Andrews et al. 2000; Andrews and Oliver 2002).
Germ cells in the testis are also arranged from anterior to posterior, with stem cells arranged around the hub at the apex; dividing cystocytes are also found within the apex, with growing spermatocytes, spermatids, and sperm arranged to the posterior (Fuller 1993). Expression of the lacZΔbp or lacZΔap reporters in the testis is weak and restricted to the apex (Figure 1) as previously reported (Andrews et al. 2000; Andrews and Oliver 2002).
To examine the effect of a female soma on reporter expression we utilized either of two constitutive transformer (tra) alleles, traHsp83.PS and trahs.PB, both of which encode the female-specific TRA protein. Constitutive expression of TRA transforms 1X animals into somatic females with ovarian tumors (Steinmann-Zwicky et al. 1989; FlyBase 2003). The precise sexual nature of the tumor cells is controversial (Oliver 2002), but they are certainly poorly differentiated. Both lacZΔap and the endogenous ovo-B promoter show clear, but weak, expression of ovo-B in 1X trahs.PB females (Andrews and Oliver 2002), but in 1X traHsp83.PS females we find moderate to strong expression of lacZΔap (Figure 1). In contrast to the ovo-B reporter lacZΔap, which is clearly expressed in 1X males transformed into females, the ovo-A reporter lacZΔbp showed no overt staining in 1X female flies (Figure 1). Thus, a female soma supports the expression of ovo-B, but not ovo-A.
To examine the effect of a 2X karyotype on ovo reporter expression, we transformed females into anatomical males using loss-of-function alleles of transformer-2 (tra-2). This results in a 2X male with a spermatogenic germline, although the number of germ cells is greatly reduced (Nöthiger et al. 1989). These 2X males showed strong expression of lacZΔap (Figure 1). Interestingly, 2X males bearing lacZΔbp also showed very strong staining (Figure 1) that exceeded the staining seen in the egg chambers of wild-type 2X females and dramatically exceeded the expression in the 2X germarium. In summary, we observed strong ovo-B expression in all 2X germ cells regardless of somatic phenotype, while we observed stronger ovo-A expression in 2X males than in 2X females. The strong expression of ovo-A in 2X females transformed into somatic males along with the weak expression in 1X males transformed into females suggests that sex-determination signals differentially regulate ovo-A vs. ovo-B promoters.
Mapping cis-regulatory domains:
To identify cis-regulatory regions directing ovo expression, we used a computational approach coupled with a deletion analysis. We compared the sequences of the ∼1-kb ovo promoter region from D. melanogaster, D. simulans, D. yakuba, D. pseudoobscura, and B. oleae and made a series of constructs in the D. melanogaster base reporters, lacZΔap and lacZΔbp, introduced them into the genome of D. melanogaster by P-element-mediated transformation, and then tested them in wild-type and sex-transformed flies (Figure 2).
Figure 2.—
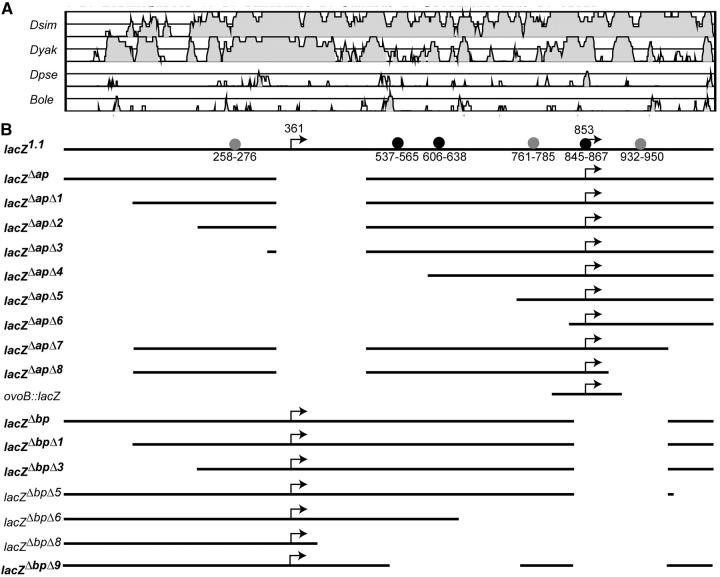
Genomic alignments and reporters for ovo expression. (A) VISTA plots showing the alignments between the 1-kb ovo control region of D. melanogaster aligned against D. simulans (Dsim), D. yakuba (Dyak), D. pseudoobscura (Dpse), and B. oleae (Bole). For each plot the lowest mapped score is 50% and the maximum is 100%. The line bisecting the plots is 75% and regions showing >80% homology are shaded. (B) D. melanogaster reporter genes are also shown. The names of the reporters are shown (left) and those showing detectable expression are indicated (boldface type). DNA present in the reporter is indicated by lines. Positions of the transcription start sites are indicated (bent arrows). On the base construct lacZ1.1, the positions of OVO footprints (the entire protected region, not simply the consensus sites) are indicated (circles). Strong footprints (solid circles) and weak footprints (shaded circles) are also indicated, along with the coordinates.
Sequences of different species of flies were compared to the D. melanogaster sequence (Figure 2A) by using VISTA plots (Mayor et al. 2000). These data show that the flies more closely related to D. melanogaster, D. simulans [∼5.4 million years ago (MYA)], and D. yakuba (∼22.8 MYA; Tamura et al. 2004) have not diverged sufficiently to highlight discrete regions. Sequence alignments of the more distantly related flies, D. pseudoobscura (∼54.9 MYA; Tamura et al. 2004) and B. oleae (80–100 MYA; Khila et al. 2003), reveal limited homology to D. melanogaster in the ovo promoter region. However, given that the B. oleae sequences in question are able to support the expression of a YFP reporter gene in the D. melanogaster ovary, it is clear that sequences critical for ovo regulation are present (Khila et al. 2003). There are two short regions of homology between B. oleae and D. melanogaster upstream of ovo-A, two more between the ovo-A and ovo-B promoters, and one between ovo-B and the exon 2 junction. There is some homology between D. pseudoobscura and D. melanogaster in the vicinity of the B. oleae and D. melanogaster homologies, but the most striking D. pseudoobscura and D. melanogaster homology is at the ovo-B transcription start site.
The deletion series built from lacZΔap revealed that important cis-regulatory regions for expression are near the ovo-B start site. Females bearing any of the six 5′ deletions (lacZΔapΔ1 through lacZΔapΔ6) showed at least some expression in the germline (Figure 2B; Figure 3, A–L; additional data not shown), indicating that no regions upstream of −39 bp from the ovo-B start site are obligatory for female germline expression. Similarly, the lacZΔapΔ8 reporter was expressed to a clearly detectable extent even though all sequences 3′ of +35 bp from the ovo-B start site were removed (Figure 2B; Figure 3, M–P). These data indicate that the −39- to +34-bp region bears information required for at least some expression of ovo-B in the appropriate spatial and temporal pattern. The −71- to + 58-bp core promoter construct ovo-B::lacZ showed no detectable expression, as previously reported (Lu and Oliver 2001), clearly indicating that the 129-bp core promoter cannot function in the absence of any flanking cis-regulatory regions. Therefore, we conclude that ovo-B expression requires either upstream or downstream elements and the core promoter.
Figure 3.—
Examples of ovo-B reporter expression in wild-type and sex-transformed flies. The sex chromosome karyotype and differentiation phenotype are shown at the top and the reporter genotype is shown at the left. (A–D) Heterozygous for ovoΔapΔ4. (E–H) Heterozygous for ovoΔapΔ5. (I–L) Heterozygous for ovoΔapΔ6. (M–P) Heterozygous for ovoΔapΔ8. (A, E, I, and M) Wild-type females. (B, F, J, and N) Wild-type males. (C, G, K, and O) Males transformed into females with traHsp83.PS. (D, H, L, and P) Females transformed into males due to absence of tra-2 [tra-2B/Df(2R)Trix].
We applied the same deletion strategy to exploring the regulation of the ovo-A promoter (Figure 2B; data not shown). The results were straightforward. The 5′ deletions in the lacZΔbp, lacZΔbpΔ1, or lacZΔbpΔ3 constructs had no effect on ovo-A expression in 2X females. Similarly, the deletion of sequences between the ovo-A and ovo-B transcription start sites (lacZΔbpΔ9) did not abolish transcription, whereas all the 3′ deletions greatly reduced or abolished expression, indicating that the region downstream of the ovo-B promoter (+126 to +230 bp from the ovo-B start site) is required for proper expression from ovo-A.
All of the ovo-B reporters (other than the core promoter construct ovoB::LacZ, which was not detectably expressed) were expressed in germ cells with either a 1X or a 2X karyotype residing in either a female or a male soma (Figure 3; additional data not shown). Similarly, 1X flies transformed from males into somatic females expressed none of the ovo-A reporters, except for the single lacZΔbpΔ9 line. Thus, the deleted reporters show a wild-type response to sex-determination signals. While there may be sequences responding preferentially to a 2X karyotype and a female soma, these are not neatly delimited modules—one responding to karyotype and another responding to somatic sexual identity.
Distinct gonadal proteins bind at the ovo-B transcription start site:
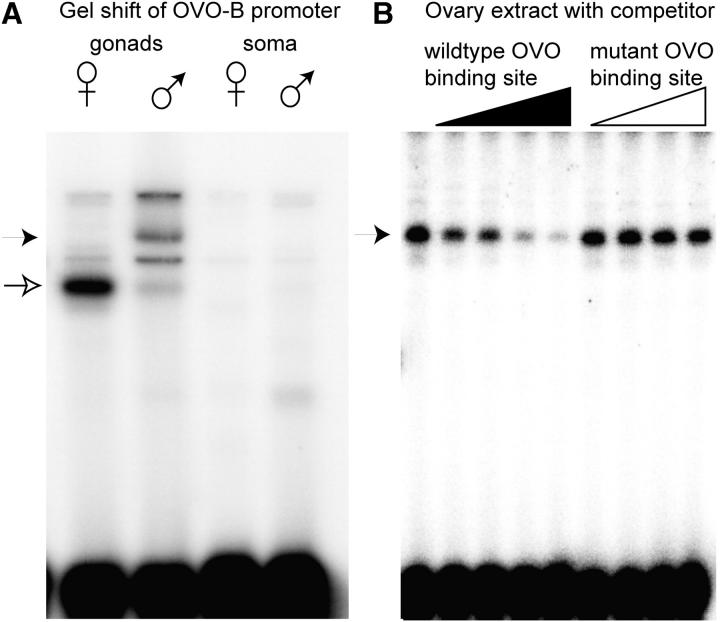
The results of the reporter deletion study suggests that the ovo-B core promoter region (the ∼60-bp region that is occupied by initiating RNA polymerase) bears regulatory information and is not simply a basal promoter depending only on more distant instruction by enhancers. This same region was previously studied in the context of the otu locus, where the ovo-B core promoter could substitute for the otu promoter, while core promoters without OVO binding sites could not (Lu and Oliver 2001). If the core promoter has some regulatory function, we can predict that it will be bound by regulatory proteins to mediate activation or derepression in the female germline. To test for such binding activities, we performed DNA mobility shift assays on the ovo-B core promoter using proteins extracted from ovaries and testes, as well as female and male nongonadal soma (Figure 4A). Striking differences were observed in these DNA mobility shift assays. A very strong shift is observed following incubation of the ovo-B core promoter with ovary extract, and there was a different pattern of strong shifts in extracts from testis. The female shifting activity is consistent with positive action at the ovo-B core promoter. The strikingly different pattern of mobility shifts using testis extracts raises the possibility that there are distinct core promoter-binding complexes in the two tissues. The testis complex may be repressive or weakly activating, while the ovarian complex is strongly activating. There was little shifting activity from extracts from either male or female soma. This is consistent with the idea that the absence of ovo-B reporter expression in the soma (data not shown) is due to the absence of activation at the core promoter, rather than repression. While we have been unable to determine if the proteins binding to the core promoter are derived from ovo (further purification has not been successful), it is clear that wild-type, but not mutant, OVO binding sites compete for the binding activity (Figure 4B). This raises the possibility that OVO binding at the ovo-B core promoter is important. Thus, looking for this site in other species could be informative.
Figure 4.—
Gel mobility shifts using the ovo-B core promoter and extracts from gonadal and somatic tissues. (A) Shifting activities from proteins extracted from female or male gonads or nongonadal soma were used. The source of protein for the gel shifts is indicated above the lane. Bands highly enriched in shifts using ovarian extracts (open arrow) or testis (solid arrow) are indicated. (B) Similar gel shifts performed with increasing amounts of unlabeled wild-type OVO binding site oligo or mutated oligo. The mobility of the shifted band in the absence of competitor is indicated (arrow).
Comparative genomic analysis of OVO binding sites:
Before the comparative genomic analysis, we refined the OVO binding site definition and made new scoring matrices on the basis of the previous footprinting and SELEX studies (Lu et al. 1998; Lee and Garfinkel 2000), augmented with analysis of OVO binding to mutated sites (Figure 5; additional data not shown). The new gel-mobility shift data confirm the importance of the central GTT core of previously identified sites. For example, if the G residue in the fourth position of the strongly binding ACCGTTACA motif is changed to C, binding is abolished and mutating either of the T's in fifth and sixth positions greatly reduces OVO binding (Figure 5A). We have now generated a list of confirmed OVO binding sites. The resulting 9-bp consensus motif is similar to those previously reported (ACNGTTACA; Figure 5B; Table 2).
Figure 5.—
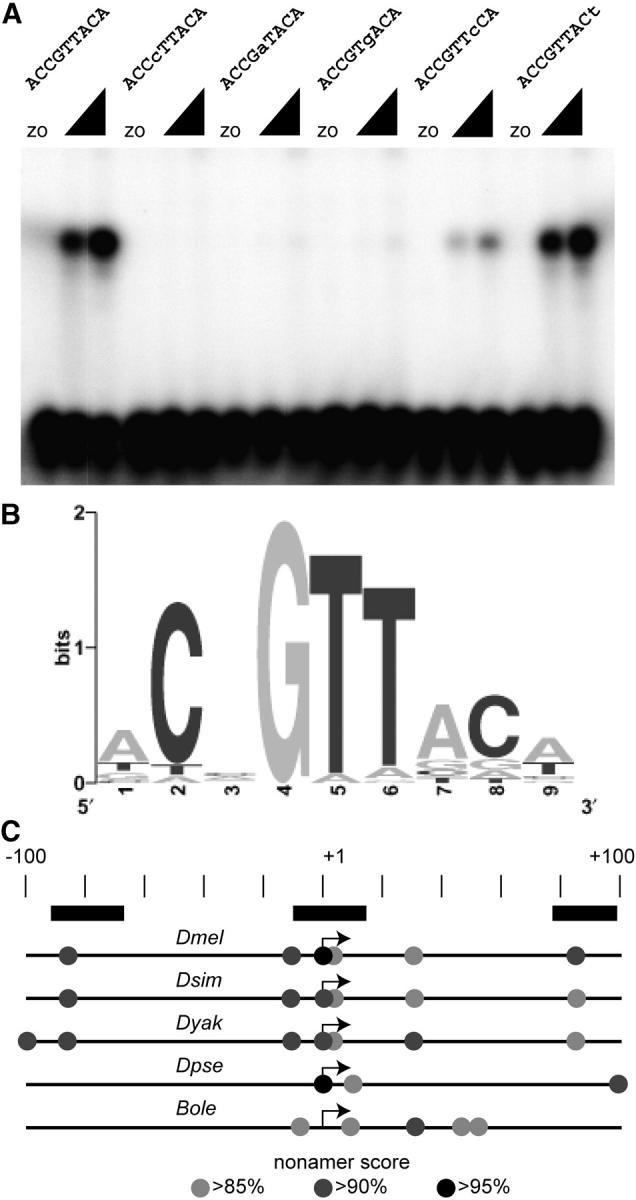
OVO binding sites. (A) Examples of gel shifts used to test for binding activity of mutated strong OVO binding sites. The sequences of the binding sites are shown. Altered residues are in lowercase type. Two concentrations of OVO DNA-binding domain were used as well as extracts from bacteria expressing a control transcript (zo). (B) Sequence Logo showing the refined OVO binding site model. (C) The position of potential OVO binding sites in D. melanogaster (Dmel), D. simulans (Dsim), D. yakuba (Dyak), D. pseudoobscura (Dpse), and B. oleae (Bole) relative to the known (Dmel and Bole) or predicted ovo-B transcription start site (scale in base pairs shown). Nonamer scores are indicated by shading (see key). The extent of OVO footprints in D. melanogaster is shown (solid rectangles).
TABLE 2.
OVO binding sequences
| Putative binding site | Method | Score (−47.5 to 13.6) |
% of maximum | Reference |
|---|---|---|---|---|
| ACTGTTACG | SELEX | 10.0 | 94.19 | Lee and Garfinkel (2000) |
| ACTGTTACT | SELEX | 11.1 | 96.01 | Lee and Garfinkel (2000) |
| ACGGTTACA | SELEX | 11.8 | 97.12 | Lee and Garfinkel (2000) |
| ACAGTTGCT | SELEX | 10.0 | 94.22 | Lee and Garfinkel (2000) |
| ACAGTTACA | SELEX | 12.8 | 98.75 | Lee and Garfinkel (2000) |
| TCTGTTAAG | SELEX | 4.6 | 85.31 | Lee and Garfinkel (2000) |
| TCGGTTGCT | SELEX | 7.2 | 89.64 | Lee and Garfinkel (2000) |
| TCGGTTTCT | SELEX | 6.1 | 87.73 | Lee and Garfinkel (2000) |
| GCTGTTCGT | SELEX | 4.8 | 85.74 | Lee and Garfinkel (2000) |
| TCCGTTACT | SELEX | 10.8 | 95.51 | Lee and Garfinkel (2000) |
| GCCGTTAGA | SELEX | 9.7 | 93.68 | Lee and Garfinkel (2000) |
| CCGGTTACG | SELEX | 7.1 | 89.46 | Lee and Garfinkel (2000) |
| GCTGTTAAT | SELEX | 6.1 | 87.77 | Lee and Garfinkel (2000) |
| TCGGTTTCT | SELEX | 6.1 | 87.73 | Lee and Garfinkel (2000) |
| ACAGTTATA | SELEX | 8.8 | 92.15 | Lee and Garfinkel (2000) |
| ACAGTTAGT | SELEX | 9.4 | 93.19 | Lee and Garfinkel (2000) |
| GCAGTTACT | SELEX | 10.5 | 94.9 | Lee and Garfinkel (2000) |
| CCCGTTACG | SELEX | 8.9 | 92.34 | Lee and Garfinkel (2000) |
| ACTGTTTCT | SELEX | 8.1 | 91.11 | Lee and Garfinkel (2000) |
| ACTGTTCGA | SELEX | 7.2 | 89.59 | Lee and Garfinkel (2000) |
| ACTGTTTCA | SELEX | 9.1 | 92.65 | Lee and Garfinkel (2000) |
| ACAGTTACA | Gel shift, footprint | 12.8 | 98.75 | Lu et al. (1998) |
| GCCGTTAAA | Gel shift, footprint | 8.5 | 91.77 | Lu et al. (1998) |
| ACAGTAACA | Gel shift, footprint | 9.0 | 92.54 | Lu et al. (1998) |
| ACAGTTAGA | Gel shift, footprint | 10.3 | 94.74 | Lu et al. (1998) |
| TTCGTTGCC | Gel shift, footprint | 3.5 | 83.59 | Lu et al. (1998) |
| ACCGTTACA | Gel shift, footprint | 13.6 | 100 | Lu et al. (1998) |
| CCTGTTATC | Gel shift, footprint | 3.3 | 83.29 | Lu et al. (1998) |
| GCCGTAGTA | Gel shift, footprint | 2.5 | 81.89 | Lu et al. (1998) |
| AACGTTACA | Gel shift | 8.6 | 91.94 | This study |
| ACCGATACA | Gel shift | 9.1 | 92.73 | This study |
| ACCGTGACA | Gel shift | 8.8 | 92.17 | This study |
| ACCGTTCCA | Gel shift | 11.2 | 96.06 | This study |
| ACCGTTACT | Gel shift | 12.6 | 98.46 | This study |
| TTAGTTGCC | Gel shift | 2.8 | 82.34 | This study |
| ACCGTTACT | Gel shift | 12.6 | 98.46 | This study |
| GCAGTTAAA | Gel shift | 7.8 | 90.52 | This study |
| ACAGTAACA | Gel shift | 9.0 | 92.54 | This study |
| TTAGTTGCC | Gel shift | 2.8 | 82.34 | This study |
| ACAGTTACA | Gel shift | 12.8 | 98.75 | This study |
| ACAGTTAGA | Gel shift | 10.3 | 94.74 | This study |
| AACGTTACA | Gel shift | 8.6 | 91.94 | This study |
| ACCGATACA | Gel shift | 9.1 | 92.73 | This study |
| ACCGTTCCA | Gel shift | 11.2 | 96.06 | This study |
To query DNA sequences for the presence of OVO binding sites, we generated a scoring matrix (Table 3). Applying this matrix to any nonamer results in a score between −47.5 and 13.6, which we express as a percentage of the maximum score. The 46 confirmed binding sites have an average score of 92.3% with a standard deviation of 5%. The minimum score of a confirmed OVO binding site is 81.9%. We determined the frequency of high-scoring nonamers in several sets of control sequences (Table 4). One set of 5000 control sequences is from random segments of the genome within each Muller element (the ovo locus is X linked). The second set of 9958 control sequences is from 5′-UTRs (some of the potential OVO binding sites are within the 5′-UTR). The third set of control sequences is −50- to +50-bp segments from a core promoter database (Ohler et al. 2002). These sequences allow us to determine the significance of high-scoring nonamers in the ovo-B core promoters of different species of Drosophila.
TABLE 3.
OVO binding site matrices
| Position
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Base | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| Position frequency matrix | |||||||||
| A | 28 | 2 | 15 | 0 | 2 | 3 | 32 | 4 | 25 |
| C | 3 | 41 | 17 | 0 | 0 | 0 | 4 | 33 | 4 |
| G | 7 | 0 | 5 | 46 | 0 | 1 | 6 | 6 | 4 |
| T | 8 | 3 | 9 | 0 | 44 | 42 | 4 | 3 | 13 |
| Position weight matrix | |||||||||
| A | 1.02 | −2.78 | 0.12 | −10.43 | −2.78 | −2.20 | 1.21 | −1.78 | 0.86 |
| C | −1.51 | 2.16 | 0.89 | −9.85 | −9.85 | −9.85 | −1.20 | 1.84 | −1.20 |
| G | −0.390 | −9.85 | −0.88 | 2.32 | −9.85 | −3.19 | −0.52 | −0.515 | −1.20 |
| T | −0.79 | −2.20 | −0.52 | −10.43 | 1.57 | 1.51 | −1.78 | −2.20 | −0.09 |
TABLE 4.
High-scoring (>85%) OVO nonamers in control sequences
| Chromosome arm |
N sampled |
Average | SD | Maximum | Median |
|---|---|---|---|---|---|
| Random 100-bp sequences | |||||
| X | 1000 | 0.67 | 0.98 | 11 | 0 |
| 2L | 1000 | 0.64 | 0.89 | 6 | 0 |
| 2R | 1000 | 0.69 | 0.87 | 5 | 0 |
| 3L | 1000 | 0.64 | 0.92 | 7 | 0 |
| 3R | 1000 | 0.66 | 0.89 | 5 | 0 |
| 5′-UTR sequencesa | |||||
| X | 1748 | 0.85 | 0.64 | 4.2 | 0.8 |
| 2L | 1757 | 0.77 | 0.61 | 4.3 | 0.7 |
| 2R | 2051 | 0.84 | 0.68 | 5.4 | 0.8 |
| 3L | 1921 | 0.80 | 0.66 | 5.2 | 0.7 |
| 3R | 2481 | 0.83 | 0.65 | 5.8 | 0.8 |
| Core promoter sequencesb | |||||
| Allc | 1941 | 1.02 | 1.05 | 6 | 1 |
Adjusted for 100-bp length, sequences <100 bp excluded.
Trimmed to 100 bp centered on the transcription start site.
Pooled to increase sample size.
If OVO binding sites at the ovo-B core promoter are important for ovo-B expression, then we expect that those sites will be enriched in that region in multiple species. We therefore asked if there was significant enrichment for high-scoring (≥85%) nonamers in the 100-bp region flanking the ovo-B transcription start sites as compared to control sequences. The ovo-B core promoter regions of D. melanogaster, D. simulans, D. yakuba, and B. oleae each have four such high-scoring nonamers in the 100-bp core promoter region (P < 0.01 vs. random sequences, <0.01 vs. 5′-UTRs, and <0.03 vs. core promoters). D. pseudoobscura shows three high-scoring nonamers (P < 0.04 vs. random sequences, <0.04 vs. 5′-UTRs, and <0.09 vs. core promoters). Furthermore, there was a nearly identical arrangement of high-scoring OVO binding sites in D. melanogaster, D. simulans, and D. yakuba (Figure 5C). In all three cases there are binding sites ∼85 bp upstream and downstream of the ovo-B start site, which correspond to two regions that are protected by OVO protein in DNA footprint experiments (Lu et al. 1998). There are also nearly identical clusters of three overlapping OVO binding sites at the transcription start site, where a very strong OVO footprint was observed (Lu et al. 1998). Thus, the organization of OVO binding sites is conserved between these species. The conserved binding sites at the predicted start site of the D. pseudoobscura ovo-B transcript are even more striking (Figure 5C) given the low overall conservation between D. melanogaster and D. pseudoobscura ovo promoter regions (Figure 2A). The region flanking the predicted ovo-B start site of B. oleae has only one nonamer scoring above 90% and does not show the −80-bp and +100-bp sites found in the other species. Perhaps the supernumerary motifs compensate for weaker binding to individual sites. It has been shown that cis-regulatory modules can retain function across species as an intact module, while still diverging to the point that chimeric modules derived from two species fail to function (Ludwig et al. 2000). It is therefore possible that function of the B. oleae ovo-B core promoter in a D. melanogaster ovary (Khila et al. 2003) is maintained using a slightly different transcription factor solution.
Response of reporters to OVO-A and OVO-B:
At least some of the OVO binding sites in the ovo promoter region are functional, but the effect of OVO expression on the ovo promoter has not been mapped to any particular OVO binding sites (Lu et al. 1998; Andrews et al. 2000). We therefore assayed for reporter expression in flies heterozygous for ovoD1, an antimorphic allele encoding an OVO-A isoform from the ovo-B promoter, and in flies bearing various copy numbers of ovoΔap, an OVO-B encoding transgene.
The ovoD1 allele resulted in nearly complete silencing of both the lacZΔap and the lacZΔbp reporter gene series (Figure 6). The lacZ1.1 reporter that bears both the ovo-B and the ovo-A start sites was repressed, as were deletion reporters. Indeed, any reporter that is expressed has several OVO binding sites (Figure 2) and all such reporters can be repressed by expression of OVO-A. Collectively, the reporters that are downregulated by ovoD1 remove all the strong OVO binding sites at the ovo locus and all but one of the weak sites (Figure 6), raising the distinct possibility that any OVO binding site can act as a silencer in conjunction with OVO-A proteins.
Figure 6.—

Examples of ovo reporter expression in an ovoD1/+ background. The genotype with respect to ovo encoding alleles is shown at the top and the reporter genotype is shown at the left. An ovariole is shown for the +/+ micrographs, and an entire atrophic ovary is shown from the ovoD1/+ flies. (A, C, and E) Ovarioles from females wild type for ovo. (B, D, and F) Ovaries from females heterozygous for ovoD1. (A and B) Heterozygous for ovoΔapΔ4. (C and D) Heterozygous for ovoΔapΔ5. (E and F) Heterozygous for ovoΔapΔ6.
We were unable to find convincing evidence for female germline positive ovo autoregulation in previous work (Andrews et al. 2000), even though the ovo-B core promoter positively responds to OVO-B when swapped into the otu regulatory region (Lu and Oliver 2001). This suggests that there are cis-sequences required for positive autoregulation present in otu, but not ovo, or that there are cis-sequences that block positive autoregulation at ovo, or both. We therefore asked if any of the deletion reporters are influenced by the dose of OVO-B produced in trans. Females bearing the lacZΔapΔ8 construct showed a striking increase of reporter expression with increasing copies of the OVO-B encoding transgene ovoΔap (Figure 7). Additionally, reducing the copy number of endogenous ovo greatly reduced the expression of the lacZΔapΔ8 reporter. The expression of other reporters was only marginally increased with the dose of OVO-B encoding transgenes in 2X females (Figure 7). Thus, OVO binding sites at ovo can respond negatively to OVO-A and positively to OVO-B.
Figure 7.—
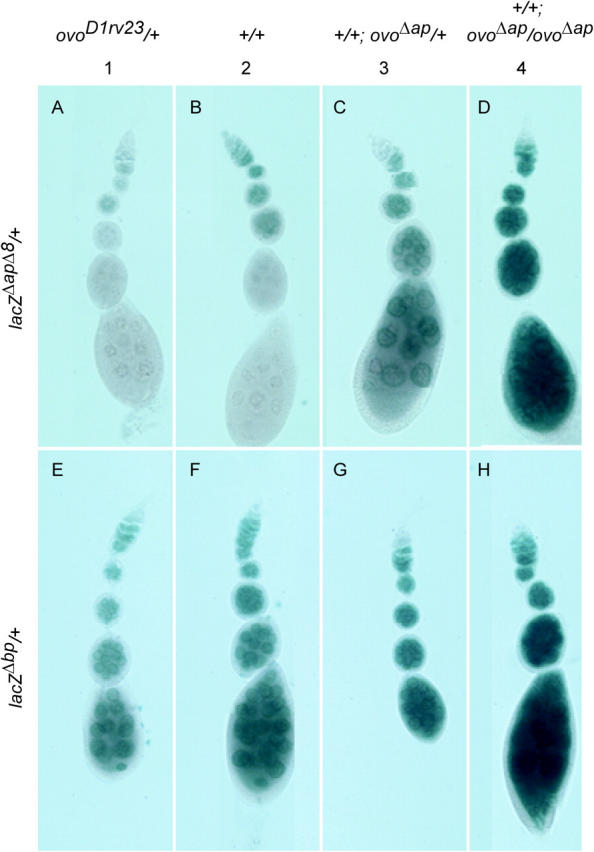
Examples of ovo-B and ovo-A reporter expression in flies with different OVO-B encoding copy numbers. The genotype with respect to ovo encoding alleles is shown at the top and the reporter genotype is shown at the left. Staining of gonads bearing the ovo-B-specific reporter (A–D) lacZΔapΔ8, and the ovo-A-specific reporter (E–H) lacZΔb, in flies with (A and E) one, (B and F) two, (C and G) three, or (D and H) four copies of ovo alleles encoding ovo-B isoforms is shown. The ovoΔap allele encodes only OVO-B, while the wild-type alleles also express OVO-A, albeit at a lower level.
DISCUSSION
Transcriptional circuits at ovo:
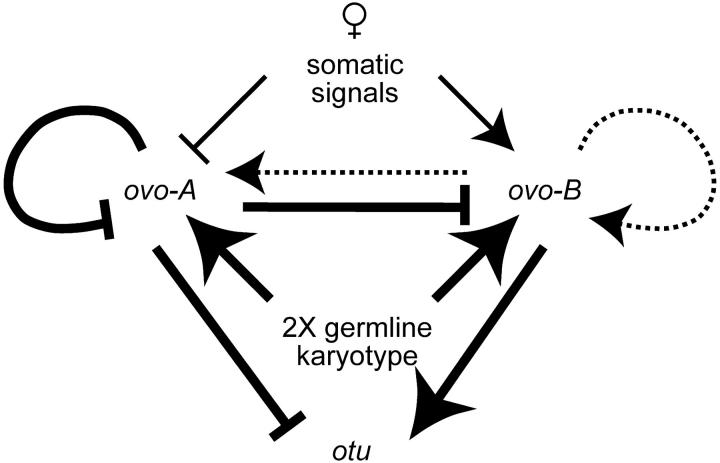
We are beginning to have a reasonable understanding of the germline pathway centered on ovo (Figure 8). OVO-A and OVO-B functions are in a delicate balance in the female germline. OVO-B is absolutely required for oogenesis and is downregulated by OVO-A. An excess OVO-A results in defective oogenesis and subsequent embryogenesis, while too little results in defective germline function in progeny (Mevel-Ninio et al. 1996; Andrews et al. 1998, 2000). Having the female soma repress ovo-A function in the germline may prevent damage to developing eggs, while the positive effect of a 2X karyotype may ensure that OVO-A protein is ultimately deposited in those eggs. We show that OVO-B can have a positive effect on the ovo-B promoter following the deletion of some promoter-proximal sequences, but negative autoregulation occurs in all reporters. This difference between response to OVO-A vs. OVO-B does not appear to be due to different inherent strengths of the two transcription factors, as the otu promoter, a direct target of ovo, is strongly positively regulated by OVO-B (Lu and Oliver 2001) in addition to being negatively regulated by OVO-A (Andrews et al. 2000). Further, this difference in response dose not appear to be due to the ovo-B core promoter sequence, as in the otu sequence milieu, the ovo-B promoter is also strongly positively regulated by OVO-B (Lu and Oliver 2001). Thus, the ovo context is likely to specifically dampen the trans effect of OVO-B, but not OVO-A, on ovo-B promoter activity.
Figure 8.—
The regulatory circuit centered on the ovo locus. Positive effects (arrows) and negative effects (blocked end) are shown. The lines represent strong (thick), moderate (thin), and weak (dashed) effects. See text for details.
The ovo-B promoter encodes the OVO-B isoforms required and sufficient for female germline development and is regulated by the number of X chromosomes in the germline cells, and the sex of the surrounding soma positively regulates ovo-B, even though neither signal is absolutely required (Andrews and Oliver 2002). For example, only 1X males fail to robustly express ovo-B in the germline, suggesting that both the intrinsic 2X signal and the extrinsic female somatic signal can upregulate ovo-B independently. Also we know that somatic signaling is not required for ovo genetic function, because 2X males have germline cells, while 2X males lacking ovo do not (Hinson and Nagoshi 1999; Andrews and Oliver 2002). This dual regulatory input ensures that ovo-B is most highly expressed in the cells that require ovo activity—wild-type female germ cells. We show that ovo-A expression is more dynamically regulated. The highest ovo-A promoter activity is in 2X males, followed by 2X females, 1X males, and 1X females. This pattern suggests that a 2X karyotype activates ovo-A, while a female soma inhibits ovo-A activity within the germline.
The combination of negative and positive autoregulation adds considerable complexity to the regulatory circuit. For example, the positive effect of a female soma on the expression of ovo-B in our working model could be due to repression of ovo-A expression by a female soma, followed by derepression of ovo-B because of lowered OVO-A levels, or a more direct positive effect of the female soma on ovo-B.
The ovo-B core promoter:
Analysis of promoters active in the germline of Drosophila suggests that they are often more compact than many of the promoters studied in somatic cells (Arnosti 2003). This may be the case for ovo-B. While the ovo-B core promoter alone is insufficient for transcription, transcriptional activity from ovo-B is remarkably resistant to deletions from either the 5′ or the 3′ direction. The lacZΔapΔ6 reporter has only 268 bp of ovo sequence but is expressed in the female germline. The overlap between the lacZΔapΔ6 and lacZΔapΔ8 reporters, both of which are expressed, is only 73 bp. This is unusually close to the transcription start site. The OVO binding site footprints overlap the transcriptional start sites of both otu and ovo-B (Lu and Oliver 2001), and we show that there are proteins or complexes in gonad extracts that bind to this core sequence. We therefore suggest that OVO alters the structure of the core promoter and promotes preinitiation complex formation (Lu and Oliver 2001). The highly conserved position of OVO binding sites at ovo-B in multiple species of flies supports the idea that OVO functions at the transcription start site. A recent study of human promoters suggests that the binding of transcription factors within 100 bp of the transcription start site may be more common than previously thought (FitzGerald et al. 2004).
The importance of the core promoter raises some interesting questions about how ovo interprets the number of X chromosomes in the germline and the sex of the surrounding soma. For example, the Sex-lethal gene counts X chromosomes in the soma by binding several transcription factors, encoded on the X chromosome, to a region rich in the corresponding binding sites. The balance toward expression of Sxl is thus tipped by a graded occupancy at a complex cis-regulatory module (Louis et al. 2003). There does not appear to be an extended cis-regulatory module that is essential for the qualitative expression of ovo. Perhaps sex-determination signals indirectly regulate ovo. The molecular nature of the karyotype and somatic signals to the germline is a major unresolved problem in germline sex determination.
Acknowledgments
We thank George Poy for sequencing, Virginia Boulais for micro-injections and maintaining fly stocks, Paul Schedl and the Bloomington Stock Center for fly stocks, Sandra Farkas for maintaining fly stocks, and Justen Andrews, Aniello Cerrato, Jurrien Dean, Vaijayanti Gupta, Alan Kimmel, Michael Parisi, Sonia Santa Anna, and the National Institutes of Health fly community for helpful discussions and/or comments on the manuscript.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andrews, J., and B. Oliver, 2002. Sex determination signals control ovo-B transcription in Drosophila melanogaster germ cells. Genetics 160: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, J., I. Levenson and B. Oliver, 1998. New AUG initiation codons in a long 5′ UTR create four dominant negative alleles of the Drosophila C2H2 zinc-finger gene ovo. Dev. Genes Evol. 207: 482–487. [DOI] [PubMed] [Google Scholar]
- Andrews, J., D. Garcia-Estefania, I. Delon, J. Lu, M. Mevel-Ninio et al., 2000. OVO transcription factors function antagonistically in the Drosophila female germline. Development 127: 881–892. [DOI] [PubMed] [Google Scholar]
- Arnosti, D. N., 2003. Analysis and function of transcriptional regulatory elements: insights from Drosophila. Annu. Rev. Entomol. 48: 579–602. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1989 Drosophila. A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell and D. L. Wheeler, 2004. GenBank: update. Nucleic Acids Res. 32: D23–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al., 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X., C. Schonbaum, L. Degenstein, W. Bai, A. Mahowald et al., 1998. The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 12: 3452–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald, P. C., A. Shlyakhtenko, A. A. Mir and C. Vinson, 2004. Clustering of DNA sequences in human promoters. Genome Res. 14: 1562–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase, 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, M. T., 1993 Spermatogenesis, pp. 71–148 in The Development of Drosophila, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Garfinkel, M. D., A. R. Lohe and A. P. Mahowald, 1992. Molecular genetics of the Drosophila melanogaster ovo locus, a gene required for sex determination of germline cells. Genetics 130: 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson, S., and R. N. Nagoshi, 1999. Regulatory and functional interactions between the somatic sex regulatory gene transformer and the germline genes ovo and ovarian tumor. Development 126: 861–871. [DOI] [PubMed] [Google Scholar]
- Khila, A., A. El Haidani, A. Vincent, F. Payre and S. I. Souda, 2003. The dual function of ovo/shavenbaby in germline and epidermis differentiation is conserved between Drosophila melanogaster and the olive fruit fly Bactrocera oleae. Insect Biochem. Mol. Biol. 33: 691–699. [DOI] [PubMed] [Google Scholar]
- King, O. D., and F. P. Roth, 2003. A non-parametric model for transcription factor binding sites. Nucleic Acids Res. 31: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., and M. D. Garfinkel, 2000. Characterization of Drosophila OVO protein DNA binding specificity using random DNA oligomer selection suggests zinc finger degeneration. Nucleic Acids Res. 28: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard, B., A. Sandelin, L. Mendoza, P. Engstrom, N. Jareborg et al., 2003. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, M., L. Holm, L. Sanchez and M. Kaufman, 2003. A theoretical model for the regulation of Sex-lethal, a gene that controls sex determination and dosage compensation in Drosophila melanogaster. Genetics 165: 1355–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., and B. Oliver, 2001. Drosophila OVO regulates ovarian tumor transcription by binding unusually near the transcription start site. Development 128: 1671–1686. [DOI] [PubMed] [Google Scholar]
- Lu, J., J. Andrews, D. Pauli and B. Oliver, 1998. Drosophila OVO zinc-finger protein regulates ovo and ovarian tumor target promoters. Dev. Genes Evol. 208: 213–222. [DOI] [PubMed] [Google Scholar]
- Ludwig, M. Z., C. Bergman, N. H. Patel and M. Kreitman, 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564–567. [DOI] [PubMed] [Google Scholar]
- Mayor, C., M. Brudno, J. R. Schwartz, A. Poliakov, E. M. Rubin et al., 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047. [DOI] [PubMed] [Google Scholar]
- Mevel-Ninio, M., R. Terracol and F. C. Kafatos, 1991. The ovo gene of Drosophila encodes a zinc finger protein required for female germ line development. EMBO J. 10: 2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel-Ninio, M., E. Fouilloux, I. Guenal and A. Vincent, 1996. The three dominant female-sterile mutations of the Drosophila ovo gene are point mutations that create new translation-initiator AUG codons. Development 122: 4131–4138. [DOI] [PubMed] [Google Scholar]
- Nöthiger, R., M. Jonglez, M. Leuthold, P. Meier-Gerschwiler and T. Weber, 1989. Sex determination in the germ line of Drosophila depends on genetic signals and inductive somatic factors. Development 107: 505–518. [DOI] [PubMed] [Google Scholar]
- Ohler, U., G. C. Liao, H. Niemann and G. M. Rubin, 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, B., 2002. Genetic control of germline sex determination in Drosophila. Int. Rev. Cytol. 219: 1–60. [DOI] [PubMed] [Google Scholar]
- Oliver, B., N. Perrimon and A. P. Mahowald, 1987. The ovo locus is required for sex-specific germ line maintenance in Drosophila. Genes Dev. 1: 913–923. [DOI] [PubMed] [Google Scholar]
- Oliver, B., J. Singer, V. Laget, G. Pennetta and D. Pauli, 1994. Function of Drosophila ovo+ in germ-line sex determination depends on X chromosome number. Development 120: 3185–3195. [DOI] [PubMed] [Google Scholar]
- Pauli, D., B. Oliver, and A. P. Mahowald, 1993. The role of the ovarian tumor locus in Drosophila melanogaster germ line sex determination. Development 119: 123–134. [DOI] [PubMed] [Google Scholar]
- Payre, F., A. Vincent and S. Carreno, 1999. ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature 400: 271–275. [DOI] [PubMed] [Google Scholar]
- Salles, C., M. Mevel-Ninio, A. Vincent and F. Payre, 2002. A germline-specific splicing generates an extended ovo protein isoform required for Drosophila oogenesis. Dev. Biol. 246: 366–376. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sandelin, A., and W. W. Wasserman, 2004. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 338: 207–215. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., 1993 Developmental genetics of oogenesis, pp. 1–70 in The Development of Drosophila, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Steinmann-Zwicky, M., H. Schmid and R. Nothiger, 1989. Cell-autonomous and inductive signals can determine the sex of the germ line of Drosophila by regulating the gene Sxl. Cell 57: 157–166. [DOI] [PubMed] [Google Scholar]
- Tamura, K., S. Subramanian and S. Kumar, 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36–44. [DOI] [PubMed] [Google Scholar]
- Waterbury, J. A., J. I. Horabin, D. Bopp and P. Schedl, 2000. Sex determination in the Drosophila germline is dictated by the sexual identity of the surrounding soma. Genetics 155: 1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]