Abstract
Escherichia coli cells expressing the tpm gene encoding the bacterial thiopurine methyltransferase (bTPMT) are shown to methylate selenite and (methyl)selenocysteine into dimethylselenide (DMSe) and dimethyldiselenide (DMDSe). E. coli cells expressing tpm from a gene library cosmid clone (harboring a Pseudomonas syringae insert of about 20 kb) also methylated selenate into DMSe and DMDSe. bTPMT is the first methyltransferase shown to be involved in the methylation of these selenium derivatives.
Selenium is an essential element, but its presence in micromolar concentrations can lead to death and deformities of wildlife (14). It can be found in enzymes under the form of selenocysteine amino acids or bound in such a way that it can be detached by denaturing or reducing agents (for a review, see reference 9). Bacteria have been shown to play a key role in the biological cycle of selenium by reduction (19) and volatilization of inorganic selenium (for a review, see references 4 and 7) in the environment. A significant contribution to the understanding of the biochemistry of this cycle was made by studies with Thauera selenatis. A dissimilatory selenate reductase enzyme complex was found in this bacterium and shown to be trimeric and periplasmic and to reduce selenate to selenite (18). This selenate reductase was shown to have a high affinity and turnover rate for selenate, much higher than the ones of the bacterial periplasmic nitrate reductases (17). The T. selenatis nitrite respiratory system was shown to reduce selenite to elemental selenium in the presence of nitrate (3). T. selenatis was isolated from sediments of the San Joaquin Valley, and its selenate-reducing capacity was recently used for the bioremediation of selenium from agricultural drainage water (1). The biochemistry and genetics of the enzymes involved in selenium methylation remain almost unexplored. Bioremediation by volatilization of selenium from contaminated sites represents an interesting alternative to the use of selenate-respiring bacteria (10).
Recently, we discovered a methyltransferase that might be involved in the methylation of inorganic selenite (2). This enzyme, named the bacterial thiopurine methyltransferase (bTPMT), was shown to catalyze the S adenosylmethylation of aromatic and heterocyclic sulfhydryl compounds like 6-mercaptopurine. It belongs to a group of methyltransferases whose presence in eukaryotes (human, rat, mouse, etc.) is very well documented (for examples, see references 15 and 20). The human TPMT (hTPMT) is essential for the metabolism of thiopurine drugs (11), which are frequently used in the treatment of human autoimmune diseases and for transplantations (5, 16). The bTPMT, overexpressed in Escherichia coli, conferred resistance to tellurite and the ability to grow at high concentrations of sodium biselenite (2). Here, we present clear evidence that bTPMT is involved in the methylation of inorganic and organic selenium into dimethylselenide (DMSe) and dimethyldiselenide (DMDSe). bTPMT is the first methyltransferase shown to be involved in the bacterial selenium methylation processes.
The effect of bTPMT on the production of DMSe and DMDSe was tested in the E. coli cellular environment. Gas chromatography-mass spectrometry (GC-MS) analyses were performed to estimate the production levels of DMSe and DMDSe. These were carried out on a Hewlett-Packard 6890 GC coupled to mass spectrometer model 5973. The chromatography column was a Supelco VOCOL (60 m by 250 μm by 1.5 μm). One milliliter of the headspace gases was injected per analysis. The split ratio was 0.1:1 μl, with a flow rate of 200 μl per min. The gas saver was set at 20 ml per min for 2 min. The carrier gas was helium (obtained from Air Liquide) and was used at a flow rate of 1.7 ml per min in the column and at a flow rate of 4.2 ml per min in the injector. Gas pressure was 180 kPa. The temperature of the injector was 210°C. Elution conditions were 2 min at 30°C and a subsequent temperature ramp of 5°C per min from 30 to 210°C. Retention times of DMSe and DMDSe were estimated by using purchased gases (from Sigma-Aldrich). To demonstrate the role of bTPMT in selenium methylation, GC-MS experiments compared the productions of DMSe and DMDSe by E. coli DH10B strains expressing or not expressing the gene (tpm) encoding the bTPMT and grown with inorganic and organic molecules of selenium [selenite, selenate, (methyl)selenocysteine, and selenomethionine]. E coli cells can naturally transform some of these selenium molecules. For example, E. coli cells growing on selenite accumulate red-orange pigments, known to be elemental selenium and the products of reduction reactions. Our results show that they transform (by an unknown mechanism that is less efficient than the bTPMT-driven one) (methyl)selenocysteine, but not selenomethionine or inorganic selenium, into volatile DMSe and DMDSe (Table 1). The significance of the bTPMT-driven production of DMSe and DMDSe was thus tested by performing Student's t tests with the Staview-SE package.
TABLE 1.
DMSe and DMDSe produced by E. coli DH10B cells harboring the tpm-encoded thiopurine methyltransferase
| tpm construct and selenium supplementa | Mean ± SDb of GC-MS spectrum peak area for production (mm2/μg of total proteinsc) of:
|
|
|---|---|---|
| DMSe | DMDSe | |
| pAV257 | ||
| No supplement | bdld | bdl |
| Potassium selenite | 13 ± 6.7 | 3.4 ± 1.3 |
| Potassium selenate | 15.6 ± 6.8 | 0.8 |
| Selenomethionine | 5.6 ± 4 | bdl |
| (Methyl)selenocysteine | 190 ± 12.8 | 6 ± 1 |
| pAV617 | ||
| No supplement | bdl | bdl |
| Potassium selenite | 4.7 ± 3.4 | 2.3 ± 1 |
| Potassium selenate | bdl | bdl |
| Selenomethionine | 2.1 ± 0.8 | 0.5 ± 0.5 |
| (Methyl)selenocysteine | 76.1 ± 15.5 | 13.1 ± 3.1 |
| pD5e | ||
| No supplement | bdl | bdl |
| Potassium selenite | bdl | bdl |
| Potassium selenate | bdl | bdl |
| Selenomethionine | bdl | 1.6 ± 1.5 |
| (Methyl)selenocysteine | 5.2 ± 0.4 | 6.5 ± 1.7 |
| E. coli DH10B | ||
| No supplement | bdl | bdl |
| Potassium selenite | bdl | bdl |
| Potassium selenate | bdl | bdl |
| Selenomethionine | 0.8 ± 0.7 | 0.2 ± 0.2 |
| (Methyl)selenocysteine | 8.9 ± 3.4 | 9.7 ± 2.1 |
E. coli cells were grown in Luria-Bertani broth (13) with 10 ppm concentrations of selenite, selenate, selenomethionine, or (methyl)selenocysteine. See the text for the GC-MS methodology used to estimate DMSe and DMDSe productions and for descriptions of plasmids.
All samples were analyzed at least twice and all analyses were at least triplicated (with the exception of the E. coli pAV257 cells grown with potassium selenate).
Total proteins were determined by using the BCA protein assay (Pierce, Rockford, Ill.).
bdl, below detectable level or absent. The detection limit was between 10 and 50 ng per liter.
pAV257 containing a Tn7 insertion in tpm.
bTPMT is encoded by the tpm genetic determinant which was previously isolated from pAV257, a tellurite-resistant (Telr) pLAFR3 library clone (containing a DNA insert of more than 20 kb) of Pseudomonas syringae pathovar pisi strain 203 (2). In this study, the pAV257 tpm gene was inactivated by using a Tn7-based insertional mutagenesis approach to further investigate its function. In vitro transposition was performed with the GPS-M mutagenesis system following the manufacturer's recommendations (New England Biolabs, Beverly, Mass.). Using this system, 190 mutagenized pAV257 cosmids subcloned into E. coli DH10B (Gibco BRL, Cergy Pontoise, France) were obtained and were then screened on 4-ppm potassium tellurite to identify tpm-inactivated Tels cosmid clones. Five Tels cosmid clones were obtained, and the positions of the Tn7 insertions were determined, in the first instance, by PCR (following Gibco BRL's recommendations) using Tn7L and Tn7R primers (New England Biolabs) and the tpm primer 317 (2). The positions of four of five of the insertions of these Tels cosmids were determined by PCR inside the tpm open reading frame, and one was located in a region likely to be part of the tpm promoter region. In the second instance, the locations of three of these insertions were determined by DNA sequencing (done by Genome Express S. A., Grenoble, France) and found to be located 278 bp downstream of the tpm start codon of cosmid pD5, 166 bp downstream for pG10, and 372 bp downstream for pB21. These data suggest that a single gene, tpm, on this cosmid conferred Telr. pD5 was used to evaluate the role of tpm (and the bTPMT) in selenium methylation.
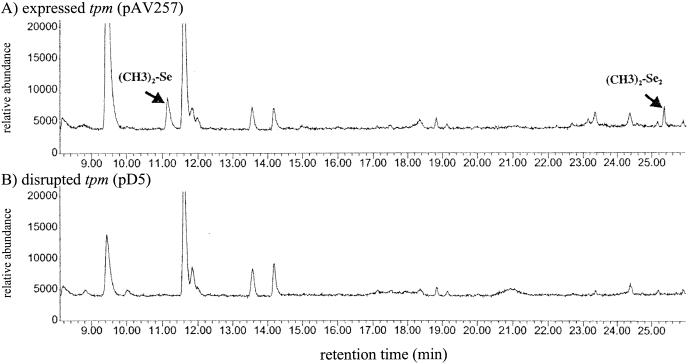
The E. coli DH10B clones harboring pAV257, pD5, and control strains were inoculated into 250-ml flasks sealed with rubber stoppers fastened by aluminum crimps and containing 50 ml of Luria-Bertani medium supplemented with various selenium compounds (Fig. 1; Table 1). A tpm pBluescript subclone, pAV617 (2), strictly harboring tpm was also used in these methylation assays. E. coli DH10B harboring the vector pBluescript SK(−) was included as a negative control in all these experiments. Concentrations of 10 ppm of potassium selenate, potassium selenite, (methyl)selenocysteine, and selenomethionine (Sigma) were added independently to these cultures after growth for 24 h at 37°C. The cultures were then incubated 4 days at 37°C on a rotary shaker (200 rpm). The headspace gases of each flask were then analyzed by GC-MS (as described above). Significant levels (P < 0.05 by comparison with the control and pD5-harboring cells) of DMSe and DMDSe were detected for the tpm-harboring E. coli strains grown with inorganic (selenite and selenate) and organic [(methyl)selenocysteine] selenium (Fig. 1; Table 1). tpm-harboring cells producing DMSe always showed (for a P value of <0.1 and excluding cells grown with potassium selenate) a less important DMDSe production. When selenite was used, DMSe and DMDSe were detected at significant levels (P < 0.05 by comparison with the control and pD5-harboring cells) for both tpm constructs tested, i.e., pAV257 (library clone) and pAV617 (harboring only tpm). When selenate was used, significant DMSe production (P < 0.05 by comparison with the control and pD5-harboring cells) could be detected only for E. coli strains harboring pAV257 (Table 1). Significant peak values (P < 0.05 by comparison with the control and pD5-harboring cells) for DMSe (but not DMDSe) were detected with the pAV257 and pAV617 constructs and (methyl)selenocysteine as a substrate but not selenomethionine. The absence of significant methylation activity observed with pD5 (inactivated tpm) (by comparison with the control cells) showed that tpm encodes the key enzyme involved with the selenate, selenite, and selenocysteine methylation process presented here.
FIG. 1.
Total ion chromatograms of the GC-MS analysis of headspace gases of E. coli DH10B cells grown in Luria-Bertani broth (13) supplemented with 10-ppm potassium selenite and harboring pAV257 (P. syringae tpm-harboring gene library cosmid clone) (A) and pD5 (pAV257 containing a Tn7 insertion in tpm) (B). See the text for a description of the methodology used.
Cournoyer et al. (2) reported the presence of metallic tellurium and selenium by tpm-harboring E. coli cells growing with tellurite and selenite. It is thus likely that oxidoreduction steps performed by E. coli are essential in this methylation process. However, the reduction of selenate to selenite did not seem to be performed by E. coli because of the lack of significant methylation activity by the strictly tpm-harboring pBluescript subclone (pAV617). Significant DMSe production (P < 0.05 by comparison with pAV617-harboring cells) with selenate could be detected only from pAV257-harboring cells. This suggests a reduction of selenate into selenite by an enzyme (or complex) encoded by pAV257 tpm-neighboring genes. A dissimilatory selenate reductase like the one reported by Schroeder et al. (18) could be involved in this process and be encoded by this library clone. Similarly, the production of DMSe was found significantly (P < 0.05) more important for pAV257-harboring cells than for pAV617-harboring cells which were grown with (methyl)selenocysteine (Table 1). This suggests that pAV257 genetic determinants other than tpm are also having an effect on the TPMT-driven methylation of (methyl)selenocysteine.
Three main observations have thus been made in this work. First, bTPMT can methylate inorganic and organic selenium; second, it can transform these molecules into DMSe and DMDSe; third, other genetic determinants on the bTPMT-encoding cosmid clone (pAV257) are likely to be involved in the methylation process of selenate and selenocysteine. It was previously suggested that the bTPMT was likely to be involved in tellurium and selenium methylation, and a pathway, adapted from the one of Doran (4), presenting a likely methylation process going from selenite to DMSe, had been proposed (2). Here, we provided pieces of evidence that are in line with this pathway but also showed that the processing of other selenium molecules [i.e., selenate and (methyl)selenocysteine] converges toward key intermediates that can be methylated by bTPMT. For selenate, it is more than likely that reduction into selenite is required prior to its volatilization. Regarding (methyl)selenocysteine, methane selenol could be the intermediate shared with the selenite or selenate methylation pathway, although none was detected here (likely because of the chromatography column used). The single cleavage of the covalent bond between the carbon and selenium can easily lead to the release of this product, which could be methylated by bTPMT into DMSe.
From this work, it is now clear that bTPMT can detoxify a broader spectrum of molecules than reported so far. This enzyme could become a key in the remediation of anthropogenically or naturally Se-contaminated sites. DMSe is 500 to 700 times less toxic than inorganic selenium species (6, 8, 12). This process is also likely to be involved in the production of dimethyl selenide in eukaryotes, including humans, because of the widely conserved nature of the methyltransferase involved (2). We are now planning to investigate further the spectrum of molecules that could be methylated by the bTPMT, including other metalloids like arsenic.
Acknowledgments
We thank the “Pôle Expertise Eau” and J. Molas of Danone France for the GC-MS analyses and their helpful expertise on selenium chemistry.
REFERENCES
- 1.Cantafio, A. W., K. D. Hagen, G. E. Lewis, T. L. Bledsoe, K. M. Nunan, and J. M. Macy. 1996. Pilot-scale selenium bioremediation of San Joaquin drainage water with Thauera selenatis. Appl. Environ. Microbiol. 62:3298-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cournoyer, B., S. Watanabe, and A. Vivian. 1998. A tellurite-resistance genetic determinant from phytopathogenic pseudomonads encodes a thiopurine methyltransferase: evidence of a widely-conserved family of methyltransferases. Biochim. Biophys. Acta 1397:161-168. [DOI] [PubMed] [Google Scholar]
- 3.Demoll-Decker, H., and J. M. Macy. 1993. The periplasmic nitrite reductase of Thauera selenatis may catalyse the reduction of selenite to elemental selenium. Arch. Microbiol. 160:241-247. [Google Scholar]
- 4.Doran, J. W. 1982. Microorganisms and the biological cycling of selenium, p. 1-32. In K. C. Marshall (ed.), Advances in microbial ecology. Plenum Press, New York, N.Y.
- 5.Elion, G. B. 1989. Nobel lecture. The purine path to chemotherapy. Biosci. Rep. 9:509-529. [DOI] [PubMed] [Google Scholar]
- 6.Franke, K. W., and A. L. Moxon. 1936. A comparison of the minimal fatal doses of selenium, tellurium, arsenic and vanadium. J. Pharmacol. Exp. Ther. 58:454-459. [Google Scholar]
- 7.Gadd, G. M. 1993. Microbial formation and transformation of organometallic and organometalloid compounds. FEMS Microbiol. Rev. 11:297-316. [Google Scholar]
- 8.Ganther, H. E., O. A. Levander, and C. A. Saumann. 1966. Dietary control of selenium volatilization in the rat. J. Nutr. 88:55-60. [DOI] [PubMed] [Google Scholar]
- 9.Heider, J., and A. Böck. 1993. Selenium metabolism in micro-organisms. In A. H. Rose (ed.), Advances in microbial physiology, vol. 35. Academic Press, London, United Kingdom. [DOI] [PubMed]
- 10.Karlson, U., and W. T. Frankenberger. 1990. Volatilization of selenium from agricultural evaporation pond sediments. Sci. Total Environ. 92:41-54. [DOI] [PubMed] [Google Scholar]
- 11.Lennard, L., J. A. Van Loon, J. S. Lilleyman, and R. M. Weinshilboum. 1987. Thiopurine pharmacogenetics in leukemia: correlation of erythrocyte thiopurine methyltransferase activity and 6-thioguanine nucleotide concentrations. Clin. Pharmacol. Ther. 41:18-25. [DOI] [PubMed] [Google Scholar]
- 12.McConnell, K. P., and O. W. Portman. 1952. Toxicity of dimethyl selenide in the rat and mouse. Proc. Soc. Exp. Biol. Med. 79:230-231. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. F. 1972. Experiments in molecular genetics, p. 190-201. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Ohlendorf, H. M., D. J. Hoffman, M. K. Saiti, and T. W. Aldrich. 1986. Embryonic mortality and abnormalities of aquatic birds: apparent impacts of selenium from irrigation drainwater. Sci. Total Environ. 52:49-63. [Google Scholar]
- 15.Otterness, D. M., and R. M. Weinshilboum. 1987. Mouse thiopurine methyltransferase pharmacogenetics: monogenic inheritance. J. Pharmacol. Exp. Ther. 240:817-824. [PubMed] [Google Scholar]
- 16.Paterson, A. R. P., and D. M. Tidd. 1975. 6-Thiopurines, p. 384-403. In A. C. Sartorelli and D. G. Johns (ed.), Antineoplastic and immunosuppressive agents II. Springer Verlag, New York, N.Y.
- 17.Sabaty, M., C. Avazeri, D. Pignol, and A. Vermeglio. 2001. Characterization of the reduction of selenate and tellurite by nitrate reductases. Appl. Environ. Microbiol. 67:5122-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroeder, I., S. Rech, T. Frafft, and J. M. Macy. 1997. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 272:23765-23768. [DOI] [PubMed] [Google Scholar]
- 19.Stolz, J. F., and R. S. Oremland. 1999. Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23:615-627. [DOI] [PubMed] [Google Scholar]
- 20.Szumlanski, C., D. Otterness, C. Her, D. Lee, B. Brandriff, D. Kelsell, N. Spurr, L. Lennard, E. Wieben, and R. M. Weinshilboum. 1996. Thiopurine methyl transferase pharmacogenetics: human gene cloning and characterization of a common polymorphism. DNA Cell Biol. 15:17-30. [DOI] [PubMed] [Google Scholar]